Functional and structural properties of stannin: Roles in cellular growth, selective toxicity, and mitochondrial responses to injury
Abstract
Stannin (Snn) was discovered using subtractive hybridization methodology designed to find gene products related to selective organotin toxicity and apoptosis. The cDNAs for Snn were first isolated from brain tissues sensitive to trimethyltin, and were subsequently used to localize, characterize, and identify genomic DNA, and other gene products of Snn. Snn is a highly conserved, 88 amino acid protein found primarily in vertebrates. There is a minor divergence in the C-terminal sequence between amphibians and primates, but a nearly complete conservation of the first 60 residues in all vertebrates sequenced to date. Snn is a membrane-bound protein and is localized, in part, to the mitochondria and other vesicular organelles, suggesting that both localization and conservation are significant for the overall function of the protein. The structure of Snn in a micellar environment and its architecture in lipid bilayers have been determined using a combination of solution and solid-state NMR, respectively. Snn structure comprised a single transmembrane domain (residues 10–33), a 28-residue linker region from residues 34–60 that contains a conserved CXC metal binding motif and a putative 14-3-3ξ binding region, and a cytoplasmic helix (residues 61–79), which is partially embedded into the membrane. Of primary interest is understanding how this highly-conserved peptide with an interesting structure and cellular localization transmits both normal and potentially toxic signals within the cell. Evidence to date suggests that organotins such as trimethyltin interact with the CXC region of Snn, which is vicinal to the putative 14-3-3 binding site. In vitro transfection analyses and microarray experiments have inferred a possible role of Snn in several key signaling systems, including activation of the p38-ERK cascade, p53-dependent pathways, and 14-3-3ξ protein-mediated processes. TNFα can induce Snn mRNA expression in endothelial cells in a PKC-ε dependent manner. Studies with Snn siRNA suggest that this protein may be involved in growth regulation, since inhibition of Snn expression alone leads to reduced endothelial cells growth and induction of COP-1, a negative regulator of p53 function. A key piece of the puzzle, however, is how and why such a highly-conserved protein, localized to mitochondria, interacts with other regulatory proteins to alter growth and apoptosis. By knowing the structure, location, and possible signaling pathways involved, we propose that Snn constitutes an important sensor of mitochondrial damage, and plays a key role in the mediation of cross-talk between mitochondrial and nuclear compartments in specific cell types. J. Cell. Biochem. 98: 243–250, 2006. © 2006 Wiley-Liss, Inc.
The mechanisms underlying selective toxicity are central to understanding cellular physiology and pathophysiology. The molecular systems responsible for selective toxicity can be categorized by those imparting protective effects and those promoting direct cellular damage. Organotin compounds are among the best characterized selective cellular toxicants that mediate cellular damage [Philbert et al., 2000]. In particular, tri-substituted organotins show tissue-specific patterns of cellular damage, which are dependent on the alkyl chain length. Substituents on the tin atom such as methyl, ethyl, and butyl groups cause rather diverse toxic effects. Triethyltin salts (TET) cause swelling of myelin and brain edema, while trimethyltin salts (TMT) trigger selective apoptosis in specific subregions of the mammalian CNS and specific subsets of immune system cells [Balaban et al., 1988; Patanow et al., 1997]. Tributyltin salts (TBT), on the other hand induce apoptosis in T-cells [Arakawa and Wada, 1993]. These specific patterns of chemical toxicity prompted us toward the identification of the molecular determinants that cause this selective cellular damage.
Initially, our lab focused on the hypothesis that cells sensitive to tri-substituted organotins might express gene product(s) that confer cellular sensitivity. In order to screen and isolate gene products expressed in neurons sensitive to TMT, we used an avidin-biotin-based subtractive hybridization method [Krady et al., 1990]. From this screening, a cDNA fragment was isolated and used to identify full-length cDNA clones, which were found to encode an mRNA of 2.7 kb with a putative ORF of 88 amino acids [Toggas et al., 1992]. Subsequent studies using in situ hybridization and Northern blot analysis supported the idea that this gene product was expressed in TMT-sensitive cells, and that TMT treatment significantly altered its expression [Toggas et al., 1993]. Additional studies were then used to characterize the cDNA and genomic sequence of the putative protein termed as Stannin (Snn) from “stannum” (latin for Tin). Chromosomal mapping revealed that Snn is located on human chromosome 16p13, and has a syntenic relationship to the murine chromosomal homolog [Dejneka et al., 1998]. Subsequently, Snn was identified in a range of organisms, with a remarkable sequence conservation among all vertebrates. Figure 1a shows the homologies of Snn sequences in all of the species sequenced to date. Note the high degree of conservation around the protein core (residues 1–61), which implies a crucial function for this small protein in cell function. Also shown in Figure 1b are some of the SNPs observed to date. Again, there are very few SNPs, notably one at residue 17 (I to V) and one at residue 88 (G to S) which produce slight changes in the amino acid sequence. Thus, the conservation, lack of SNPs, and expression in vertebrates, suggest that Snn plays a key role in cellular function.
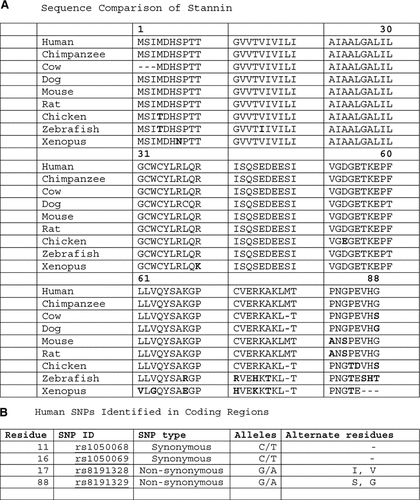
Sequence homology of Snn. Panel A shows the high degree of conservation between vertebrate species. Note that human and chimpanzee Snn are identical. The missing residues in the cow sequence may be reflective of an earlier version of sequence alignment. Panel B shows the few non-synonomous SNPs characterized for the human Snn sequences to date, underscoring the highly conserved nature of Snn.
GENOMICS AND PROTEOMICS OF SNN
While the role of Snn in mediating toxicity has been provided, the determination of Snn's physiological role is still elusive. Specifically, organotin toxicants may activate parallel biological pathways that are foreign to the normal biological function of Snn. Moreover, the lack of shared structural features or possible homology to other proteins has limited predictions for the possible biological function of Snn. In addition to this, the conserved nature of the protein has led to considerable difficulty in generation of specific, high-affinity antisera. As a result, a series of genomic and proteomic analysis were necessary to determine putative pathways that regulate gene expression and possible binding partners for Snn.
In humans, genomic cloning of Snn revealed a large gene with two exons; other vertebrates express Snn as a single transcript [Dejneka et al., 1997]. The coding region in humans, however, is contained entirely in the second exon. There are numerous predicted promoter sites located downstream from the initiation sites that could explain the high levels of expression of Snn in both the CNS and immune system, and the increased levels of expression observed during early embryonic development. Both the Icarus and ARNT sites have potential regulatory roles in lymphoid tissues, particularly during development [Dejneka et al., 1998]. Of note, Horrevoets et al. [1999] performed a series of studies in endothelial cells and found that TNFα caused a rapid and robust upregulation of Snn, suggesting that Snn was part of the response to the inflammatory process. Using these initial studies as a guide, we found that TNFα induced Snn mRNA in HUVEC and Jurkat T-cell lines, and that induction was rapid (1.5–3 hr) and robust (3–9 fold increase). Moreover, studies using siRNA and PKC inhibitors demonstrated that PKC-ε was critical for TNFα-induced expression of Snn [Reese et al., 2005]. Treatment of HUVECs with TMT also induced Snn mRNA levels as well, suggesting that organotin toxicity may involve both direct and indirect effects on Snn function and expression levels.
These studies in HUVEC were extended into microarray analysis, using TNFα and Snn-specific siRNA [Reese et al., 2006]. The design allowed for comparisons between TNF-Snn and Snn or TNF alone. Using these approaches, 96 genes were differentially expressed when compared between TNFα and TNFα/siRNA against Snn. Analysis of siRNA inhibition of Snn alone revealed that COP-1, a negative regulator of p53, was significantly lowered. Several important growth-regulatory genes were differentially upregulated between the two conditions, and several of these included genes such as MDM4, IL-4, WTI/PRKC, and HRas-like suppressor. These genes act on cyclin D1 and/or p53, and thus, perturbations were predicted to alter passage through the cell cycle. Indeed, altering Snn levels affected growth rates and progression through the G1 cell-cycle boundary. Thus, based on genomic and microarray analysis, Snn can be upregulated by both TNFα and by TMT. The cellular levels of Snn, in turn, alter the expression of TNF responsive genes. Reduction of Snn in the absence of TNF signaling reduced levels of COP1, another protein which regulates p53. Thus, Snn is likely to play a regulatory role in both the TNF/inflammatory response, and possibly a role in regulation of the cell-cycle progression. Several recent papers have supported the idea that Snn levels are increased during inflammation; Snn increased in rat dorsal root ganglia following carageenan-induced inflammatory responses [Belkowski et al., 2005]. It was further observed that Snn levels increased in human peripheral blood monocytes following SARS infection [Reghunathan et al., 2005].
Analysis of the linker region of Snn (residues 34–60) revealed a conserved CXC metal binding motif (residues 32–24), and a putative 14-3-3 binding site (RISQSED; residues 39–45). Studies using co-immunoprecipitation assays indicated that 14-3-3ξ was the specific isoform bound to Snn. These studies were carried out using a C-terminal flag-tagged Snn which was over-expressed in PC-12 cells [Davidson et al., 2005]. Overexpression of Snn in these cells increased activation of p38/MAPK and concomitantly decreased activation of the MAPK/ERK pathways. This is consistent with the idea that Snn/14-3-3 interaction may sequester the scaffolding actions of 14-3-3 away from parts of the MAPK/ERK pathway. The conserved CXC domain is a likely site for metal coordination. Buck et al. [2003] tested this hypothesis using a nine amino acid peptide derived from the Snn sequence, and showed that the CXC motif binds TMT and catalyzes its demethylation into DMT. Mass spectroscopic and CD analysis revealed that the organotin moiety remained bound to this region. The CXC domain is just upstream from the 14-3-3ξ site, and raises the prospects that TMT binding and demethylation could alter 14-3-3 binding.
Another distinguishing feature of Snn is that it is localized to the membranes of mitochondria and other organelles [Davidson et al., 2004]. A careful series of experiments using flag-tagged Snn coupled with subcellular fractionations and triple-label confocal microscopy were used to confirm the cellular localization of Snn (Fig. 2). Using GRP-75 as a marker for mitochondria, and GRP-78 as a marker for endoplasmic reticulum, we concluded that Snn is expressed in both the mitochondria and endoplasmic reticulum. The mechanisms for insertion are not obvious, but may be similar to the tail-anchored proteins (TA-proteins) or via post-translational insertion.
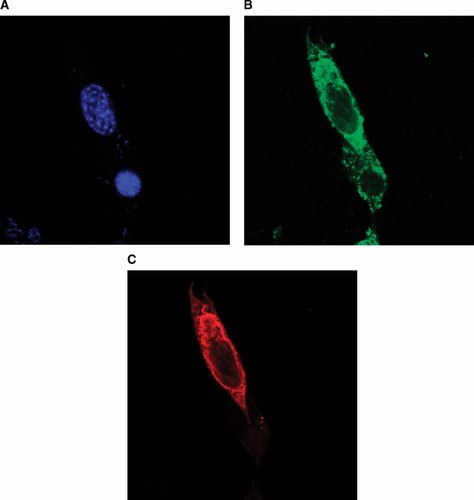
Laser confocal microscopy of flag-tagged Snn indicates that Snn is localized in the mitochondria of NIH 3T3 cells. Panel A shows a DAPI nuclear stain of NIH 3T3 cells transfected with Snn. Panel B shows the pattern of staining with the mitochondrial protein GRP-75. Panel C shows staining of flag-tagged Snn. The cellular localization pattern of Snn was further confirmed using subcellular fractionation experiments.
Furthermore, overexpression of Snn in NIH-3T3 cells was found to sensitize the cells to both TMT and DMT [Davidson et al., 2004]. Hence, the location of Snn in mitochondria could explain the alterations in Ca+2 flux observed after organotin intoxication. The mitochondrial pathway to apoptosis has been well-characterized, and consists of a series of protein interactions (Bcl-2/Bax family), outer membrane permeability changes, and the ultimate release of proteins from the intermembrane space into the cytosol, triggering caspase cascades [Spierings et al., 2005]. Mitochondrial targeting of the organotin compounds could lead to a local microenvironment in which TMT interacts with Snn, is demethylated to DMT and binds irreversibly to Snn, compromising the integrity of the mitochondrial membrane, initiating apoptosis. Thus, the genomic and proteomic analysis to date has led to several enticing mechanistic roles for Snn. In particular, Snn may be involved in the modulation of growth-related and inflammatory responses, possibly by affecting key components of the p38 and TNFα signaling paths. A model for this signaling is shown in Figure 3.
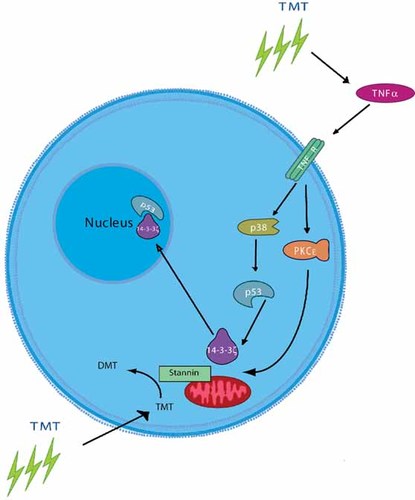
Signaling pathways of Snn regulation. This schematic reflects the current hypothesis that Snn is localized to the mitochondrial and endoplasmic reticulum membranes. Substantial evidence indicates that Snn dealkylates TMT via the CXC motif, and coordinates DMT. Snn's interactions with organotins may consequently affect the binding of 14-3-3ξ, and alter the p38/MAPK system as a result.
THE STRUCTURE OF SNN
Protein structure is an indispensable requisite to understand protein function. This is particularly true for Snn, which presents a unique sequence that has no analogy with other proteins in the databases. The relatively small size of Snn makes this protein an ideal candidate for utilizing the emerging structural tools offered for membrane proteins by solution and solid-state NMR.
The full backbone assignment of Snn was determined in detergent micelles using standard 2D and 3D heteronuclear NMR methodologies [Wuthrich, 1986; Cavanagh et al., 1996]. From the NMR structure, Snn is composed of a single transmembrane helix (residues 10–33) and a cytoplasmic helix (residues 61–79) separated by a long unstructured linker [Buck-Koehntop et al., 2005]. The metal binding CXC motif (Cys-32–Cys-34) is located at the lipid/solvent interface making it available for interaction with organotin compounds. The putative 14-3-3 binding region (residues 39–45) is further located in the unstructured linker region, distal to the conserved CXC motif. Backbone dynamics analysis of Snn in detergent micelles indicated that the transmembrane domain was fairly immobile, while the long unstructured linker exhibits dynamics on a µs–ms time scale, suggesting the linker may play a role in protein recognition. The backbone dynamics also indicated that the cytoplasmic helix was more mobile than the transmembrane domain, but less mobile than the unstructured linker region. Thus, we hypothesized that the cytoplasmic domain may be interacting with the surface of the micelle.
To verify the solution NMR structural results in the micelle system, and to determine if the cytoplasmic helix is interacting with the membrane surface, the orientation of uniformly 15N labeled Snn was determined in synthetic lipid bilayers by solid-state NMR. In addition to the full-length protein, two truncated versions of Snn corresponding to the transmembrane (Snn1–48) and cytoplasmic (Snn30–88) domains were also characterized individually by solid-state NMR. Using solid-state NMR, the 15N chemical shift can be directly correlated to the angle of the protein with respect to the lipid bilayer [Bechinger et al., 1999; Marassi, 2002]. The solid-state NMR studies on Snn indicated that when the full-length protein or the individual domains were oriented in lipid bilayers, the transmembrane, and cytoplasmic helices form angles of 20° and 80° ± 4° with respect to the membrane normal, respectively. Thus, the cytoplasmic helix is indeed interacting with the lipid bilayer surface and may have a role in mediating the toxicity of organotin compounds. The structure of Snn oriented in a lipid bilayer is represented in Figure 4.
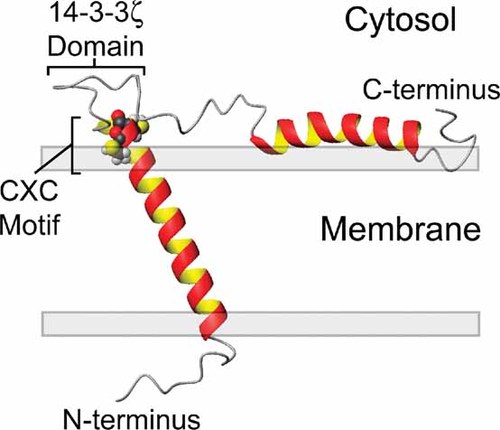
Model of SNN oriented in a lipid bilayer highlighting the CXC alkyltin metal binding motif residing at the lipid/solvent interface (residues Cys-32 and Cys-34) and the putative 14-3-3 binding domain between residues 39–45. Adapted with permission from J Mol Biol, 2005, 354, 652–665.
In summary, the proposed structure of Snn as determined in micelles and lipid bilayers suggests several features which could explain, in part, the mechanisms of organotin toxicity. The linker region is dynamic and structural, or dynamical alterations caused by interactions with organometals could recruit binding partner proteins such as 14-3-3, initiating the apoptotic cascade. In addition, the C-terminal tail of Snn, which has the greatest sequence divergence amongst species, interacts with the bilayer surface, and is available for interactions with other cytoplasmic targets.
POSSIBLE FUNCTIONAL PERSPECTIVES ON SNN
There are clear gaps in our understanding of Snn's role in mediating organotin toxicity, which leads to both opportunities and speculative hypotheses. First, the entire cascade from organotin entry into cells, binding to Snn, disruption of signaling cascades, release of cytokines, further induction of Snn, and ultimate changes in cell growth, and apoptosis has not been fully characterized in one cellular system. A comprehensive series of experiments are needed to demonstrate all of these predicted effects in a target cell. Based on our studies, we suggest that cells from the immune system, such as Jurkat T-cells, may afford the best cellular system for addressing these issues in a comprehensive manner. Jurkat cells respond to a variety of inflammatory cytokines, release cytokines, and show calcium-dependent T-cell receptor signaling. Most importantly, Snn is normally present in these cells and apoptotic signals can be measured directly. The major limitation to studying Snn in a target cell line is the lack of an available high-affinity antibody reagent against Snn due to its high sequence homology. The use of flag-tagged Snn has been used in other systems, but development of a high-affinity antibody reagent is crucial.
Second, we need to better understand how Snn's interaction with putative target proteins such as 14-3-3 induce significant downstream signaling events. It is possible that other proteins also interact directly with either the linker region or the C-terminal region of Snn. Additional studies using Snn binding assays or related biophysical technologies could prove useful for characterizing additional Snn protein binding partners involved in the apoptotic cascade. Finally, in a recent book “Power, Sex, Suicide: Mitochondria and the Meaning of Life” by Nick Lane [2005], mitochondria have been anthropomorphized as the clandestine rulers of the cellular world. Given the integral role of the mitochondria in the elaboration of Bcl/Bax-mediated apoptosis, it is possible that the vertebrate protein Snn is another example of a protein that helps to regulate communications between the clandestine mitochondria and the more orderly world of the nucleus. Such key questions of life and death clearly await more information, and Snn may be an integral link in the pathway.




