NFAT expression in human osteoclasts
Abstract
Nuclear factor of activated T-cells cytoplasmic (NFATc) is a family of transcription factors originally identified in T-cells. The gene family is currently known to have four members (NFATc1 through NFATc4) which have roles both within and outside the immune system. We show that NFATc1 is the major induced NFAT in human osteoclasts, with expression greatly exceeding that of NFATc2 through NFATc4. In macrophage-like cells in culture, NFATc1 through NFATc4 are expressed at similar low levels. NFATc1 is comprised of five mRNA transcript variants known to encode three different protein isoforms. The mRNA encoding isoform C (mRNA variant 3) was the most expressed with 38 copies per nanogram followed by isoform B (mRNA variant 5) with 17 copies per nanogram of total RNA. Isoform A (mRNA variant 1) and mRNA variants 2 and 4 made up less than 1% of the total NFATc1 expressed. NFATc1 is activated by calcineurin after calcium-calmodulin signalling. The induction of NFATc1 in osteoclasts was not altered in the presence of cyclosporin A, an inhibitor of calcineurin, suggesting that NFATc1 does not participate in autoregulatory activation of its own promoter. The NFATc1 variants expressed by human osteoclasts are not those normally expressed by effector T-cells but are similar to those seen in naïve T-cells. © 2005 Wiley-Liss, Inc.
Nuclear factor of activated T-cells cytoplasmic (NFATc) is a widely expressed transcription factor family which is dependant on Ca2+ signalling for activation through the action of calmodulin and calcineurin. NFATc is crucial for normal T-cell cytokine production and is required for the differentiation and function of several cell types with no immune function [Horsley and Pavlath, 2002]. The NFATc gene family has four members designated NFATc1 through NFATc4. Although NFATc was first identified in T-cells these factors have major roles in non-immune cell types. NFATc1 was shown recently to be induced during both mouse [Ishida et al., 2002; Takayanagi et al., 2002; Hirotani et al., 2004] and human [Day et al., 2004] osteoclast development. Osteoclasts differentiate from mononuclear precursors under the action of the cytokine RANKL (receptor activator of NFκB ligand), resulting in large specialised multinuclear cells that stain positive for tartrate resistant acid phosphatase (TRAP), are positive for bone degradation and express marker genes such as calcitonin receptor. Inhibition of NFATc by cyclosporin A (CsA) blocked osteoclast formation from both human and mouse precursors, resulting in mononuclear TRAP positive cells [Ishida et al., 2002; Day et al., 2004]. In human cells, CsA inhibited calcitonin receptor expression while in mice NFATc over expression resulted in induction of calcitonin receptor indicating that NFATc is crucial for calcitonin receptor expression.
Three protein isoforms of NFATc1 have been identified in human and mouse T-cells, isoforms A, B and C [Chuvpilo et al., 1999; Sherman et al., 1999]. These variants differ on both the N and C terminus and are driven from two different promoters [Chuvpilo et al., 2002]. In the mouse, NFATc1 mRNA variants are expressed preferentially by different cell types; the mouse variant alpha is induced upon activation in T-cells and mast cells while the beta variant is expressed basally in T-cells but is up-regulated in activated mast cells [Sherman et al., 1999]. In the human, NFATc1 isoform A is expressed at high levels in effector T-cells where it enhances cell function through production of IL-2 and inhibition of apoptosis [Chuvpilo et al., 1999, 2002]. NFATc1 isoforms B and C are expressed preferentially by naïve T-cells [Chuvpilo et al., 1999].
In this study, we examine the expression of the four members of the NFATc family and the specific mRNA variants of NFATc1 in human osteoclasts. We also tested whether NFATc1 was regulated by NFATc family members by blocking activation of NFATc using cyclosporin A.
METHODS
Cell Culture, RNA Isolation, cDNA Synthesis
Human peripheral blood mononuclear cells (PBMC) were isolated from donor blood and differentiated into bone-resorbing osteoclasts after treatment for 21 days with RANKL (40 ng/ml) and M-CSF (25 ng/ml) in MEM supplemented with 5% foetal calf serum (Invitrogen, Carlsbad, CA), as described previously [Day et al., 2004]. Parallel cultures were treated identically, without RANKL, in order to differentiate macrophage like cells. RNA was isolated from 3-week-old cultures using CsCl ultra-centrifugation and converted to cDNA using Superscript reverse transcriptase, as previously described [Day et al., 2004]. Experiments were repeated on three separate occasions. For each experimental repeat, new cultures of osteoclasts and macrophage-like cells were established using fresh samples of blood from the same donors. Gene expression data was established independently for each experimental repeat. The reproducibility of gene expression was assessed by comparison of nineteen different gene assays determined in the three independent replicate experiments. The average coefficient of variation in cycle threshold in real time quantitative PCR in replicate experiments, across all assays was 4.2% with a standard error of 0.6%. Error bars in figures are the standard deviation of the data from the three independent replicate experiments. Analysis of variance and Student's t-test were used to establish significance. Western blots were done as described previously [Aitken et al., 2004] using the 7A6 anti-NFATc1 monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA). A representative result is presented.
Quantitative Real-Time PCR (Q-PCR) Analysis of Gene Expression Analysis
Q-PCR was performed using the Bio-Rad iQ iCycler system and SYBR Green I Supermix. Assays were standardised using known quantities of target and all assays showed high linearity, similar efficiency (around 100%) and absence of artefact. Gene specific primers (Table I) were developed from the genomic sequence obtained through the BLAT server at the University of Santa Cruz (http//:genome.ucsc.edu). Primers for NFATc1, c2, c3 and c4 were designed to amplify across short introns in the 3′ regions and to have no cross reactivity. Primers for NFATc1 variants were similarly selected for the specificity of product (Table II). All PCR products were run on acrylamide gels to verify specificity.
| Gene assay | Forward | Reverse |
|---|---|---|
| c-fos | CTGAGGTTGATGTTCTCGTC | CTAAGGCCACTGTCATCACA |
| c-jun | CGTTAGCATGAGTTGGCAC | GCATGAGGAACCGCATCGC |
| NFATc1 (total) | GCATCACAGGGAAGACCGTGTC | GAAGTTCAATGTCGGAGTTTCTGAG |
| NFATc2 | GATGGAAGCCACGGTGGAT | GCGGATATGCTTGTTCCGAT |
| NFATc3 | CTGAGTCCCTGGATTTAGGA | AGTTGGAAACCCAAGGTCCA |
| NFATc4 | CAGTCCTACCCAGAGTTTCA | GTGGTGAGAAGTCCATGTCA |
| NFATc1 variant mRNAs | ||
| Variant 1 | CTGTGTGATGTCCCGTTAGT | CCGTGACTATCTGAGTATGCA |
| Variant 3 | CCACGGTCCGCAGGGACGAGT | TCTTCAGGCTCTCGCTTGACC |
| Variant 3 and 5 | GACCCGCCATGACGGGGCTGGAGGA | CTCGTCGCGCTGGTTAAACTCGA |
| Variants 1, 2 and 4 | CAGAGCGAGACTCAGAGGCTCCGA | CTAGGACCTGCGCGGTGGCTCCGA |
- All primers are written 5′–3′.
| Exon | Occurrence in RNA variant | Assay | ||||
|---|---|---|---|---|---|---|
| V1 | V2 | V3 | V4 | V5 | ||
| 5–6 | + | + | + | + | + | Total NFATc1 mRNA |
| 10 | + | V1 assay (isoform A) | ||||
| 11 | + | V3 assay (isoform C) | ||||
| 2 | + | + | V3 + V5 assay (isoform B and C) | |||
| 1 | + | + | + | Other than V3 + V5 assay | ||
- Table shows exons in which primers are located and the occurrence of that sequence in variant mRNAs (designated V1 through V5). Assay indicates the variant mRNAs detected and the cognate isoforms.
Quantitation was performed relative to known standards made from purified PCR products ranging from 10 to 1010 copies per Q-PCR. Total NFATc1 mRNA was calculated using an assay derived from exons 5 and 6, which are in common between all mRNA variants (Table II). Specific assays were available for mRNA variants 1 and 3, using primers in exons that are specific for these mRNAs. NFATc1 mRNA variant 5 (isoform B) copy number was calculated by subtracting copies of variant 3 (isoform C) from the total for the combined 3 and 5 copy number. The copies for variants 2 and 4 were calculated by the same method with variant 1 being subtracted from the combined total.
RESULTS
Expression and Regulation of NFATc1, c2, c3 and c4
Comparison of NFAT regulation in human osteoclasts versus macrophage-like cells, after 3 weeks treatment with RANKL and M-CSF, demonstrates that NFATc1 is the most potently induced by RANKL (Fig. 1). NFATc2, NFATc3 and NFATc4 were expressed at considerably lower levels (11, 11 and 7-fold less, respectively) than NFATc1 in osteoclasts. In addition to the lower level of expression, NFATc2, NFATc3 and NFATc4, were all weakly regulated by RANKL, with an approximate twofold increase, although these changes were not statistically significant (P > 0.05 in each case). In marked contrast, NFATc1 was induced 28.5-fold by RANKL (P = 0.001). These data verify that NFATc1 is the most abundantly expressed NFAT in human osteoclasts and is the most potently induced NFAT during osteoclast differentiation. Although we have not examined protein levels, the data verify that NFATc2, NFATc3 and NFATc4, are expressed in mature osteoclasts, albeit at a much lower level than NFATc1.
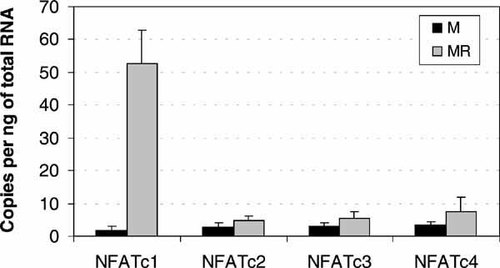
Expression of nuclear factor of activated T-cells cytoplasmic (NFATc) genes in macrophage-like cells compared to mature osteoclasts. mRNA expression was measured after 21 days treatment with M-CSF (M, black columns) or M-CSF and RANKL (MR, grey columns) to produce macrophage-like cells and osteoclasts, respectively. NFATc1 expression is sevenfold greater than NFATc4 expression and more than 11-fold greater than that of either NFATc2 and NFATc3. Error bars are the standard deviation of three replicate experiments.
NFATc1 Isoform Expression
NFATc1 is expressed in five different mRNAs from two different promoters (P1 and P2), yielding three identified protein isoforms designated A, B and C (see the NCBI website at www.ncbi.nlm.nih.gov). Protein isoforms A, C and B are coded for by mRNA variants 1, 3 and 5 respectively [Chuvpilo et al., 1999]. Variant 2 possesses the 5′ end of variant 1 and the 3′ end of variant 5 indicating it would be driven off the P1 promoter. To test the expression of NFATc1 isoforms variant specific primers were designed. Only variants 1 and 3 could have specific primers designed so primers that amplified all sequences in common between variants 1, 2 and 4 and variants 3 and 5 were designed. Fortunately, the major variant was variant 3, for which a specific assay was possible (Fig. 2). Variant 1 was of extremely low abundance and this number accounted for all the amplification observed in assays that amplified variants 1, 2 and 4, meaning that variants 2 and 4 were essentially absent. Variants 2 and 4 were expressed at 0.081 copies per ng of total RNA after subtraction of the expression of variant 1 from the total of 1, 2 and 4. Isoforms B and C (mRNA variants 5 and 3) made up around 99% of the sum of total NFATc1 mRNA expressed by 3 week PBMC derived osteoclasts (Fig. 2A).
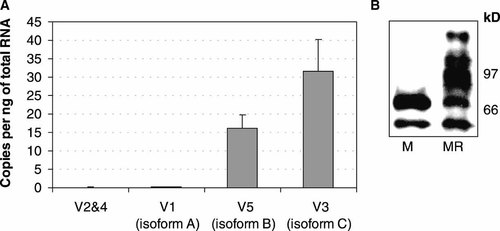
NFATc1 variant expression. A: NFATc1 variant mRNA expression in mature osteoclasts per nanogram of total RNA. Variants encoding isoform B and C are highly expressed, making up more than 99% of the total expressed NFATc1 mRNA. Cells were treated with M-CSF and RANKL for 21 days. Error bars are the standard deviation of three replicate experiments. B: Representative Western blot of total protein from macrophage-like cells (M) and osteoclasts (MR), probed using monoclonal NFATc1 antibody 7A6. Cell extracts were prepared from 14 day cultures. kDa indicates kilo Daltons.
The NFATc1 isoforms that are involved in RANKL mediated osteoclast differentiation must be those that show specificity of expression and induction in osteoclasts compared to macrophages. Western blots were used to detect those NFAT variants that are specifically up-regulated in osteoclasts compared to macrophage-like cells. The antibody used (7A6) detects all forms of NFATc1, including all phosphorylation states and therefore cannot discriminate between different isoforms of NFATc1. Although the identity of bands has not been established, the banding patterns on Western blots from 14 day osteoclast cultures were largely compatible with high molecular weight NFATc1 variants being specifically up-regulated in osteoclasts (Fig. 2B). In contrast, a low molecular weight band was not differential between osteoclasts and macrophages and a band compatible in size with NFATc1 isoform A decreased between macrophage and osteoclast (Fig. 2B). High molecular weight forms of NFATc1 were the only isoforms prominently induced in osteoclasts.
NFATc1 expression during osteoclast formation has been linked to c-fos [Matsuo et al., 2004], NFATc2 and c-jun [Ikeda et al., 2004]. We tested the hypothesis that the induction of NFATc1 would be blocked by CsA, since NFATc1 expression can be induced by itself and other NFATc proteins. The levels of expression of mRNAs for c-fos, c-jun and NFATc1 were measured in cells that had been treated with either M-CSF alone (resulting in macrophage like cells), RANKL and M-CSF (MR, resulting in osteoclasts) and CsA, RANKL and M-CSF (MR + CsA, resulting in mononuclear TRAP positive cells) (Fig. 3). In real time PCR analysis, NFATc1 was less than 0.2 cycle different between MR and MR + CsA treatment, resulting in a non-significant (P = 0.4) change of less than 10 copies per ng of total RNA. Considering that NFATc1 is induced 28.5-fold, this effect would seem minor. Both c-fos and c-jun were induced approximately twofold by RANKL (P = 0.03 and P = 0.02, respectively). Expression of c-fos and c-jun was not affected significantly by the presence of CsA, meaning that AP-1 induction by RANKL is unaffected by CsA.
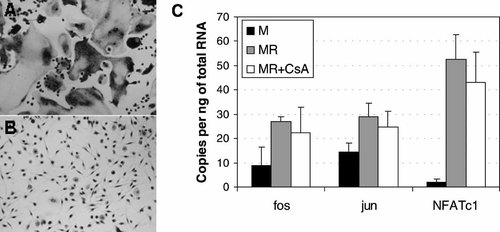
Effect of cyclosporin A (CsA, 1,000 ng/ml) on cell differentiation and expression of NFATc1 and AP-1 components. A: Microscopy of mature osteoclasts derived from PBMC treated with M-CSF and RANKL. B: Inhibition of osteoclast formation by CsA; no large multinuclear cells were observed in cultures treated with M-CSF, RANKL and CsA. C: Expression of c-fos, c-jun and NFATc1 in macrophage like cells (M, black columns), osteoclasts (MR, grey columns) and in osteoclasts suppressed with CsA (MR + CsA, white columns). CsA treatment has only a minor effect on the RANKL mediated induction of c-fos, c-jun and NFATc1. All cultures were treated for 21 days. Error bars are the standard deviation of three replicate experiments.
DISCUSSION
Of NFATc genes, NFATc1 was the most responsive to RANKL and was most abundantly expressed in human osteoclasts (Fig. 1). Furthermore, of NFATc1 protein isoforms, the majority of NFATc1 transcripts comprised mRNA for isoforms B and C (Fig. 2). This pattern of isoform expression is similar to that seen in naïve T-cells. The functional consequences of this level of NFATc1 transcript specificity is not known, however, it seems reasonable to assume that these variants are responsible for inducing characteristic osteoclast specific genes. NFATc1 is known to bind the promoters of GM-CSF, IL-2, IL-3, IL-4, IL-5, IL-10 and TNF-α in T-cells and mast cells [Park et al., 1996; Chuvpilo et al., 1999].
The potential targets of NFATc1 in osteoclasts are yet to be fully determined, however NFATc response elements are reported in the promoters of osteoclast marker genes cathepsin K, TRAP and calcitonin receptor. The TRAP promoter NFATc response element was transactivated best by NFATc4 when compared to NFATc1, c2 and c3 [Matsuo et al., 2004]. NFATc1 activates the cathepsin K promoter in combination with MiTF, PU.1 and p38 MAP Kinase [Matsumoto et al., 2004] however the total activity attributable to NFATc1 was not dramatic (1.7-fold) [Matsumoto et al., 2004]. Calcitonin receptor promoters from a number of species have NFATc response elements [Anusaksathien et al., 2001]. Matsuo et al. [2004] showed that NFATc1 could transactivate the calcitonin receptor promoter and that NFATc1 was critical for calcitonin receptor expression. The human calcitonin receptor is induced by RANKL and this induction is highly sensitive to CsA [Day et al., 2004]. Under CsA blockade of osteoclast differentiation, cathepsin K and TRAP activities were not strongly affected [Day et al., 2004] suggesting that calcitonin receptor is a prime candidate for regulation by NFATc1, but that cathepsin K and TRAP are less likely candidates.
After NFATc1, NFATc4 is the next most abundant NFATc in terms of mRNA, however the difference in mRNA abundance between the other NFATc mRNAs is minor (between 5 and 7.5 copies per ng total RNA). The roles of NFATc2, NFATc3 and NFATc4 in osteoclasts is unknown, however our data eliminate the possibility that other NFATc are involved in regulating NFATc1, since CsA treatment did not result in blockade of RANKL mediated NFATc1 induction. We conclude that RANKL mediated induction of NFATc1 is not sensitive to calcineurin inhibition by CsA.
The expression of different mRNA variants of NFATc1 was calculated in order to determine which variant is the most abundant and therefore which cognate protein isoform may be important to osteoclast function. NFATc1 isoforms in the human differ on both the N and C terminus of the protein. Isoform A is 716 amino acids in length with the Rel homology domain (RHD) and calcineurin binding site at the C terminus of the protein. Isoform A is the shortest protein sequence reported and is 92% homologous to the mouse isoform A (analysis not shown). Isoform B is the second longest at 812 amino acid residues and is 92% homologous to isoform B of the mouse (analysis not shown). Isoform C is the longest variant with 930 amino acids. Isoform C differs from isoform A by the first 39 amino acids in the N terminus and is also extended in the C terminus when compared to both isoforms A and B. Of the five transcript variants presented on NCBI only the three encoding published isoforms were found to be expressed at greater than one copy per nanogram of total RNA in human osteoclasts. The sum of the mRNAs for these three isoforms equalled the total sum of all mRNA copies of NFATc1 detected when using a primer that amplified all five possible transcript variants. Although we have not measured protein levels directly, mRNAs for isoforms B and C made up 99% of the expressed NFATc1 transcripts (Fig. 2). If mRNA content reflects protein levels, isoforms B and C (the highest molecular weight forms of NFATc1) would represent the major induced NFATc1 in osteoclasts. These observations were consistent with Western blots done on 14 day cultures, in that high molecular isoforms of NFATc1 are specifically induced in osteoclasts compared to macrophages. Although Western blots also detected lower molecular weight forms of NFATc1, these were either decreased in osteoclasts or unchanged compared to macrophage-like cells. In particular, low molecular weight forms were not induced as would be expected for the autoregulation of NFATc1 isoform A. The limitation of the Western blot data is that the identity of the various bands has not been established and the experiments were done on 14 day cultures compared with 21 day cultures. Analysis of 14 day mRNA was in agreement with 21 day mRNA, indicating isoforms B and C were the most prominent (data not shown).
The NFATc1 gene contains a strong inducible P1 promoter with tandem NFATc1 response elements, providing for autoregulation by NFATc1 and by autologous regulation by other NFATc proteins. This inducible promoter is responsible for the large quantity of NFATc1 isoform A found in effector T-cells. Expression of NFATc1 isoforms B and C is controlled by the P2 region of the NFATc1 promoter, which is not self regulated by NFATc [Chuvpilo et al., 2002] and is weak in effector T-cells. The B and C isoforms of NFATc1 are expressed in naïve T-cells [Chuvpilo et al., 1999] which then switch to high levels of isoform A on activation. Naïve T-cells express reduced amounts of IL2 and IL4 when compared to activated effector T-cells; cells that express a majority of isoform A [Chuvpilo et al., 1999]. Chuvpilo et al. [1999] also demonstrated that isoform B and C did not activate the IL2 and IL4 promoters equally as potently as does isoform A.
CsA blocks the upregulation of isoform A in T-cells [Chuvpilo et al., 2002] by preventing an autoregulatory loop that drives the NFATc1 P1 promoter. The NFATc1 P2 promoter drives the expression of the B and C isoforms and is not reliant on NFAT for activation [Chuvpilo et al., 2002]. CsA was unable to block the RANKL mediated induction of NFATc1 in osteoclasts. The lack of regulation of NFATc1 in the presence of CsA is consistent with low levels of isoform A in osteoclasts. From the information available it is likely that AP-1 or a factor down stream of AP-1 is responsible for NFATc1 induction in osteoclasts as loss of c-fos expression or induction of dominant negative c-jun eliminates NFATc1 expression in osteoclast like cells [Ikeda et al., 2004; Matsuo et al., 2004]. This indicates that NFAT is a secondarily activated pathway in osteoclast formation and is not the primary driving force in differentiation of osteoclasts.
We have shown that, in human osteoclasts, NFATc1 is the most potently induced member of the NFATc family. The other NFATs (c2, c3 and c4) are present at lower mRNA copy number and are only weakly induced by RANKL. Of the NFATc1 transcripts induced by RANKL, the vast majority is comprised of mRNAs for NFATc1 isoforms B and C. The biological significance of these differences remains to be determined. We also show that NFATc1 expression is not altered in the presence of CsA, providing evidence that the NFATc1 isoforms expressed in osteoclasts are not influenced by autoregulatory loops as described for the potent induction of NFATc1 isoform A in effector T-cells. Since the NFATc1 P2 promoter drives expression of isoforms B and C, further analysis of the P2 promoter in osteoclasts is warranted in order to understand the mechanism of induction of NFATc1 by RANKL.
Acknowledgements
Christopher Day is supported by a scholarship from the ARC. Michael Kim is supported by a scholarship from Griffith University.




