DNA sequences acting as binding sites for NM23/NDPK proteins in melanoma M14 cells
Abstract
We isolated and analyzed by chromatin immunoprecipitation (ChIP) in viable M14 cells DNA sequences bound to the antimetastatic protein nucleoside diphosphate kinase (NM23/NDPK) to shed some light on the nuclear functions of this protein and on the mechanism by which it acts in development and cancer. We assessed the presence of selected sequences from promoters of platelet-derived growth factor A (PDGF-A), c-myc, myeloperoxidase (MPO), CD11b, p53, WT1, CCR5, ING1, and NM23-H1 genes in the cross-linked complexes. Quantitative PCR (Q-PCR) showed a substantial enrichment of the correlated oncosuppressor genes p53, WT1, ING1, and NM23-H1 in the immunoprecipitated (IP) DNA. This suggests that NM23/NDPK binding is involved in the transcription regulation of these genes. These results reveal new interactions that should help us to disclose the antimetastatic mechanism of NM23. J. Cell. Biochem. 98: 421–428, 2006. © 2006 Wiley-Liss, Inc.
NM23/nucleoside diphosphate kinase (NM23/NDPK) is a ubiquitous enzyme, produced by expression of the NM23 genes in mammals. Several human isoforms are known. The most abundant of these are NDPK A and NDPK B, encoded by NM23-H1 and NM23-H2, respectively. In the cytoplasm, these proteins transform nucleoside diphosphates into triphosphates by an ATP-dependent reaction [Lacombe et al., 2000]. NM23/NDPK enzymatic activity arises from its hexameric quaternary structure [Lascu et al., 1993]. The NM23/NDPK proteins have many other functions that are not necessarily connected with their enzymatic activity. NDPK A is involved in tumor metastasis [Steeg et al., 1988], whereas NDPK B acts as a transcription factor for c-myc [Postel et al., 1993].
Although there have been many recent studies of NM23/NDPK, the role of this metastasis suppressor is still not clear and many different functions have been attributed to it. Interactions with various other proteins have been proposed [Otero, 1997; Roymans et al., 2002; D'Angelo et al., 2004; Rochdi et al., 2004]. This protein binding appears to affect a variety of functions such as cell proliferation, differentiation, cell motility, vesicle internalization, apoptosis, and metastasis [Fan et al., 2003; Gallagher et al., 2003; Salerno et al., 2003].
NDPK A and NDPK B are present also in the cell nucleus where they may act as transcription regulators. They also appear to have a nuclease activity, which may be important in apoptosis and DNA repair [Ma et al., 2002, 2004]. Several studies have demonstrated the interaction of these proteins with specific gene promoters [Postel, 2003], mainly by nm23 gene transfection and/or electrophoretic mobility shift assays (EMSA). However, in vivo evidence of binding is still lacking.
This article describes the results obtained in vivo with M14 cells, a melanoma cell line with intermediate metastatic potential [van Muijen et al., 1991; Meije et al., 2002]. An inverse correlation between the aggressiveness of the tumor and NDPK A expression was first demonstrated in murine melanoma tumor by analysis of its RNA level [Steeg et al., 1988]. Other studies have shown the same correlation in human melanoma [Hartsough and Steeg, 2000].
The assessment of the DNA sequences interacting with NM23/NDPK is obviously interesting for designing cancer chemotherapy drugs. If the NM23/NDPK protein binds directly to a DNA sequence, a cross-linking agent can be used to produce a covalent bond in vivo, and thus the DNA–protein complex can be isolated and recognized. We have already demonstrated by this technique, the in vivo binding of NDPK to the 5′S1 nuclease hypersensitive (5′SHS) element of the platelet-derived growth factor A-chain (PDGF-A) promoter in K562 cells [Cervoni et al., 2003]. Therefore, we have continued studying NDPK binding of DNA sequences by in vivo cross-linking and chromatin immunoprecipitation (ChIP) in another human cell line, namely M14, which is a moderately aggressive melanoma line. This treatment does not affect the living cells over the time course of the experiment, and the subsequent analysis of DNA fragments bound to NDPK allows us to “photograph” the interactions occurring in the cell without manipulating or changing the growth conditions.
The genes we chose to investigate contain in their promoters a G-rich sequence related to a non-B DNA conformation [Postel, 2003]. Some of these genes have already been found in vitro to interact structurally, and in some cases functionally [Postel et al., 2000], with NM23/NDPK. The other genes were chosen on a rational basis for their involvement in metastasis or apoptosis (Table I).
| Gene | Primers | PCR product (bp) | Probes (5′→ 3′) |
|---|---|---|---|
| 5′SHS PDGF-A | Fw:AGGACAAGGCCTGGCTTTAA | 300 | AGAGACGTGGGGAGGGGGCCTGCAGGTGTGT |
| Rv:CTAGGCCAGCTCTAGCATTT | |||
| NHE PDGF-A | Fw:GGGGCTTTGATGGATTTAGC | 207 | AGAGGGGGCGGGGGCGGGGGCGGGGGGAGT |
| Rv:GGCGGGGAGAGGGTTATAG | |||
| c-myc | Fw:AGGCGCGCGTAGTTAATTCA | 214 | ATGGGGAGGGTGGGGAGGGGTGGGGAAGGTG |
| Rv:TCGCATTATAAAGGGCCGGT | |||
| MPO | Fw:TGTGTGTACCTTCCAACCCA | 174 | TCCTCTAGCGGGGGTGGGGTGAGGTAGAGG |
| Rv:GTCATCCAGCTTCCAAGGAC | |||
| CCR5 | Fw:CCCGTGAGCCCATAGTTAA | 170 | GTGTGGGGGTTGGGGTGGGATAGGGGATACG |
| Rv:TAGAGGGGGATCCTGGACTT | |||
| CD11b | Fw:ACCCAAGAAACAAGTGGGTG | 145 | GCTGGGGAGGAAGGGTGGGCAGGCTGTGGG |
| Rv:TCAGTGAGCACATAGCCCTG | |||
| p53 | Fw:GGATCCAGCTGAGAGCAAAC | 160 | GCTTTCTTCCTTCCACCCTTCATATTTGAC |
| Rv:CGACCTGGTGCCGTAGATA | |||
| WT1 | Fw:AGCAAGAGCCAGACTCAAGG | 159 | GAGCCTACCTGCCCCTCCCTCCAAACCACT |
| Rv:TGGGAGTAGAGATGGGGTTG | |||
| ING1 | Fw:ACGCCACAGGAAACAAAACT | 241 | TTTTATCCCAAGGGGTGGGCTAAAAGTTTT |
| Rv:GTGCGCTGGGGATACAGTAG | |||
| nm23-H1 | Fw:TCTGGCCTTTTCTTCACAGC | 214 | CCTATACTCCCATCCCTCCCAGAAGCTCCA |
| Rv:GCTCCCGCTTTGTGTTTATT | |||
| exon 2 p53 | Fw:TGGAAGTGTCTCATGCTGGA | 219 | |
| Rv:CTTCCCACAGGTCACTGCTA | |||
| exon 1a ING1 | Fw:GTTAGGTCCTGGTCGGGTTT | 201 | |
| Rv:GCCTTGCCTGTCACTTTTTC | |||
| exon 10 MPO | Fw:AGCCCTCTTCTCGAATCCTC | 222 | |
| Rv:CTGCAATTTGGTTCTGACGA |
The antibodies we used recognize both NDPK A and NDPK B isoforms, which are naturally present in the cell as heterohexamers.
Our results show that both the A and B forms of the protein interact with DNA, and that the in vivo interaction is not necessarily related to upregulation or downregulation of the examined gene. We also found that in M14 cells, the promoters of p53 and other related genes are all natural ligands of NM23.
MATERIALS AND METHODS
M-280 sheep anti-rabbit IgG Dynabeads were from Dynal. Rabbit IgG was from Sigma. Proteinase K and cis-diammine dichloro platinum II (cis-DDP) were from Sigma, Benzonase was from Merck, and DNase-free RNase A was from Boehringer. The oligonucleotides used were synthesized by MWG (Florence, Italy) and by PRIMM (Milan, Italy).
Recombinant NDP kinase A and B were purified as described [Lascu et al., 1997; Gonin et al., 1999] on Q-Sepharose columns and stored as precipitates in saturated ammonium sulfate solutions.
Antibodies
Polyclonal antibodies against NDP kinase were raised in rabbits immunized with NDP kinase B by Davids (Regensburg, Germany) and were purified by affinity chromatography using a HiTrap NHS-activated column (Amersham-Pharmacia) [Phang-Ba et al., 1998]. These antibodies cross-react with the highly homologous NDP kinase A, thus allowing simultaneous detection of both isoforms, which migrate differently on SDS–polyacrylamide gel electrophoresis.
M14 Cells
The cells were grown in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum, 5 mM glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, and 1 mM Na pyruvate, in humidified atmosphere containing 5% CO2, and collected by centrifugation. The cells were washed twice with PBS buffer (10 mM Na2HPO4, 1.76 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, pH 7.5).
cis-DDP Cross-Linking and Preparation of Chromatin
106 cells were suspended in 1 ml cis-DDP buffer (1 mM cis-DDP, 10 mM Na2HPO4, 1.76 mM KH2PO4, 3 mM MgSO4, 135 mM CH3COONa, pH 7.5). After 60 min of incubation at 37°C, 1/200 volume of 1 M thiourea solution was added to stop the cross-linking reaction [Ferraro et al., 1992, Cervoni et al., 2003]. The cells were harvested by centrifugation at 2,000g for 5 min at 4°C. The cell pellet was washed in cis-DDP buffer with 5 mM thiourea. The final cell pellet was re-suspended in 5 volumes of nuclei buffer (10 mM Hepes pH 8, 10 mM KCl, 1.5 mM MgCl2, with protease inhibitor cocktail) and incubated for 10 min on ice [Dignam et al., 1983]. After centrifugation at 2,000g for 10 min at 4°C, the cell pellet was re-suspended in 2 volumes of lysis buffer 1 (nuclei buffer with 0.05% v/v Triton X-100, with protease inhibitor cocktail) by passage through a hypodermic needle (26 gauge, ½ in.) four times. After centrifugation at 2,000g for 10 min at 4°C, the nuclei were re-suspended in nuclei buffer and washed twice. The nuclei were then re-suspended in lysis buffer 2 (10 mM Tris-HCl pH 8, 1 mM EDTA, with protease inhibitor cocktail). A sonicator (Soniprep150, MSE) was used to disrupt the nuclear membranes and to fragment the chromatin. DNA fragments, each between 0.6 and 0.8 kb, were generated with ten 25-s pulses at a power setting of 5. After each pulse, the sample was allowed to cool for 1 min in ice. The chromatin solution was adjusted to 0.5% SDS, mixed for 10 min at room temperature, and centrifuged at 10,000g for 10 min. The supernatant (whole chromatin) was then stored at −80°C.
Immunoprecipitation of Chromatin
All the experiments were repeated on three different batches of cross-linked cells. Sheep anti-rabbit IgG-conjugated Dynabeads (100 µl) were washed three times with cold PBS containing 5 mg/ml bovine serum albumin (BSA) and re-suspended in 5 ml of cold PBS. Rabbit polyclonal antibody against NDP kinase (10 µg), or rabbit IgG (10 µg) for a control mock precipitation, was added to the mixture and incubated at 4°C overnight. After collecting the beads with the magnet and washing them three times with cold PBS containing 5 mg/ml BSA, the beads were re-suspended in 100 µl of cold PBS containing 5 mg/ml BSA. They were then ready for immunoprecipitation. Whole chromatin (1 µg) [Ren and Dynlacht, 2003] was adjusted to 0.1% Triton X-100, 0.1% Na deoxycholate, 1 mM PMSF, and mixed with 100 µl of magnetic beads pre-bound to the antibodies. The mixture was incubated at 4°C overnight. The beads were then collected using a magnet and washed five times with RIPA buffer (50 mM Tris-HCl pH 8, 1 mM EDTA, 0.1% Na-deoxycholate, 1% Triton X-100, 140 mM NaCl, 0.1% SDS, with protease inhibitor cocktail) and once with TE buffer (10 mM Tris-HCl pH 8, 1 mM EDTA, with protease inhibitor cocktail). The beads were re-suspended in 100 µl TE containing 50 µg/ml DNase-free RNase A and incubated for 30 min at 37°C. After washing twice with TE, the beads were re-suspended in 50 µl of elution buffer (50 mM Tris-HCl pH 8, 10 mM EDTA, 1% SDS) and incubated at 65°C for 10 min. Supernatant (50 µl )(immunoprecipitated (IP) and IgG samples) was removed and mixed with 50 µl of TE.
Purification of DNA
The proteins from the IP and IgG samples were removed by incubation with 500 µg/ml Proteinase K for 2 h at 37°C. The solution was then incubated for 30 min at 50°C in 1.5 M thiourea to reverse the cross-linking. The chromatin Input control was prepared by subjecting 100 µg of whole chromatin to the same digestion with Proteinase K and treating it with thiourea to reverse the cross-linking. DNA was then extracted twice from all the samples with phenol/chloroform/isoamyl alcohol (25:24:1) and precipitated with ethanol.
Purification of Proteins
DNA in the IP and IgG samples was removed by incubation with 0.25 U/µl Benzonase overnight at 37°C. After addition of the loading buffer (62.5 mM Tris-HCl pH 6.8, 2% SDS, 10 mM glycerol, 5 mM dithiothreitol) containing 0.1 M thiourea, the samples were heated for 30 min at 90°C to reverse the cross-linking.
DNA Quantification
The amount of DNA recovered from IP, IgG, and Input samples was detected by ethidium bromide fluorescent quantification according to Sambrook et al. [1989].
PCR Amplification of IP DNA
We confirmed the presence of specific DNA sequences by subjecting the IP DNA to PCR, using the primers listed in Table I. PCR was carried out in a 50 µl volume in the presence of 5 ng of IP DNA, using a Perkin Elmer GeneAmp PCR System 2400. We ensured that the PCR was still in the exponential phase to allow for quantitative interpretation of the results by analyzing control reactions with increasing amounts (1, 5, 10 ng) of DNA Input in parallel. An aliquot of the reaction mixture was electrophoresed through 2% agarose gel and the fragments were revealed by ethidium bromide staining. The fragments were identified by Southern blotting and hybridization with [33P]-labeled oligonucleotides (Table I) according to Cervoni et al. [2003].
Real-Time PCR
Quantitative PCR (Q-PCR) analysis was carried out on a 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA). The primers, listed in Table II, were designed using Primer Express software (Applied Biosystems) and were synthesized by PRIMM. The reactions were carried out in a 25 µl volume, in 96-well plates, using SYBR Green PCR Master mix (Applied Biosystems). A specific standard curve for each gene was performed in parallel to the analyses. Each sample was analyzed in triplicate. PCR conditions were 10 min at 94°C, followed by 40 cycles of 15 sec at 94°C and 1 min at 60°C. Results were analyzed by using SDS software (Applied Biosystems).
| Gene | Primers |
|---|---|
| p53 | Fw:AGGATCCAGCTGAGAGCAAA |
| Rv:AGAGCTGTGAGGGCAGAATT | |
| WT1 | Fw:TTCTGCGCTTTCCTGAAGTTC |
| Rv:GAATGCGGTGGGAGTAGAGATG | |
| ING1 | Fw:GCTTGATCTTCTAAGTTTGTTCC |
| Rv:GCTGGGGATACAGTAGTGAA | |
| nm23-H1 | Fw:TTTGCTTATTTGCTTCTTGTTTTCC |
| Rv:CCTAGGGATAAAACAGTATGCTCTCTTG | |
| exon 1a ING1 | Fw:CTTTGCATTTTGCAGTGCTATTTT |
| Rv:TTGGCAGGACTCAACATGGTT |
RESULTS
M14 melanoma cells were subjected to in vivo cross-linking with cis-DDP. IP proteins were recovered from their isolated nuclei and purified. The pool of bound proteins, after DNA digestion, was shown by Western blotting to contain both NDPK A and NDPK B (Fig. 1).

Western blotting of proteins present in the immunoprecipitated (IP) sample after cross-linking with cis-DDP. The polyclonal anti-NDP kinase antibodies used react with both NDP kinases A and B. Lane 1: M14 IP proteins; (lane 2) recombinant NDPK A standard, 10 ng; (lane 3) recombinant NDPK B standard, 20 ng. The molecular mass of standard markers is indicated on the right.
The chromatin was subjected to immunoprecipitation with antibodies against NDPK. We analyzed the DNA fragments from the immunoprecipitate by PCR and Southern blotting. We looked only at genes containing in their promoters a G-rich sequence related to a non-B DNA conformation [Postel et al., 2000, 2003]. These sequences can form a G-quadruplex structure, which is a repressive element involved in silencing gene expression and which is a putative binding site for NM23 [Siddiqui-Jain et al., 2002].
The sequences chosen (Table III) are present in the promoters of the myeloperoxidase (MPO) gene, some oncogenes (PDGF-A, c-myc, CCR5, CD11b), and tumor/metastasis suppressor genes (p53, ING1, WT1, NM23-H1). These genes are all expressed in different ways in the melanoma M14 cell line [Su et al., 2002]. We subjected three different exons to the same treatment as a negative control. These DNA fragments were located in the p53 exon 2, the ING1 exon 1a, and the MPO exon 10. There were no G-rich sequences inside these regions.
| −1,416 AGACGTGGGGAGGGGGCCTGC −1,396 | PDGF-A 5′SHS silencer |
| −72 GGGGCGGGGGCGGGGGCGGGG −52 | PDGF-A NHE promoter |
| −115 ATGGGGAGGGTGGGGAGGGTG −135 | c-myc promoter |
| −38 TAAGCGGGGGTGGGGTGAGGT −58 | MPO promoter |
| −529 GGGGTTGGGGTGGGATAGGGG −509 | CCR5 promoter |
| −126 GGAGGAAGGGTGGGCAGGTCC −106 | CD11b promoter |
| −234 ATATGAAGGGTGGAAGGAAG −254 | p53 promoter |
| −376 GTTTGGAGGGAGGGGCAGGTA −396 | WT1 promoter |
| −2,014 ATCCCAAGGGGTGGGCTAAAA −1,994 | ING1 promoter |
| −410 AGCTTCTGGGAGGGATGGGA −430 | nm23-H1 promoter |
Hybridization control of PCR products by Southern blotting was always positive (not shown). Figure 2 shows representative examples of PCR products separated on agarose gel by electrophoresis. We used the same quantity of DNA from the ChIP tests for the PCR (5 ng) allowing a visual estimate of any differences found. Controls were prepared in the same manner as the samples, except that the antibody used in the immunoprecipitation procedure was a non-specific IgG. Semi-quantitative densitometry analysis (not shown) showed that NM23/NDPK was bound to all the chosen genes in vivo, whereas no binding to the three control exons was observed (IP/IgG = 1.12 ± 0.28 average value from three different samples for each exon). We then focused on the oncosuppressor p53 gene and the correlated ING1, WT1, nm23-H1 genes to obtain a quantitative evaluation of the binding to nm23/NDPK. These were further examined using real-time PCR (Fig. 3). Analysis of the products (Fig. 4) clearly showed enrichment of the IP sample DNA for each of the four promoters, no binding to the control exon, confirming in vivo binding to NM23.
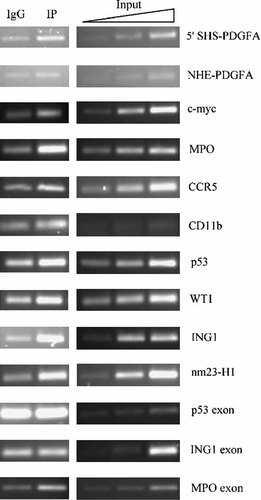
Analysis of IP DNA after PCR amplification. PCR product from IgG control sample (obtained by ChIP with a non-specific antibody) and IP sample (obtained by ChIP with polyclonal antibody against NDP kinase) were examined by 2% agarose gel electrophoresis followed by staining with ethidium bromide. Control reactions with increasing amounts (1, 5, 10 ng) of DNA Input were analyzed in parallel for each gene to verify that the PCR was still in the exponential phase.
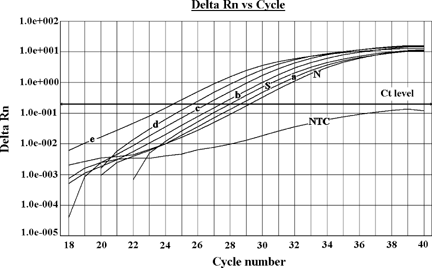
Representative real-time PCR logarithmic plot. Curves show the amplification of standards (a = 1.25 µg, b = 2.5 µg, c = 5 µg, d = 10 µg, and e = 20 µg), negative control (N), and analyzed sample (S), using p53 primers. The curve under the threshold line (black horizontal line) is “no template control” (NTC). Samples were analyzed in triplicate; each curve represents the average of the three values.
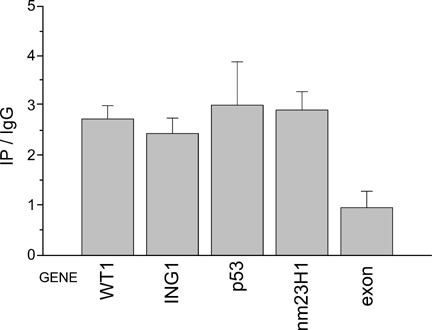
Gene enrichment in immunoprecipitate DNA calculated from real-time PCR data. Results are expressed as the ratio of DNA (IP) to control DNA (IgG). Each column represents the average of three different experiments. +1 standard deviation is shown on top.
DISCUSSION
A recent study [Zhao et al., 2004] on the expression of genes associated with NM23, based on the comparison between a high metastatic carcinoma cell line and the same line transfected with nm23-H1, suggested that the antimetastatic mechanism of NM23 proteins involved the downregulation of genes associated with cell adhesion and motility.
It would be very useful to know the DNA sequences that interact with NM23/NDPK, both to define the action of this protein in the nucleus and to allow new antimetastatic drugs to be developed. Therefore, we tried to obtain new information using our already successful method of in vivo cross-linking followed by ChIP.
This study was limited to M14 cells, a moderately metastatic melanoma cell line, and to interactions between nm23/NDPK proteins and several genes containing in their promoters G-rich sequences related to a non-B DNA conformation [Postel, 2003; Siddiqui-Jain et al., 2002].
We found by semi-quantitative PCR that all of the chosen sequences were linked to NM23/NDPK after cross-linking. Real-time PCR showed a more than 2.5 times enrichment of the p53 gene and the correlated ING1, WT1, nm23-H1 genes in the immunoprecipitate, and no enrichment in the control exon 1a ING1. All of these have been described as oncosuppressors, but p53 [Chang et al., 1993; Zhang et al., 2002] and ING1 are overexpressed in M14, whereas WT1 and nm23-H1 in the same cell line are underexpressed [Su et al., 2002]. The physical association between p53 and the product of ING1 transcription (p33ING1) detected by immunoprecipitation [Garkavtsev et al., 1998], indicates that this two-protein complex is involved in the negative regulation of cell proliferation. Our results show that NDPK binds to both p53 and ING1 genes, suggesting that this protein may act as antimetastatic factor by favoring their transcription. The transcription of NM23-H1 was found to be regulated by p53 [Chen et al., 2003], with different regulation mechanisms in different cell lines, which further complicates things.
WT1 is a tumor suppressor gene that encodes for a 52–54 kDa nuclear zinc-finger protein [Rauscher et al., 1993]. WT1 promotes the transcription of genes involved in growth and cellular proliferation, such as EGR1 and IGF2. A physical and functional association between WT1 and p53 genes was identified. It has also been shown that in baby rat kidney (BRK) and human melanoma cells, WT1 can act as a transcriptional repressor by interacting with p53 or other cellular proteins [Maheswaran et al., 1993; Rodeck et al., 1994]. Our results show that NM23 binds to WT1 and we suggest that this interaction inhibits its transcription.
We demonstrate that NM23 also binds to the promoter of NM23-H1, suggesting the presence of a feedback regulation system for the protein level in the cells.
However, we are unable to draw any conclusions on the inhibitory or enhancing effect of the binding on transcription or on specific functions determined from other proteins binding the DNA–NM23 complex because the bound genes that we examined can be either upregulated or downregulated in M14. This shows the importance of the cellular context and of multiple interactions with other nuclear proteins in affecting the outcome of the binding. We plan to use our ChIP-based technique on other melanoma cell lines with different metastatic potential. This may help to clarify the regulatory activity of NM23/NDPK binding on the expression of the interacting genes.
In this work, we made no distinction between the binding of NDPK A and NDPK B. It is not known if DNA binds to the hexameric form of the protein that can contain both types of subunits. It was shown that G-rich single-stranded oligonucleotides had greater affinity for a dimeric mutant of NPDK in vitro [Agou et al., 1999]. Until we know the quaternary structure of the protein upon DNA binding, defining different affinities for the two types of subunits is of no use. A recent study [Bosnar et al., 2004] on the localization of NDPK A and NDPK B in tumor cells could only show that both enter the nucleus at the same point in the cell cycle. However, some unexplained quantitative differences were observed.
The interesting result is that both proteins are present in our IP complexes.
No definite proof of a direct DNA-NM23/NDPK bond in vivo has yet been obtained. Although cis-DDP can also produce protein–protein cross-linking, an indirect bond mediated by unknown protein(s) seems unlikely in our in vivo cross-linking conditions [Banjar et al., 1985; Ferraro et al., 1992]. Moreover, some of the sequences found in the coimmunoprecipitated DNA were already shown to bind NM23/NPDK by in vitro EMSA experiments [Postel et al., 2000]. In any case, we clearly show that the protein binds to or is in proximity of the analyzed promoters. We still need to determine what other genes are natural ligands of NM23/NDPK. We are currently carrying out studies in this direction.
Acknowledgements
This work was supported by a 2003 grant from Associazione Italiana per la Ricerca sul Cancro (AIRC), Milano, Italy.




