Expression Analysis of LacZ gene placed in the locus of Cnot7 exhibits its activity in osteoblasts in vivo and in mineralized nodules in vitro
Abstract
CCR4-NOT complex 7 (Cnot7) was identified as a regulator of gene expression in yeast and evolutionally conserved in mammals. Cnot7 deficient male mice exhibit abnormality in spermatogenesis. As these mice contained construct to express LacZ, we followed the expression patterning in these animals. LacZ was expressed in osteoblasts located in the primary spongiosa in adult mice. Cellular analysis indicated that LacZ is expressed in osteoblasts but not in osteoclasts. In the mineralized nodules formed in the culture of bone marrow cells obtained from Cnot7 +/− mice, LacZ expression was mainly observed in the cells forming mineralized nodules but not in un-mineralized area scattered around the periphery of the nodules. LacZ blue positive cells were gradually depositing minerals along its time course of the in vitro mineralization assay. Cnot7 expression was enhanced by the treatment with BMP. These data suggest that Cnot7 is expressed in osteoblasts and is associated with mineralization. J. Cell. Biochem. 99: 538–544, 2006. © 2006 Wiley-Liss, Inc.
Bone metabolism is controlled by many cytokines and hormones, which coordinately regulate the differentiation, and proliferation of mesenchymal cells to become bone forming cells, namely osteoblastic cells [Manolagas, 2000]. However, the mechanisms underlying such osteoblastic differentiation are not fully understood yet.
Osteoblastic differentiation is under the control of a set of transcription factors [Burdan, 2005; Dusso et al., 2005; Kobayashi and Kronenberg, 2005; Komori, 2005]. Among those, two of the critical transcription factors are Runx2 and Osterix whose deletion resulted in the total loss of osteoblastic cells in vivo [Komori, 2002; Nakashima et al., 2002; Komori, 2003; Komori, 2005]. In addition to these core transcription factors, osteoblasts also express transcription factors whose function have been either identified or yet to be identified. These transcription factors include ATF4 and AP-1 as well as nuclear receptors for vitamin D and PPAR-gamma [Haussler et al., 1997; Steinmuller et al., 2001; Kawaguchi, 2004; Yang et al., 2004; Dusso et al., 2005]. As to these categories of nuclear receptors, RXR has been observed to be expressed in osteoblastic cells. RXRs form heterodimers with nuclear receptors known to bind to specific ligands as described above (vitamin D, retinoic acid) and bind to response elements in the regulatory regions of genes encoding osteoblastic phenotype-related molecules. RXRs have also been known to bind to gene expression regulators, which are not classified as nuclear receptors [MacDonald et al., 1993; Ferrara et al., 1994; Haussler et al., 1997].
CCR4-NOT complex 7 (Cnot7) is a component of CCR4-NOT complex [Bai et al., 1999]. Within this complex, Cnot7 is involved in gene expression in yeast and is conserved from yeast to human [Albert et al., 2000]. In mammals, Cnot7 appears to be involved in the spermatogenesis as Cnot7 deficient male mice were found to be sterile [Berthet et al., 2004; Nakamura et al., 2004]. In these animals, Cnot7 is shown to interact with RXR-beta and it has been proposed that Cnot7 would function as a coregulator of the transcriptional factor [Nakamura et al., 2004]. As RXRs act as a partner for vitamin D receptor to regulate osteoblastic function [Haussler et al., 1997], we examined the expression of LacZ gene, which is replaced with the exon 2 of Cnot7 gene in the process of producing Cnot7 deficient mice. We observed that LacZ expression under the control of the regulatory system to express Cnot7 gene could be found in osteoblastic cells and the expression was associated with the formation of mineralized the nodules in culture.
MATERIALS AND METHODS
Animals
Cnot7 deficient mice were produced as described previously [Nakamura et al., 2004]. The construct used for the homologous recombination contained an nls-LacZ + neomycin cassette inserted into exon 2 of the Cnot7 gene. Genotyping of the offspring used polymerase chain reaction (PCR) of tail DNA with the appropriate primers to identify either the intact exon 2 or the LacZ insertion as described [Nakamura et al., 2004]. The Cnot7 deficient mice and wild-type mice were subjected to the following experiments. All the animal experiments were approved by the animal welfare committee of our institute.
LacZ Staining
Bone tissues were fixed in solution (0.2% glutaraldehyde, 0.1 M NaPB, 5 mM EGTA, 2 mM MgCl2, and 0.8% formaldehyde) for 45 min at room temperature. After rinsing twice for 30 min at room temperature in rinse buffer (0.1 M NaPB, 2 mM MgCl2, 0.1% sodium deoxycholate, and 0.2% NP40), they were incubated overnight at 37°C in X-gal staining solution (Rinse buffer, 1 mg/ml X-gal, 5 mM K4Fe(CN)6, 5 mM K3Fe(CN)6), washed twice for 30 min in cold PBS, postfixed in 4% paraformaldehyde in PBS overnight at 4°C and embedded in paraffin. Histological sections of paraffin-embedded stained tissues were counterstained with nuclear fast red. Wild-type mice were used as negative control.
β-Galactosidase activity was measured by using chemiluminescent substrate Galacto-PLUS kit according to the manufacturer's instructions (Tropix). β-Galactosidase activity was normalized by its total amount of protein.
Bone Marrow Cell Culture
Bone marrow cells were flashed out from the long bones and plated in 4-well plates at the density of 1 × 106 cells/ml. For mineralized nodule assay, the bone marrow cells were cultured in alpha minimal essential medium supplemented with 10% fatal bovine serum and antibiotics–antimycotics in the presence of ascorbic acid (50 µg/ml) and β-glycerophosphate (10 mM). Medium was changed every 2–3 days. After 5, 10, or 15 days cultures, the cells were fixed in fix solution for 5 min, stained with X-gal solution for 3–4 h and rinsed with PBS. The cells were re-fixed with PFA and stained with alizarin red to visualize mineralization of nodules.
For osteoclast formation assay, the bone marrow cells were cultured for 7 days in alpha minimal essential medium supplemented with 10% fatal bovine serum, antibiotics–antimycotics, vitamin D3 (10−8 M), and dexamethasone (10−7 M). Medium was changed every 3 days. After the fixation and X-gal staining as described above, the cells were re-fixed with PFA and stained with TRAP staining buffer containing 100 mM sodium acetate, 50 mM sodium tartrate, AS-MX, FRV-LB. We recognized the cells containing more than three nuclei and TRAP activities as osteoclasts.
Isolation of Total RNA and RT-PCR Analysis
For RT-PCR analysis, we used osteoblastic cell line, MC3T3E1 cells (obtained from Hiroaki Kodama, Oh-u University, Koriyama, Japan). MC3T3 cells were maintained in alpha-MEM supplemented with 10% FBS in 10 cm2 dish. The cells in sub-confluency were treated with TGF-beta (10 ng/ml), BMP2 (200 ng/ml), or vehicle (control), and cultured for 3 days. Total RNA was extracted from the cells, according to acid guanidium thiocyanate–phenol–chloroform method [Chomczynski and Sacchi, 1987]. Reverse transcription was carried out using 1 µg of total RNA, 0.2 µg of oligo (dT) primer, dNTP mix (10 mM/L), and MMLV-RT (100 units) in a final volume of 20 µl. RNA and primers were incubated together at 65°C for 10 min and then cooled rapidly on ice before addition of other reagents. RT was carried out at 37°C for 1 h. Complementary DNA was amplified by PCR according to the “hot-start” method in a 25-µl reaction volume containing 2.5 mM dNTP mix, 10 µM Cnot7 primers, and rTaq DNA polymerase (1 unit). After an initial denaturation at 94°C for 5 min in a Gene Amp PCR System 9700 (PE Biosystems), amplifications were performed at 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. Reaction products were analyzed by electrophoresis in 1.5% agarose gels containing ethidium bromide. The Cnot7 primers used for our RT-PCR experiments are 5′-CTCGGACTGACCTTTATGA-3′ and 5′-AGCAACTTCCTGTAATCCAC-3′. The GAPDH primers are 5′-ACC ACA GTC CAT GCC ATC AC-3′ and 5′-TCC ACC ACC CTG TTG CTG TA-3′. The bands were quantified based on densitometry (ONE-D scan, Scanalytics Inc.).
Statistical Analyses
The data were analyzed based on analyses of variance. P values less than 0.05 were regarded as statistically significant.
RESULTS
Expression Pattern of LacZ in Bone Tissues of Cnot7 Heterozygous Mice
To address the activity of regulatory region of Cnot7 in bone tissue, tibia of Cnot7 heterozygous mice (Cnot7 +/− mice), which exhibited similar phenotype to wild-type mice, was subjected to LacZ staining followed by section. The results indicated that LacZ positive signals were observed in osteoblastic cells localized in epiphyseal trabecular bone region (Fig. 1a). Higher magnification of the histological section exhibited that osteoblastic cells sitting on the bone matrix were positive for the LacZ staining (Fig. 1b).
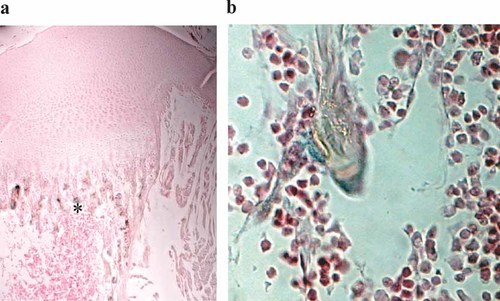
LacZ expression in osteoblasts in vivo. The microscopic images of sections of tibiae from 1-week-old mice subjected to LacZ staining. *a: Proximal end of tibia, b: primary spongiosa.
In order to follow the expression of Cnot7 in osteoblast, we obtained bone marrow cells from the femur of Cnot7 +/− mice. The cells were cultured in the presence of ascorbic acid and beta-glycerophosphate to induce differentiation of osteoblastic cells, which could form mineralized nodules. These cultures were subjected to the LacZ staining. As shown in Figure 2a, the LacZ staining positive cells were observed in and around the nodules. These cells could create mineralized matrix, which was stained positive for alizarin red. Thus, these cells were regarded as the cells in osteoblastic cell lineage. We also examined Cnot7 expression by using LacZ staining method in osteoclasts, which were formed in bone marrow cells cultured in the presence of vitamin D and dexamethasone. As shown in Figure 2b, multinucleated TRAP positive cells were developed in cultures. However, these osteoclast-like cells were negative for LacZ staining. These data showed that LacZ positive cells were specifically observed in the cells of osteoblastic lineage but not in those of osteoclastic lineage indicating certain specificity of Cnot7 expression in osteoblasts.
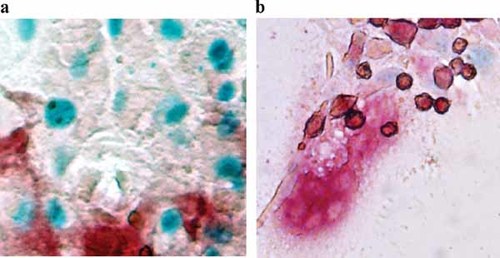
LacZ expression in osteoblasts and osteoclasts in culture. a: Osteoblasts developed in the bone marrow cell culture in osteogenic medium expressed LacZ signals. b: Osteoclasts developed in bone marrow cells cultured in the presence of vitamin D and dexamethasone. These cells did not express LacZ signals.
In the culture of osteoblastic cells, one of the functional evidences is to form mineralized nodules in vitro. Therefore, we cultured bone marrow cells obtained from the Cnot7 +/− mice to form the mineralized nodules in the cultures using the osteogenic medium. When the cells were cultured in the presence of beta-glycerophosphate and ascorbic acid, nodules were observed under the phase-contrast microscope as shadowed materials. Alizarin red staining conformed the deposition of mineral in the shadowed materials. Positive LacZ staining was detected in the cells, which surrounded well-mineralized matrix in the nodules produced in the culture (Fig. 3 lower panels). In these cultures, some cells did not indicate deposition of mineralized materials even in the same well and most of these cells did not exhibit LacZ positive blue signals (Fig. 3 upper panels).
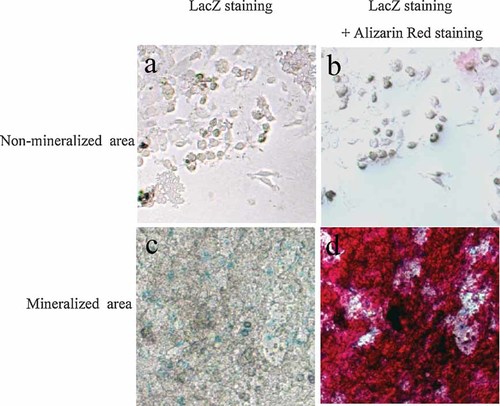
LacZ expression in the cells subjected to mineralized nodule formation assay. Nodules were formed in the bone marrow cells of Cnot7 +/− mice cultured in the presence of ascorbic acid and beta-glycerophosphate. Cnot7 expression was monitored by LacZ staining. a: LacZ expression was hardly observed in non-mineralized area. b: Cultures shown in (a) were subjected to Alizarin red staining. c: LacZ expression was observed in mineralized area. d: Mineralization in (c) was visualized by alizarin red staining.
Time course experiments provided evidence that LacZ positive cells were associated with the mineralized nodule formation. In earlier cultures such as day 5, LacZ positive cells were scattered all over the culture dish without association with mineralized materials (Fig. 4a). By day 10 in culture, nodule formation with mineralized materials was observed in the culture of bone marrow cells. When the cells were stained with both alizarin red and X-gal, the red and blue staining were associated together. The result indicated that the LacZ positive cells were depositing moralizing matrix. By day 15, LacZ positive cells were totally surrounded by the highly moralized materials (Fig. 4a). The levels of the formation of mineralized nodules were similar in these Cnot7 heterozygous cells and wild-type cells (Fig. 4a).
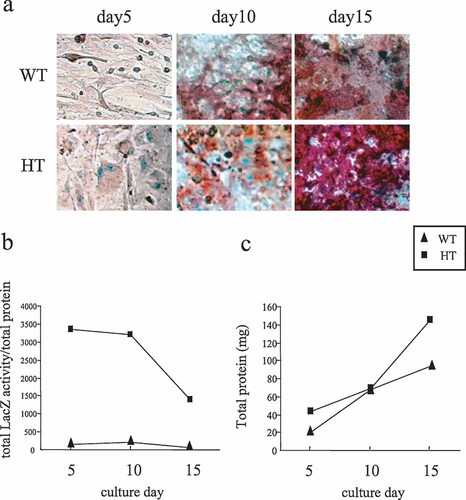
Time course of the formation of mineralized nodule and expression of LacZ. a: Nodules were formed in the bone marrow cells obtained from Cnot7 +/+ (WT) and +/− (HT) mice. These nodules were stained with X-gal and alizarin red after 5, 10, and 15 days culture. b: LacZ activities per protein c: total protein content per well. Cnot7 +/+ (WT); triangle and Cnot7 +/− (HT); square.
Such LacZ expression was also monitored by based on the biochemical assay. On day 5 and day 10, the levels of the LacZ activity were similar. However, by day 15, due to the increase in the protein content per culture dish, total LacZ activity normalized against protein was reduced (Fig. 4b).
Since matrix protein was known to contain BMP and such BMP signal may be related to nodule formation in culture, we treated the osteoblastic cell line, MC3T3E1 cells, with BMP. The BMP treatment enhanced the expression of Cnot7 mRNA (Fig. 5). Such increase was not observed when TGF-beta was treated with the cells suggesting the specificity of BMP to modulate the expression levels of Cnot7.
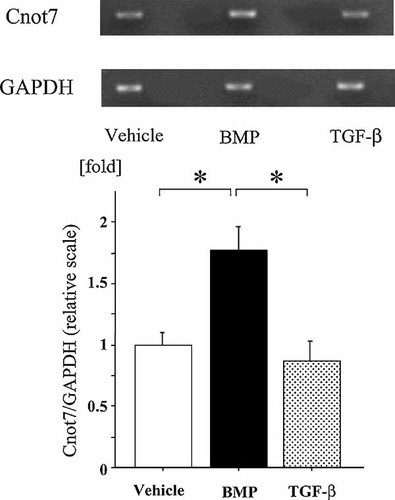
BMP treatment enhances CNOT7 mRNA expression. Osteoblastic MC3T3E1 cells were cultured in the presence of vehicle, 200 ng/ml BMP or TGF-beta for 3 days. Total RNA was extracted from these cells and the expression levels of Cnot7 were examined based on RT-PCR. Cnot7 mRNA expression in osteoblasts was enhanced by BMP treatment. Asterisks indicate that the difference is statistically significant.
DISCUSSION
We have shown that the expression of LacZ gene placed in Cnot7 locus was observed in the osteoblastic cells located in mineralized nodule formed in culture. From the histological examination, we observed that LacZ expression was detectable in osteoblastic cells sitting on the trabecular bone. Furthermore, in vivo analysis on the histology also indicated that the LacZ activity was not associated with osteoclasts. These observations suggested that Cnot7 activity would be specifically confined in osteoblastic cells.
When bone marrow cells obtained from Cnot7 +/− mice were cultured in the presence of osteogenic medium, LacZ expression was observed in bone forming cells. In contrast, when the bone marrow cells were cultured in the presence of vitamin D and dexamethasone to produce multinucleated osteoclast like cells, LacZ expression was not observed to associate with the expression of TRAP, a maker for osteoclastic cells. These observations clearly point to the specific expression of LacZ in the cells of the osteoblastic lineage in both in vitro and in vivo.
Time course study of the nodule formation in culture also supported the association of the expression of LacZ gene with the formation of alizarin red positive nodule in culture. This is corresponding to the observation on the association of LacZ gene activity in osteoblasts in vivo. When we followed a long term culture for the mineralization (15 days), LacZ expression was kept to be high enough to be detectable in the cells embedded in highly mineralized nodule.
Osteoblastic cells are considered to differentiate from immature cells, which are present in bone marrow environment. In the bone marrow environment, it has been claimed that stem cell type population could exist even in adult animals. Expression of LacZ gene placed in Cnot7 locus was also clearly observed in the cells, which were not positive for the alkaline phosphatase at least the beginning of the bone marrow cell culture. These observations suggest that the Cnot7 expression may be commonly observed in the relatively early stages of osteoblastic cells as well as the mature osteoblasts, which are involved in the formation of mineralized nodules. Since BMP treatment enhanced the expression of Cnot7, it is possible that Cnot7 may be expressed as a common marker of immature and mature osteoblastic cells during the differentiation though this point needs further elucidation.
In conclusion, we identify that expression of Cnot7 was detected in osteoblastic cells in vivo as well as in culture. Culture experiments revealed that Cnot7 was specifically observed in the osteoblastic cells and such Cnot7 activity was clearly associated with the relatively young osteoblastic cells as well as mature osteoblastic cells producing mineralized nodule.
Acknowledgements
This research was supported by the grants-in-aid received from the Japanese Ministry of Education (21st Century Center of Excellence (COE) Program, Frontier Research for Molecular Destruction and Reconstitution of Tooth and Bone, 14207056, 16659405, 16027215, 16022221), Grants from Japan Space forum, NASDA, and Japan Society for Promotion of Science (JSPS Core to Core Program on Advanced Bone and Joint Science (ABJS), Research for the Future Program, Genome Science).