Imprinting centers, chromatin structure, and disease
Abstract
Two regions that best exemplify the role of genetic imprinting in human disease are the Prader–Willi syndrome/Angelman syndrome (PWS/AS) region in 15q11-q13 and the Beckwith–Wiedemann syndrome (BWS) region in 11p15.5. In both regions, cis-acting sequences known as imprinting centers (ICs) regulate parent-specific gene expression bidirectionally over long distances. ICs for both regions are subject to parent-specific epigenetic marking by covalent modification of DNA and histones. In this review, we summarize our current understanding of IC function and IC modification in these two regions. © 2005 Wiley-Liss, Inc.
Genetic imprinting, the mechanisms that lead to parent-specific differential expression of a subset of mammalian genes, plays an important role in the pathogenesis of human disorders of growth and neurologic development. The two regions of the human genome that best exemplify the role of imprinting in human disease are the Prader–Willi syndrome/Angelman syndrome (PWS/AS) region in 15q11-q13 (reviewed by Nicholls and Knepper [2001]), and the Beckwith–Wiedemann syndrome (BWS) region in 11p15.5 (reviewed by Weksberg et al. [2003]). In both of these regions, cis-acting sequences referred to as imprinting centers (ICs) regulate parent-specific gene expression bidirectionally over long (up to 1 Mb) distances (Fig. 1). For both the PWS/AS region and the BWS region, the IC is subject to parent-specific epigenetic marking by covalent modification of DNA and histones; in both cases, the maternal IC, which is marked by CpG methylation and by histone H3 Lys9 dimethylation, is inactive and functionally equivalent to an IC deletion, while the paternal IC, which is marked by histone H3 Lys4 methylation, is active and produces the paternal pattern of gene expression and epigenetic modification throughout the imprinted domain [Xin et al., 2001; Higashimoto et al., 2003]. In this review, we will summarize our current understanding of IC modification and IC function in the PWS/AS and BWS regions, and discuss mechanisms by which unmethylated ICs may regulate imprinted gene clusters.
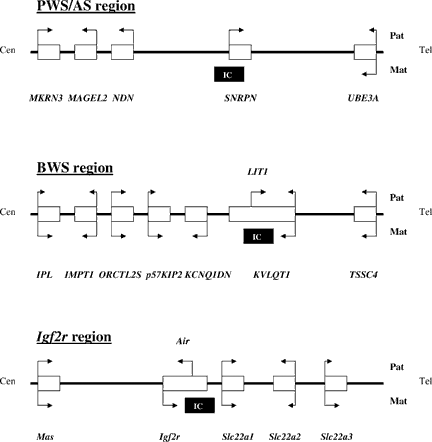
Schematic diagrams of imprinted gene clusters: Prader–Willi syndrome/Angelman syndrome (PWS/AS) gene cluster from human chromosome 15q11-q13; BWS LIT1/KIP2 gene cluster from human chromosome 11p15.5; and mouse Igf2r gene cluster. Arrows above and below boxes indicate relative expression levels from paternal and maternal alleles, respectively. Arrows of equal lengths above and below box indicate lack of imprinting; single arrow above or below box indicates monoallelic expression; and arrows of unequal length above and below box indicate preferential expression from paternal or maternal allele (in some cases, this indicates tissue-specific imprinting). Locations of imprinting centers (ICs) that are CpG-methylated and histone H3 Lys9 methylated on the maternal alleles are shown by black bars.
PWS/AS REGION
Deletions of a ∼4 Mb region from chromosome 15q11-q13 produce either of two distinct clinical syndromes, depending on the parental origin of the deleted chromosome. Deletion of the paternal chromosome causes PWS, characterized by infantile hypotonia, mild-to-moderate developmental delay, childhood-onset hyperphagia and obesity, and genital underdevelopment. Deletion of the maternal chromosome causes a completely different clinical syndrome, AS, characterized by severe mental retardation, lack of speech, seizures, and easily-provoked smiling and laughter. The PWS/AS deletion region contains at least six imprinted genes (Fig. 1). Four of these genes (SNRPN, NDN, MAGEL2, and MKRN3) are expressed exclusively from the paternal chromosome, and loss of the active paternal alleles of these genes causes PWS. Two genes in the region show tissue-limited maternal-specific expression. One of these genes, UBE3A, is imprinted only in certain brain regions [Albrecht et al., 1997] and is imprinted in neurons but not in glial cells cultured from prenatal mouse brains [Yamasaki et al., 2003]. Lack of a functional maternal allele of UBE3A causes AS [Kishino et al., 1997; Matsuura et al., 1997]. The other gene that shows tissue-specific imprinting is ATP10A [Meguro et al., 2001].
The PWS/AS region can exist in either of two mutually exclusive states of gene expression and epigenetic modification (referred to as “epigenotypes”): the paternal state and the maternal state [Buiting et al., 1995]. Establishment and maintenance of the paternal state requires a 4.3-kb DNA segment that overlaps the SNRPN promoter, referred to as the PWS-IC, in cis [Ohta et al., 1999; Bielinska et al., 2000]. Establishment of the maternal state during oogenesis requires a 0.9-kb DNA segment ∼35 kb centromeric to the PWS-IC referred to as the AS-IC, unless the PWS-IC is deleted from the chromosome [Buiting et al., 1999].
In somatic cells of human and mouse, the PWS-IC is heavily CpG methylated on the maternal chromosome and is almost completely unmethylated on the paternal chromosome [Glenn et al., 1996; Shemer et al., 1997]. Schweizer et al. [1999] showed that the PWS-IC contains two prominent nuclease-hypersensitive sites flanking SNRPN exon 1 on the paternal allele, but is completely inaccessible to nucleases on the maternal allele. Other sites of parent-specific CpG methylation in the region include the promoters of MKRN3 [Driscoll et al., 1992] and NDN [Lau et al., 2004], both of which are methylated on the silent maternal allele and unmethylated on the active paternal allele, as well as intron 7 of SNRPN, which is methylated on the paternal allele and unmethylated on the maternal allele [Glenn et al., 1996]. No parent-specific CpG methylation of the 5′ region of UBE3A has been found in either human lymphocyte DNA [Lossie et al., 2001] or mouse brain DNA (Kishino and Wagstaff, unpublished data). The PWS-IC is differentially methylated in mouse oocytes and sperm (hypermethylated in oocytes, unmethylated in sperm), and maintenance of the gamete-specific CpG methylation patterns can account for the methylation patterns in somatic cells [Shemer et al., 1997]. In humans, there have been conflicting reports regarding the CpG methylation state of the PWS-IC in oocytes: El-Maarri et al. [2001] found the region to be unmethylated whereas Geuns et al. [2003] found heavy methylation of the PWS-IC in human oocytes. The different results from these investigators probably reflect the technical difficulties of performing bisulfite genomic sequencing on very small numbers of oocytes available from human females.
Recently, several groups have examined histone modifications of the PWS-IC and of other sites within the PWS/AS region in order to gain further understanding of how parent-specific imprints are established, maintained, and spread throughout this large region. This analysis has been facilitated by the availability of antibodies specific for covalently modified histones in chromatin immunoprecipitation assays, and by the availability of cells or cell lines from individuals lacking either the paternal PWS/AS region (and therefore affected by PWS) or the maternal region (affected by AS). Two groups detected hyperacetylation of the N-terminal tails of histones H3 and H4 in the paternal PWS-IC region, which contains the promoter of the active SNRPN allele [Saitoh and Wada, 2000; Fulmer-Smentek and Francke, 2001]. Xin et al. [2001] detected specific association of dimethyl Lys9 histone H3 with the maternal PWS-IC region in cultured human lymphoid cells; this modification is generally associated with silenced or heterochromatic regions [Jenuwein and Allis, 2001]. They did not detect parent-specific association of this modified histone with the promoters of any of the other imprinted genes in the PWS/AS region (MKRN3, MAGEL2, NDN, UBE3A, ATP10A) or with the AS-IC. They also detected paternal-specific association of methyl Lys4 histone H3 with the PWS-IC and with the NDN promoter. This association of methyl Lys4 histone H3 with the promoters of active SNRPN and NDN alleles is consistent with its general association with transcriptionally active loci. Association of dimethyl Lys9 H3 with the maternal PWS-IC and of methyl Lys4 H3 with the paternal PWS-IC has also been detected in mouse [Fournier et al., 2002; Xin et al., 2003].
- (1)
transcription from the PWS-IC/SNRPN promoter regulates expression of all imprinted genes in the PWS/AS region;
- (2)
a molecular alteration (e.g., DNA modification, histone modification, histone variant, nonhistone chromosomal protein) “spreads” from the PWS-IC throughout the PWS/AS region; or
- (3)
sites throughout the PWS/AS region interact directly with the PWS-IC by DNA looping to generate the paternal pattern of imprinted gene expression.
Runte et al. [2001] have shown evidence for paternal-specific transcription spanning the >500 kb between the PWS-IC and UBE3A, in antisense orientation to UBE3A. They have hypothesized that this transcript may function to repress UBE3A sense-strand expression from the paternal allele. However, a cause-and-effect relationship between the antisense transcript and UBE3A imprinting has still not been demonstrated conclusively. This hypothesis also does not provide any insight into the mechanisms by which the PWS-IC regulates imprinting of NDN, MAGEL2, and MKRN3, located ∼1 Mb upstream from the PWS-IC. There is no evidence at present for spreading of any molecular alteration from the PWS-IC in contiguity, or for looping to bring widely-separated promoters into contact with the PWS-IC.
The PWS-IC carries parent-specific CpG methylation patterns reflective of gametic CpG methylation patterns in the mouse, and it is the only sequence in the PWS/AS region that has been shown to carry a parent-specific H3 Lys9 methylation mark. What is the relationship between these two epigenetic modifications of the PWS-IC? Recent evidence has pointed to a dependency of cytosine methylation on H3 Lys9 methylation in several species. Tamaru and Selker [2001] showed that CpG methylation in Neurospora is dependent on the function of the H3 Lys9 methyltransferase encoded by dim-5. Subsequently, Jackson et al. [2002] showed that CpNpG methylation in Arabidopsis thaliana requires function of the Kryptonite (Kyp) H3 Lys9 methyltransferase. Lehnertz et al. [2003] showed that mouse ES cells homozygously mutated for Suv39h1 and Suv39h2 (which encode closely-related histone H3 Lys9 methyltransferases specific for centromeric heterochromatin) have reduced DNA methylation of pericentric satellite repeats, but not of other repeat sequences. By contrast, H3 Lys9 methylation at pericentric heterochromatin was not impaired in ES cells lacking the major maintenance DNA methyltransferase (Dnmt1 −/−) or the two major de novo DNA methyltransferases (Dnmt3a −/− Dnmt3b −/−).
Xin et al. [2003] studied mouse ES cells and embryos homozygous for an inactivated allele of G9a, which encodes the major H3 Lys9 methyltransferase in euchromatic regions of the nucleus. G9a can methylate in vitro-synthesized H3 in the absence of other histones [Tachibana et al., 2001, 2002]. G9a −/− ES cells showed reduced association of dimethyl Lys9 H3 with the Snrpn promoter, and they lost all CpG methylation of the Snrpn promoter. They also showed loss of imprinting (i.e., biallelic expression) of Snrpn, assayed by RNA-FISH. Dnmt1 −/− ES cells, by contrast, showed normal association of dimethyl Lys9 H3 with the Snrpn promoter, absent CpG methylation of the Snrpn promoter, and monoallelic expression of Snrpn. Surprisingly, embryonic day 9.5 G9a −/− embryos showed normal methylation of the Snrpn promoter; bisulfite analysis showed almost complete CpG methylation of ∼50% of molecules, as seen in wild-type embryos. These results suggest that maintenance of PWS-IC DNA methylation in ES cells is dependent on H3 Lys9 methylation and that maintenance of Snrpn imprinting in ES cells requires H3 Lys9 methylation but not DNA methylation, while maintenance of PWS-IC DNA methylation in postimplantation embryos is not dependent on H3 Lys9 methylation.
BWS REGION
BWS is characterized by prenatal and postnatal overgrowth, enlarged tongue (macroglossia), and anterior abdominal wall defects. Additional, but variable, complications include enlargement of kidneys, adrenals, and liver; hypoglycemia in infancy; hemihypertrophy; genitourinary abnormalities; and, in ∼10% of affected children, embryonal tumors (most frequently Wilms' tumors). All known causes of BWS result from genetic or epigenetic changes within the chromosome 11p15.5 region. The most common etiology of BWS, found in ∼50% of cases, is imprinting defect, with loss of CpG methylation of the maternal differentially CpG-methylated region (DMR)-LIT1. Less common etiologies include mosaic paternal uniparental disomy (UPD) of 11p15.5, paternal duplication of 11p15.5, maternally-inherited mutations of the KIP2 (p57KIP2; CDKN1C) gene, and maternal chromosome rearrangements involving 11p15.5 (reviewed by Weksberg et al. [2003]).
The imprinted region at 11p15.5 involves approximately 1 Mb and includes two independently regulated domains: KIP2/LIT1 and IGF2/H19. Each domain is controlled by its own IC. Current evidence indicates that the organization and imprinting mechanisms of the two domains are quite different: the KIP2/LIT1 region involves bidirectional regulation of multiple genes over long distances by a maternally-methylated IC; the IGF2/H19 region involves short-range interactions between two genes and their regulatory regions, regulated by a paternally-methylated IC. Here we will focus on the KIP2/LIT1 region, because of its parallels with the PWS/AS region. The IGF2/H19 region is the subject of numerous excellent reviews [Delaval and Feil, 2004; Lewis and Murrell, 2004].
In the KIP2/LIT1 domain, LIT1 is the only gene expressed exclusively from the paternal allele [Lee et al., 1999]; other imprinted genes in the region, IPL, IMPT1, KIP2, KCNQ1DN, and KvLQT1, are expressed preferentially or exclusively from the maternal allele (Fig. 1). The DMR at the 5′ CpG island of the noncoding LIT1 transcript, namely DMR-LIT1, is a functional IC for the KIP2/LIT1 domain and is normally methylated on the maternal allele and unmethylated on the paternal. The mouse homolog of DMR-LIT1 was shown by Fitzpatrick et al. [2002] to regulate the neighboring imprinted genes within the domain in cis. In mice, targeted deletion of DMR-Lit1 on the paternal chromosome resulted in biallelic expression of genes that are normally silent on the paternal chromosome. Targeted deletion of human DMR-LIT1 using microcell hybrids produced the same result; when the paternal DMR-LIT1 was deleted, genes normally expressed preferentially from the maternal chromosome, such as KIP2, KCNQ1DN, and KvLQT1, were derepressed on the paternal chromosome [Horike et al., 2000]. The evidence indicates that DMR-LIT1 is an IC for the KIP2/LIT1 domain and that an unmethylated paternal DMR-LIT1 acts in cis to silence maternal-specific genes on the paternal chromosome in both species. Niemitz et al. [2004] have recently reported a human microdeletion of the entire LIT1 gene, including the DMR. Surprisingly, this deletion caused no abnormal phenotype when inherited on the paternal chromosome but caused BWS and diminished expression of KIP2 when inherited maternally, suggesting that sequences within the deleted region are required for activation of KIP2 expression from the maternal allele. This phenotype is clearly different from that produced by deletion of DMR-LIT1 alone. Further studies of naturally-occurring human deletions of this region and of induced mouse deletions will be required to understand this intriguing observation.
Mouse DMR-Lit1 is the only gametically methylated region in the KIP2/LIT1 domain that has been shown to be methylated in oocyte but not in sperm with maternal-specific methylation maintained in somatic cells [Yatsuki et al., 2002]. The differential methylation is associated with parent-specific nuclease hypersensitivity and histone modification. Several DNase I hypersensitive sites exist at the unmethylated paternal DMR-LIT1 but not at the methylated maternal locus [Yatsuki et al., 2002]. Parent-specific histone modification patterns of DMR-LIT1 are also present: H3 and H4 acetylation and methylation of H3 Lys4 are seen at the paternal DMR-LIT1, but maternal-specific dimethylation of H3 Lys9 is found in both mouse and human DMR-LIT1 [Higashimoto et al., 2003]. H3 Lys9 dimethylation is lost together with CpG methylation of the maternal DMR-LIT1 in imprinting defect BWS patients, suggesting either that one of these modifications is dependent on the other or that both are dependent on some other molecular determinant.
Among imprinted genes within the KIP2/LIT1 domain, KIP2, encoding a CDK inhibitor, is a critical gene for the BWS phenotype because 5%–10% of humans with BWS have point mutations of KIP2 [Hatada et al., 1996] and mice with targeted deletion of Kip2 show some features of BWS, including abdominal muscle defects, renal medullary dysplasia, and adrenal cortical hyperplasia and cytomegaly [Zhang et al., 1997]. Imprinting defects leading to absence of CpG methylation and loss of H3 K9 methylation on the maternal DMR-LIT1 allele cause diminished KIP2 expression [Diaz-Meyer et al., 2003]. It is plausible that the imprinting defect with loss of CpG methylation and loss of H3 Lys9 dimethylation causes a change of epigenotype of the KIP2/LIT1 region from maternal to paternal, reducing KIP2 expression. The relatively inactive paternal KIP2 promoter in normal human cells is not associated with either CpG methylation or H3 Lys9 methylation, implying that other epigenetic mechanisms must be involved in paternal KIP2 silencing [Chung et al., 1996; Higashimoto, unpublished results]. (However, it should be noted that the mouse Kip2 CpG island does show paternal-specific CpG methylation and H3 Lys9 dimethylation [Hatada and Mukai, 1995; Higashimoto, unpublished results].)
Umlauf et al. [2004] have recently shown that the maternal DMR-Lit1 is associated not only with dimethyl Lys9 H3 but also with trimethyl Lys27 H3 in 9.5 day p.c. mouse embryos. They and another group [Lewis et al., 2004] showed that promoters of mouse genes in the Kip2/Lit1 cluster that are imprinted only in placenta are associated with both dimethyl Lys9 H3 and trimethyl Lys27 H3 on the repressed paternal allele in placenta, without DNA methylation of the promoters, but are not associated with dimethyl Lys9 H3 or trimethyl Lys27 H3 in embryos. The two groups proposed that histone-methylation-based repression is established early in development and is maintained in the placenta, but that in the embryo imprinting is stably maintained only at genes that have parent-specific promoter DNA methylation.
SUMMARY, SYNTHESIS, AND EXTENSION TO OTHER IMPRINTED REGIONS
The PWS/AS region in human 15q11-q13 and the KIP2/LIT1 domain of the BWS region in 11p15.5 provide two of the best examples of the role of genetic imprinting in human disease. The regulation of imprinted gene expression in these regions is complex and poorly understood. The regions share features of structural organization and epigenetic modification that distinguish them from simpler and better understood imprinted regions, such as the IGF2/H19 domain. The major organizational features of the PWS/AS region and the KIP2/LIT1 domain resemble those of another intensively studied imprinted region, the Igf2r region in mouse (Fig. 1) [Zwart et al., 2001; Fournier et al., 2002] (in humans, IGF2R and nearby genes are not imprinted in most individuals [Riesewijk et al., 1996]).
- (1)
A cis-acting IC that functions bidirectionally over long distances (up to 1 Mb for the PWS-IC) to regulate imprinted gene expression.
- (2)
Maternal CpG methylation of the IC that is established in the germ-line (at least in mouse) and that is associated with dimethylation of H3 Lys9 in somatic cells (H3 methylation has not been examined in gametes for technical reasons); other regions of parent-specific CpG methylation are established after fertilization and are not associated with dimethyl Lys9 H3.
- (3)
Consequences of deletion of the IC are the same as those of CpG methylation and H3 dimethylation of the IC.
- (4)
ICs contain promoters of noncoding transcripts in the case of DMR-LIT1 and the Igf2r imprinting control element, and of a transcript encoding SNRPN and SNURF proteins in the case of the PWS-IC; in all three regions, effects of antisense transcription cannot account for all observed imprinting phenomena.
The coincidence between germ-line-established maternal-specific CpG methylation and the presence of dimethyl Lys9 H3 at all three ICs is striking, and raises the possibility that there may be a causal relationship between establishment of these epigenetic marks in the germ-line. These epigenetic marks appear to inactivate the ICs, rather than to modulate the function of the ICs; for all three ICs, the effect of an IC deletion is the same as the effect of a maternal CpG-methylated and H3 Lys9-methylated IC.
- (1)
Unmethylated ICs act as enhancer blockers that protect imprinted genes from effects of distant enhancers. This hypothesis requires that each imprinted gene regulated by an IC has an enhancer on the other side of the IC. Mancini-DiNardo et al. [2003] showed that an unmethylated DMR-Lit1 does not act as enhancer blocker, which blocks transcription only when placed between enhancer and promoter, but rather acts as silencer, which blocks transcription even when not placed between enhancer and promoter. This hypothesis is also clearly not applicable to the PWS/AS region, where most imprinted genes are paternally-active, in cis with an unmethylated PWS-IC.
- (2)
Noncoding RNAs act in cis to regulate gene expression. Sleutels et al. [2002] showed that, in the mouse Igf2r imprinted cluster, truncation of the noncoding Air RNA that is transcribed from its promoter in the IC causes loss of imprinting for all genes in the cluster. However, in the PWS/AS region, a deletion extending from Snrpn intron 1 to Ube3a intron 1 removes almost all of the paternally-expressed region downstream from the PWS-IC, but paternal deletion of this region has no effect on imprinted expression of Ndn [Tsai et al., 1999].
- (3)
Unmethylated ICs act as silencers and establish a repressive chromatin structure bidirectionally over long distances, until boundary elements are reached that prevent further spread of the repressive chromatin structure. More generally, an unmethylated IC on a paternal chromosome acts to establish a modified chromatin structure different from that on the maternal chromosome which has a methylated, nonfunctional IC. According to this hypothesis, the transcriptional activity of individual genes is a function of the interaction of promoters/enhancers with modified chromatin structure. For some genes, a paternal-specific chromatin structure may be activating; for other genes, the same chromatin structure may be repressive. At this point, the only evidence for contiguous spread of a modified chromatin state through an imprinted region comes from the mouse Kip2/Lit1 domain in placenta, which shows paternal-specific association with dimethyl Lys9 H3 and trimethyl Lys27 H3 in promoters, genes, and intergenic regions [Umlauf et al., 2004]; this contiguous region of altered chromatin structure on the paternal chromosome is not present in 9.5 day p.c. embryos.
Whether the apparent similarities of structural organization and epigenetic marking of the IC between these imprinted regions actually reflect common mechanisms by which ICs affect gene expression bidirectionally over long distances will only become clear as the mechanisms of gene regulation in the PWS/AS region, the KIP2/LIT1 domain of the BWS region, and the mouse Igf2r region are elucidated. Current data for the Igf2r region are compatible with a mechanism of RNA-mediated regulation in cis similar to Xist-mediated X chromosome inactivation. However, there is no published evidence as yet for association of the noncoding Air RNA with this region, and the roles (if any) of noncoding RNAs in imprinting of the PWS/AS and KIP2/LIT1 regions remain to be explored.
The concept of ICs as sites from which modified chromatin structures are propagated bidirectionally and maintained is attractive as a general explanation for the epigenetic phenomena observed in these three regions. Future research on parent-specific association of noncoding RNAs, histone variants, modified histones, and nonhistone chromosomal proteins with these regions promises to shed light on the complexities of these imprinted clusters and their roles in human disease.




