Localization and regulation of phospholipase D2 by ARF6
Abstract
Phospholipase D (PLD) and ADP-ribosylation factor 6 (ARF6) have been implicated in vesicular trafficking and rearrangement of the actin cytoskeleton. We have explored the co-localization of rat PLD1b and rat PLD2 with wild type and mutant forms of ARF6 in HeLa cells and studied their activation by ARF6 and the role of the actin cytoskeleton. GFP-tagged PLD1 had a similar pattern to multivesicular and late endosomes and the trans-Golgi apparatus, but not to other organelles. When wild type or dominant negative ARF6 and PLD1 or PLD2 were co-expressed, they had a similar localization in cytosolic particles and at the cell periphery. In contrast, dominant active ARF6 caused cell shrinkage and had a similar localization with PLD1 and PLD2 in dense structures, containing the trans-Golgi apparatus and actin. Disruption of the actin cytoskeleton with cytochalasin D did not induce the formation of these structures. To determine, if ARF6 selectively activated PLD1 or PLD2, wild type and mutant forms of the ARF isoform were transfected together with PLD1 or PLD2. Wild type ARF6 did not affect either PLD isozyme, but dominant active ARF6 selectively activated PLD2 and dominant negative ARF6 selectively inhibited PLD2. In contrast, dominant active ARF1 or Rac1 stimulated both PLD isozymes but the ARF1 effect on PLD2 was very small. Cytochalasin D did not affect the activation of PLD by phorbol ester. The localizations of PLD and ARF6 were also analyzed by fractionation after methyl-β-cyclodextrin extraction to deplete cholesterol. The results showed that all PLD isoforms and ARF6 mutants existed in the membrane fraction, but only wild type ARF6 was dependent on the presence of cholesterol. These experiments showed that wild type ARF6 had a similar location with PLD isoforms on cell staining, but it did not colocalize with PLD isoforms in fractionation experiments. It is proposed that activated ARF6 translocates to the cholesterol independent microdomain and then activates PLD2 there. It is further concluded that PLD2 is selectively activated by ARF6 in vivo and that disruption of the actin cytoskeleton does not affect this activation. © 2005 Wiley-Liss, Inc.
Abbreviations used:
PLD, phospholipase D; ARF, ADP-ribosylation factor; DMEM, Dulbecco's modified Eagle's medium; PMA, phorbol 12-myristate 13-acetate.
Phospholipase D (PLD) catalyzes the hydrolysis of phosphatidylcholine (PC), the major phospholipid of membranes, to phosphatidic acid (PA) and choline. It is believed to play a role in signal transduction in many cell types because it is a ubiquitous enzyme that is regulated by a great variety of hormones, neurotransmitters, growth factors, cytokines, and other molecules involved in extracellular communication [Exton, 1997]. There is evidence that PA plays a role in vesicle transport from the endoplasmic reticulum to the Golgi [Bi et al., 1997a; Siddhanta et al., 2000] and in the budding of secretory vesicles from the trans-Golgi network [Bi et al., 1997b; Siddhanta and Shields, 1998]. Further, it has been suggested that PA stimulates the assembly of COPI-coated vesicles in the Golgi [Ktistakis et al., 1996] and also regulates the recruitment of the AP2 adaptor proteins required for clathrin coat assembly on the Golgi [West et al., 1997]. There is also much evidence that PLD is involved in regulated exocytosis [Jones et al., 1999b; Siddhanta et al., 2000; Vitale et al., 2001]. Thus PLD is implicated in various forms of vesicle trafficking.
Two mammalian PLD isozymes have been cloned [Hammond et al., 1995, 1997; Colley et al., 1997]. PLD1 exists in two splice variants (a and b) [Hammond et al., 1997]. Both have low basal activity in vitro and can be activated by small GTP-binding proteins of the ADP-ribosylation factor (ARF) and Rho families and also protein kinase C (PKC) [Hammond et al., 1997]. The PLD2 isoform has high basal activity and is insensitive to activation by ARF, Rho, and PKC in vitro [Colley et al., 1997]. Both isozymes are dependent for activity on PIP2.
In regard to the subcellular location of PLD1, there are many studies of the overexpressed enzyme [Liscovitch et al., 1999] and one report of the endogenous enzyme, which localized it mainly to the Golgi [Freyberg et al., 2001]. GFP-and HA-tagged PLD1 have been localized mainly to vesicular structures in the perinuclear region [Colley et al., 1997; Sugars et al., 1999; Emoto et al., 2000; Kam and Exton, 2001; Vitale et al., 2001] but some reports have indicated their presence in early endosomes [Hughes and Parker, 2001] and the plasma membrane [Kim et al., 1999b]. PLD2 has a different subcellular distribution. Most studies have shown a predominantly plasma membrane localization [Colley et al., 1997; Emoto et al., 2000; Park et al., 2000; Chahdi et al., 2003; Sarri et al., 2003].
ADP-ribosylation factors (ARFs) are members of the Ras superfamily and have been well characterized as regulators of vesicular transport [Moss and Vaughan, 1998]. The six ARF isoforms are divided into three classes. Class I ARFs (ARFs1–3) regulate trafficking in the secretory pathway and in endosomes. However, very little is known about the role of Class II ARFs 4 and 5. The sole member of Class III ARF, ARF6 functions in the endosomal-plasma membrane trafficking system [Chavrier and Goud, 1999]. ARF6 localizes at endosomes and the plasma membrane in a manner dependent on its GTP-binding state to deliver the targeted recycling endosomal vesicles to the plasma membrane [Radhakrishna and Donaldson, 1997; D'Souza-Schorey et al., 1998; Gaschet and Hsu, 1999]. Q67L-ARF6, which is a mutant form of ARF6 in which GTP is constitutively bound, accumulates at the plasma membrane. T27N-ARF6, which is a dominant negative, GDP-bound mutant form, localizes on perinuclear recycling endosomes, and inhibits recycling to the plasma membrane [Radhakrishna and Donaldson, 1997].
PLD seems to function downstream of ARF6. For example, the M3 muscarinic receptor expressed in COS7 cells activates PLD through an ARF6-dependent pathway to PLD1 or PLD2 [Mitchell et al., 2003]. PLD2 has also been implicated in the actions of platelet-derived growth factor and angiotensin II on PLD activity in vascular smooth muscle cells [Shome et al., 2000]. Furthermore, ARNO, which is an ARF6-specific guanine-nucleotide exchange factor (GEF), activates PLD and induces membrane ruffling in epithelial cells [Santy and Casanova, 2001]. Further, ARF6 often co-localizes with PLD1 and 2 at ruffling membranes [Honda et al., 1999; Powner et al., 2002] and has been implicated in remodeling cortical F-actin [Schafer et al., 2000] and adherens junction turnover [Palacios et al., 2001]. PLD has also been suggested to play a role in actin-fiber formation induced by thrombin, sphingosine 1-phosphate and lysophosphatidic acid [Ha and Exton, 1993; Cross et al., 1996; Kam and Exton, 2001; Porcelli et al., 2002]. PLD directly associates with G-actin in vitro and its catalytic activity is inhibited by this binding [Lee et al., 2001], but F-actin has the opposite effect [Kusner et al., 2002]. These results led us to examine if ARF6 regulates PLD localization and activation, and if actin is necessary for PLD activation.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Monoclonal anti-ARF6 antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal anti-mannosidase II and anti-EEA1 antibodies were purchased from B.D. Transduction Labs (Lexington, KY). Monoclonal anti-CD63 antibody was obtained from Cymbus Biotechnology Ltd. (Southampton, UK). Wheat germ agglutinin (WGA)-TexasRed, phalloidin-TexasRed, and secondary antibodies were from Molecular Probes (Eugene, OR). Cytochalasin D, methyl-β-cyclodextrin, and phorbol 12-myristate 13-acetate (PMA) were purchased from Sigma (St. Louis, MO). Anti-myc monoclonal antibody was obtained from BD Biosciences Clontech (Palo Alto, CA). [9,10-3H]Myristic acid was from Perkin-Elmer Life Sciences (Boston, MA).
Constructs for Cell Expression
For wild type (WT)-ARF6-6His construction, PCR was done using primers containing NheI and HindIII restriction enzyme sites and 6× CAT repeats at the C-terminus. Then the PCR product was inserted into pcDNA3(−) plasmid at NheI and HindIII restriction sites. For dominant negative (T27N;DN)- and dominant active (Q67L;DA)-ARF6, a mutagenesis kit (Stratagene, La Jolla, CA) was used. cDNAs for PLD1 and PLD2 were cloned from a rat brain cDNA library as described earlier [Park et al., 1997; Xie et al., 2002]. For GFP-PLD1, N-terminally deleted rat PLD1b was inserted into pEGFP-C3 plasmid (Clontech) at PstI and Smal sites. The N-terminal region was generated by PCR using primers containing ScaI and XhoI restriction sites and then ligated into the previous plasmid. GFP-rat PLD2 was made by the PCR method using primers containing KpnI and XbaI restriction sites and pEGFP-C3 plasmid. Myc-tagged dominant active and negative Rac1 in pEF-BOS were kindly given by Dr. Tadaomi Takenawa (University of Tokyo). Wild type Rac1 was generated with a quikchange site-directed mutagenesis kits (Stratagene). PLD and ARF6 double expression vectors were made based on the PLD-pEGFP-C3 constructs. For WT-ARF6, ARF6-6His in pcDNA3(−) was digested with BglII and BciVI. For DN- and DA-ARF6, ARF6-6His/V5 in pBudCE4.1 was digested with DrdI and NheI. For all ARF1 constructs, ARF1 in pcDNA3(−) was digested with BglII and BciVI. For all Rac1 constructs, Rac1 in pEF-BOS was digested with AseI and ApaLl. The fragments containing small G proteins and promoter were blunted with Klenow large fragment and then ligated into pEGFP-C3 or PLDs-pEGFP-C3 which were dephosphorylated with calf intestinal alkaline phosphatase (New England Biolabs, Beverly, MA) after digestion with AseI and then blunting. The EF promoter region of pBudCE4.I was used for control.
Cell Culture, Transfection, and Immunocytochemistry
HeLa cells were maintained in 10% FCS-Dulbecco's modified Eagle's medium (DMEM) (Sigma). The cells were transfected with either PLD1, PLD2 or ARF6 using Fugene-6 (Roche, Indianapolis, IN) and the constructs described above. Five hours later, the culture medium was changed to serum-free medium. The next day, cells were fixed with 3.7% formaldehyde in phosphate-buffered saline (PBS) for 15 min at room temperature. After three washes with PBS, the cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min and then washed with PBS three times. The permeabilized cells were blocked with 10% horse serum and 1% BSA in PBS for 60 min and then incubated with the primary antibody in PBS containing 3% BSA for 60 min at room temperature. Fluorescein- or TexasRed-labeled secondary antibodies were incubated with the cells for 60 min. Phalloidin-TexasRed, WGA-TexasRed and concanavalin A (ConA)-TexasRed were then mixed with secondary antibody. Membrane ruffling was induced by incubating the cells with 100 nM PMA for 30 min before the fixation. To collapse actin fibers, the cells were treated with 1 μM cytochalasin D for 10 min. The cells were observed using a confocal laser scanning microscope (Model MC4 1024, Bio-Rad Laboratories, Hercules, CA).
In Vivo PLD Assay
HeLa cells were seeded onto 6-well plates at 20 × 104 cells/well and the next day the cells were transfected with the relevant plasmids using Fugene-6. After 5 h, the medium was changed to DMEM containing 1 μCi/ml [3H]myristic acid. After overnight incubation, the cells were washed with PBS once and incubated with 0.3% BSA–DMEM for 40 min. Where appropriate, cytochalasin D was added 10 min before butanol addition and 100 nM PMA was added 5 min before butanol. PLD activity was then measured as described by Xie et al. [2002]. Briefly, cells were incubated in 0.3% 1-butanol for 40 min. They were washed with ice-cold PBS and the reaction was stopped with methanol. Lipids were extracted, and the phosphatiylbutanol product was resolved by thin layer chromatography. Bands co-migrating with a phosphatidylbutanol standard (Avanti, Alabaster, AL) were quantitated by scintillation counting.
Cell Fractionation
HeLa cells were seeded on 100-mm dishes at 30% confluency and the next day the appropriate plasmids were transfected using Fugene-6, and 5 h later the culture medium was changed to starvation medium. For methyl-β-cyclodextin treatment, the culture medium was changed to DMEM containing the indicated concentration of methyl-β-cyclodextrin and then the cells were incubated for 60 min. For cytochalasin D treatment, the cells were treated with 1 μM cytochalasin D for 10 min. The cells were washed with ice cold PBS and then washed twice with lysis buffer containing 20 mM HEPES (pH 7.2), 10% glycerol, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and protease inhibitors. The cells were harvested by scraping and the collected cells were suspended in 150 μl lysis buffer. The cells were disrupted by passing through a 27-G needle 15 times and the lysate was centrifuged at 500g for 10 min to remove unbroken cells and nuclei. The cell lysate was then centrifuged at 120,000g for 40 min to separate membrane and cytosol fractions. The supernatant was used as the cytosol fraction and the pellet was used as the membrane fraction. The fractions were applied to 4%–20% Tris-glycine gels (Invitrogen, Carlsbad, CA).
Immunoblotting
To confirm the expression levels of PLDs and small G proteins in the in vivo PLD assays, the total lysate was applied to 4%–20% Tris-glycine gels. Proteins were transferred to PVDF membranes and the membranes were blotted with the appropriate antibodies. Expression was detected by ECL (Amersham Pharmacia, Piscataway, NJ).
RESULTS
Localization of GFP-PLDs
Cellular localization of GFP-tagged PLD1 and PLD2 was examined in detail prior to testing the effects of cytochalasin D treatment and ARF6 co-transfection. GFP-PLDs plasmids were transfected into HeLa cells and an anti-CD63 antibody was used to identify multivesicular endosomes and late endosomes [Kobayashi et al., 2000]. In most cells GFP-PLD1 co-localized with anti-CD63 staining in a perinuclear location (Fig. 1A-a,b,c), but GFP-PLD2 did not localize to these endosomes (Fig. 1B-a,b,c). We further examined if GFP-PLDs localized in lysosomes. However, neither PLD1 nor PLD2 localized in those structures that were stained with anti-LAMP-1 (data not shown). It was important to examine whether GFP-PLDs localized in the Golgi apparatus. This structure was identified using WGA-conjugated Texas-Red, which is a marker of trans-Golgi apparatus, and anti-mannosidase II antibody, which is a marker of cis- and medial-Golgi apparatus. GFP-PLD1 co-localized with WGA-Texas-Red in about 20% of the cells (Fig. 1A-d,e,f), but not with mannosidase II (data not shown). These results show some localization of GFP-PLD1 in the trans-Golgi apparatus in accordance with a role for this PLD isozyme in Golgi function. GFP-PLD2 did not localize in any component of the Golgi apparatus (Fig. 1B-d,e,f). In line with previous reports [Colley et al., 1997; Palacios et al., 2001; Santy and Casanova, 2001], GFP-PLD2 was more prominently localized to the cell periphery than was GFP-PLD1 (Fig. 1B cf. Fig. 1A).
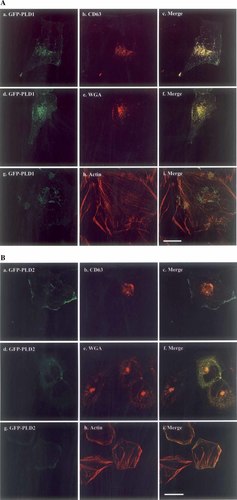
Localization of GFP-phospholipase D (PLD)1 and GFP-PLD2. A: To examine GFP-PLD1 localization, PLD1 in pEGFP-C3 was transfected into HeLa cells using Fugene-6. After fixation with 3.7% formaldehyde-PBS, the cells were treated with 0.2% Triton-X 100 and then blocked with 10% horse serum and 1% BSA–PBS for 1 h. Anti-CD63 (b) was used as a marker for multivesicular and late endosomes and then anti-mouse IgG-TexasRed was used as a secondary antibody. WGA-TexasRedX (e) and phalloidin-TexasRedX (h) were incubated with the secondary antibody for 1 h to stain trans-Golgi and actin, respectively. The stained cells were observed by confocal microscopy. GFP-PLD1 is shown in a, d, and g and the merge is shown in c, f, and i. B: To examine GFP-PLD2 localization, PLD2 in pEGFP-C3 was transfected into HeLa cells. The other procedures were as described in A. GFP-PLD2 is shown in a, d, and g and the merge is shown in c, f, and i. Scale bar: 25 μm. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
To examine PLDs localization in endoplasmic reticulum (ER), ConA-conjugated Texas-Red was used to stain the ER. Neither GFP-PLD1 nor GFP-PLD2 localized in ER (data not shown). The early endosome antigen 1 (EEA1) was used as an early endosome marker. GFP-PLD1 did not localize in the punctuate structures which were stained with anti-EEA1 antibody (data not shown), GFP-PLD2 also did not localize in these early endosomes (data not shown). We next tested for possible effects of PLD overexpression on actin fiber formation, overexpression of either PLD isozymes did not affect actin filament formation (data not shown). Neither PLD isozyme co-localized with actin fibers (Fig. 1A-g,h,i and Fig. 1B-g,h,i).
Localization of PLDs With ARF6
We examined the intracellular localization of ARF6 and PLD. This is because it has been reported that PLDs co-localize with ARF6 in membrane ruffles induced by ARNO transfection or antigen-stimulation of RBL-2H3 cells [Powner et al., 2002]. GFP-PLD1 was transfected with wild type and mutant forms of ARF6 into HeLa cells and then stained with anti-ARF6 monoclonal antibody. In wild type (WT)-ARF6 and GFP-PLD1 coexpressing cells, ARF6 and GFP-PLD1 had a similar localization to some extent on the plasma membranes, but mainly on cytosolic particles especially near the nucleus (Fig. 2A-a,b,c). In dominant negative (DN)-ARF6 expressing cells, the proteins also had a different localization with PLD1 on these particles (Fig 2A-d,e,f). Dominant active (DA)-ARF6 and GFP-PLD1 localized in dense structures (Fig. 2A-g,h,i). Expression of dominant active ARF6 also caused a decrease in cell size.
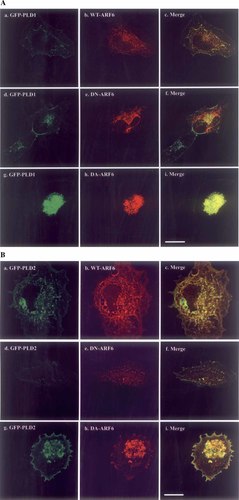
PLDs localization with ADP-ribosylation factor 6 (ARF6). A: To observe ARF6 effects on PLD1 localization, wild type (WT)-, dominant-negative (DN)-, and dominant active (DA)-forms of ARF6 were co-transfected with GFP-PLD1 into HeLa cells. Twenty-four hours later, cells were stained with anti-ARF6 antibody and anti-mouse IgG-TexasRedX. GFP-PLD1 is shown in a, d, and g. The merge is shown in c, f, and i. B: To observe ARF6 effects on PLD2 localization, the same forms of ARF6 were co-transfected with GFP-PLD2. GFP-PLD2 is shown in a, d, and g. The merge is shown in c, f, and i. Scale bar: 25 μm. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
When GFP-PLD2 and WT-ARF6 were co-expressed, they had a similar localization on the plasma membrane and also cytosolic particles (Fig. 2B-a,b,c). However, GFP-PLD2 had a different localization pattern from DN-ARF6 (Fig. 2B-d,e,f). GFP-PLD2 had a similar localization with DA-ARF6 in the dense structures similar to that seen with GFP-PLD1 (Fig. 2B-g,h,i).
Dominant Active ARF6 Localization
In DA-ARF6 expressing cells, GFP-tagged PLD1 and PLD2 were recruited into the dense structures (Fig. 2A,B-g,h,i). To examine the location where the PLDs were recruited, DA-ARF6 was transfected into HeLa cells and the cells were stained with ARF6 antibody and subcellular marker antibodies. For CD63 staining, DA-ARF6 and GFP-PLD1 were co-transfected because the antibodies against CD63 and ARF6 were monoclonal. The dense structure was not coincident with mannosidase II staining (data not shown), but it was coincident with WGA staining (Fig. 3-a,b,c), which identifies the trans-Golgi apparatus. The dense structure also corresponded with phalloidin-TexasRed staining (Fig. 3-g,h,i), but not with the structures stained with CD63 antibody (Fig. 3-d,e,f), EEA1 antibody (data not shown) or concanavalin A (data not shown). The actin staining also demonstrated the presence of some ARF6 at the plasma membrane (Fig. 3-g,h,i). Using transfection of Xpress-tagged and non-tagged PLDs, we confirmed that these PLDs also located in the dense structure (data not shown) i.e. the localization was not due to the GFP-tag. It is known that ARF6 induces actin remodeling at the cell periphery and destroys actin fiber structure [Song et al., 1998; Boshans et al., 2000; Schafer et al., 2000] and our results indicate that the collapse of actin fibers induced by DA-ARF6 induces the accumulation of PLD isoforms in the trans-Golgi apparatus.
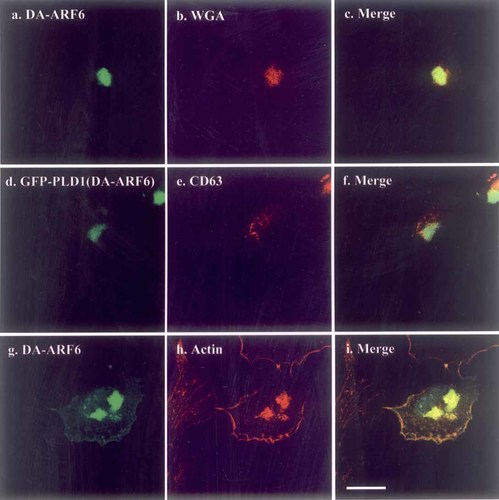
Dominant-active (DA)-ARF6 localization. The DA-ARF6 plasmid was transfected into HeLa cells and the cells were stained with anti-ARF6 (a and g), WGA-TexasRed (b), and phalloidin-TexasRed (h). For CD63 staining (e), GFP-PLD1 was co-transfected with DA-ARF6 because the CD63 antibodies were monoclonal as well as the anti-ARF6 antibody. Anti-mouse IgG-TexasRed was used as a secondary antibody for these. GFP-PLD1 is shown in d. The merge is shown in c, f, and i. Scale bar: 25 μm. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
PLD Activation by ARF6
As described above, GFP-PLD2 localized with WT-ARF6 to a greater extent than GFP-PLD1 on the plasma membrane. We, therefore, examined if ARF6 could activate PLD2 to a greater extent than PLD1. GFP-PLDs and wild type and mutant forms of ARF6 were transfected into HeLa cells using a double expression vector system and then in vivo PLD activity was measured by the transphosphatidylation reaction. ARF1 and Rac1 were used as comparative controls. GFP-PLD1-expressing cells had increased PLD activity and GFP-PLD2-expressing cells showed greater PLD activity in control experiments without G proteins (Fig. 4A) consistent with PLD2 having higher constitutive activity. WT-ARF6 had little effect on the activity of either PLD, whereas DN-ARF6 suppressed PLD2 activity but not that of PLD1. DA-ARF6 increased the activity of PLD2, but not PLD1 (Fig. 4A). WT-ARF1 did not affect PLD1 or PLD2 activity, whereas DN-ARF1 suppressed both PLD1 and PLD2 activity and DA-ARF1 activated both PLD1 and PLD2. WT-Rac1 and DN-Rac1 had no effect on PLD1 and PLD2 activity, whereas DA-Rac1 activated both PLDs. Thus, these results indicate ARF1 and Rac1 activate both PLDs, whereas ARF6 has a selective stimulatory effect on PLD2. The expression levels of the two GFP-tagged PLD isoenzymes and of the G proteins of these experiments are shown in Figure 4B. PLD1 and PLD2 were well expressed and the expression was not altered by the G proteins. On the contrary, co-expression with PLD1 and PLD2 reduced the expression of DA-ARF6 and DA-Rac1 compared with co-expression with GFP alone. However, these G proteins were still clearly expressed in the presence of the PLD isozymes.
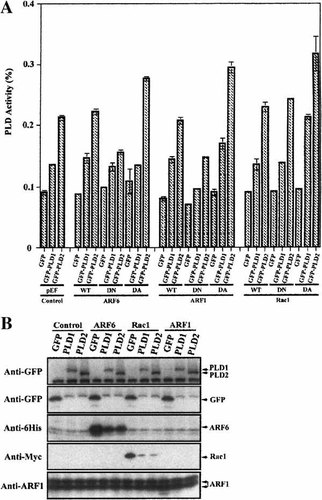
PLD activation by ARF6, ARF1, and Rac1. A: WT-, DN-, and DA-ARF6,-ARF1, and-Rac1 were co-transfected with pEGFP-C3, PLD1-pEGFP-C3, and PLD2-EGFP-C3 using double expression vectors and Fugene-6. Five hours later, the culture medium was changed to [3H]-myristic acid-containing DMEM and the cells were cultured for 10 h further. The cells were then incubated in 0.3% BSA–DMEM containing 0.3% 1-butanol for 40 min and then total lipids were extracted with cold methanol. The lipids were separated as described in Experimental Procedures. The data were calculated as cpm incorporated into phosphatidylbutanol as a percentage of cpm incorporated into total lipids. B: Shows the expression levels of the PLD isozymes and G proteins in a representative experiment.
Effect of Cytochalasin D on Localization of PLDs and Actin
Figure 2 showed that expression of DA-ARF6 results in condensation of actin and PLDs into the trans-Golgi apparatus after disruption of the actin cytoskeleton. We tested if disruption of actin polymerization by cytochalasin D could lead to the same result. Cytochalasin D treatment at 1 μM for 10 min induced disruption of the actin cytoskeleton and accumulation of actin in the cytosol fraction (Fig. 5-b,h cf. Fig. 1A,B-g,h,i), but it did not cause the appearance of the dense structure induced by DA-ARF6 (Fig. 5 cf. Fig. 2). After cytochalasin D treatment, PLD1 and PLD2 were partly distributed in actin dots at the periphery (Fig. 5-c,i) and partly with WGA-staining (Fig. 5-f,l). Cytochalasin D, therefore, had very different effect from DA-ARF6, which not only had an effect on actin fibers but also on PLD localization and/or activation.
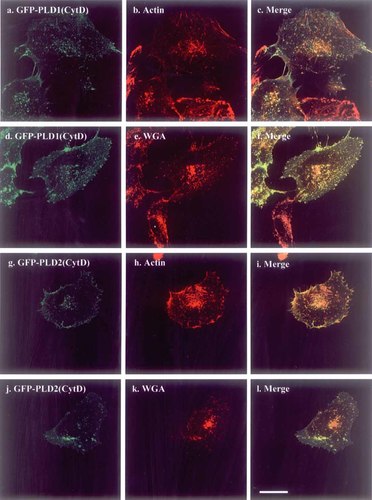
Cytochalasin D effect on the localization of PLD1 and PLD2. HeLa cells were transfected with GFP-PLD1 and 2, using Fugene-6 and then treated with 1 μM cytochalasin D for 10 min. After fixation, cells were stained with phalloidin-TexasRedX (b and h) and WGA-TexasRed (e and k). The merged images are shown in c, f, i, and l. Scale bar: 25 μm. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
WT-ARF6 Localizes in Cholesterol-Containing Microdomains
Cell staining experiments showed that WT- and DA-ARF6 had a similar localization with PLD isoforms (Fig. 2). To further explore their location, we undertook cell fractionation after treatment of cells with cytochalasin D and methyl-β-cyclodextrin. Cytochalasin D destroyed actin filaments, but did not induce the dense structure (Fig. 5). We considered whether actin filament disruption affected the localization of ARF6 or PLDs in the membrane fraction. Figure 6A shows that cytochalasin D did not change the association of WT-ARF6 and PLD isoforms with the membrane fraction. It was also examined whether the localization of ARF6 mutants was changed by methyl-β-cyclodextrin, which removes cholesterol from the plasma membrane, especially lipid rafts. All forms of ARF6 were located in the membrane fraction and DN-ARF6 and DA-ARF6 were resistant to methyl-β-cyclodextrin treatment (Fig. 6B). On the other hand, the level of WT-ARF6 in the membrane fraction was reduced by methyl-β-cyclodextrin treatment (Fig. 6B,C). Thus WT-ARF6 is located in a domain sensitive to cholesterol depletion, but not the other two types of ARF6. This is consistent with the different cellular location of the different forms of ARF6 (Fig. 2). It was further examined whether the membrane association of ARF6 and PLDs was changed in cells treated with PMA. HeLa cells transfected with WT-ARF6- or GFP-PLD were stimulated with 100 nM PMA for 30 min. Membrane levels of WT-ARF6 were reduced by methyl-β-cyclodextrin pre-treatment but were not altered by PMA stimulation for 30 min (Fig. 6C). PLD1 and PLD2 were exclusively in the membrane fraction and were not altered by treatment with methyl-β-cyclodextrin or PMA. This suggests that the location of the PLDs in membranes is not dependent on the presence of cholesterol. It has been reported that phosphorylated PLD1 exists in caveolae [Kim et al., 2000]. However, it is possible that the extent of phosphorylation was low in our experiments. These data indicate that PLDs and WT-ARF6 are located in different membrane domains. Our findings are consistent with reports that ARF6 exists in caveolae. Since it was still unclear whether ARF6 directly associates with PLD2, we attempted to detect the association by coimmunoprecipitation assay without success.
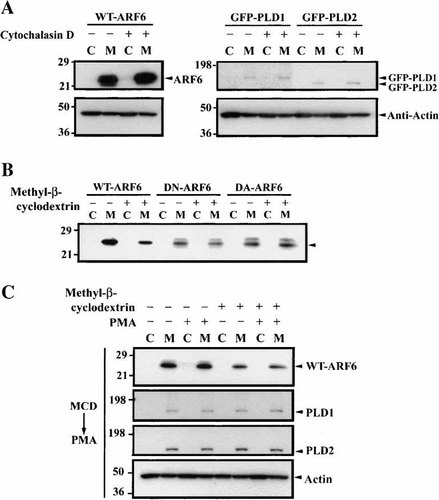
WT-ARF6 localizes in cholesterol-containing microdomains. A: WT-ARF6 or GFP-PLDs were transfected to HeLa cells and then the cells were treated with 1 μM cytochalasin D for 10 min. The cells were fractionated to cytosolic and membrane fractions and then the portions were applied to 4%–20% Tris-Glycine gel and then proteins were transferred to PVDF membranes. The membranes were immunoblotted with appropriate antibodies. C means cytosol fraction and M means membrane fraction. Actin was used as a control. B: Three kinds of ARF6 including WT-, DN-, and DA-forms are transfected in HeLa cells. The cells were treated with 5 mM methyl-β-cyclodextrin for 60 min and then the cells were fractionated. The cytosolic and membrane fractions were applied to 4%–20% Tris-glycine gels and then proteins were transferred to PVDF membranes. The membrane was immunoblotted by appropriate antibodies. C: GFP-PLD 1 and 2 or WT-ARF6 were transfected into HeLa cells. The cells were pre-treated with 5 mM methyl-β-cyclodextrin for 60 min and then with 100 nM phorbol 12-myristate 13-acetate (PMA) for 30 min. Actin was used as a control. MCD, methyl β-cyclodextrin.
Actin Cytoskeleton Is not Required for PLD Activation by Phorbol Ester
As described above, overexpressed-PLDs colocalized with actin dots but not with actin fibers. The observation that DA-ARF6 collapsed the actin cytoskeleton but still activated PLD led us to explore the effects of cytochalasin D on the activation of PLD. HeLa cells were pretreated with cytochalasin D and then stimulated with PMA. Although the actin filaments were destroyed, cytochalasin D did not inhibit PLD activation by PMA (Fig. 7). Thus actin fiber organization is not required for PLD activation by PMA or ARF6.
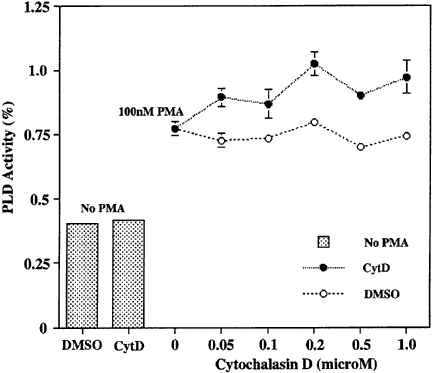
Cytochalasin D effect on PMA-induced PKC activation. HeLa cells were treated with cytochalasin D (0, 0.05, 0.1, 0.2, 0.5, and 1 μM) for 10 min and then with PMA for 5 min to activate PLD before 1-butanol addition. Cells were incubated in 0.3% BSA–DMEM containing 0.3% 1-butanol for 40 min and then total lipids were extracted with cold-methanol. An in vivo PLD assay was done as described in Experimental Procedures, and the data were calculated as described in the legend to Figure 4. DMSO was used as the control solvent.
DISCUSSION
In the present study, GFP-PLD1 appeared to localize mainly to a perinuclear site including the trans-Golgi apparatus, multivesicular endosomes, and late endosomes, but rarely early endosomes (data not shown). It was also not localized to the cis-Golgi, lysosomes, and endoplasmic reticulum. The perinuclear location agrees with most previous studies of tagged PLD1 [Colley et al., 1997; Kim et al., 1999b; Sugars et al., 1999; Toda et al., 1999; Emoto et al., 2000; Kam and Exton, 2001; Vitale et al., 2001]. However, there is no consistent agreement about the organelles with which PLD1 is associated in the perinuclear region. This appears to be due to differences in cell type and in the markers used to identify the organelles. There is more agreement with the present study about the plasma membrane localization of PLD2 [Colley et al., 1997; Emoto et al., 2000; Park et al., 2000].
In agreement with other reports [Radhakrishna and Donaldson, 1997; D'Souza-Schorey et al., 1998; Song et al., 1998; Radhakrishna et al., 1999; Al-Awar et al., 2000], our studies indicate a distribution of wild type and dominant negative ARF6 between the plasma membrane and perinuclear vesicles (recycling endosomes). This pattern was little changed by co-expression of PLD1 or PLD2. However, coexpression of these isozymes with (WT)-ARF6 induced some overlapping of images, especially in the perinuclear region.
Expression of (DA)-ARF6 induced profound changes. There was formation of dense structures in which PLD1 and PLD2 overlapped with collapsed actin fibers and the trans-Golgi apparatus. In other studies, (DA)-ARF6 has been localized to the plasma membrane, including membrane ruffles, folds and sacs [D'Souza-Schorey et al., 1998; Boshans et al., 2000; Palacios et al., 2001; Schweitzer and D'Souza-Schorey, 2002]. However, there have also been reports that ARF6 alters the actin cytoskeleton and causes loss of adherens junctions [Song et al., 1998; Boshans et al., 2000; Schafer et al., 2000; Palacios et al., 2001].
DA-ARF6 overexpression in HeLa cells caused translocation of PLD1 and 2 into the dense structure which can be stained with WGA-TexasRed-X and phalloidin-TexasRed-X. These results also showed that, after actin filament degradation by ARF6, PLDs, and actin appeared to accumulate in the trans-Golgi apparatus. The actin cytoskeleton is important for the regulation and organization of both exocytotic and endocytotic functions. Actin, actin-binding proteins, and myosin are bound to Golgi membranes and Golgi-derived vesicles [Heinmann et al., 1999; De Matteis and Morrow, 2000; Valderrama et al., 2000]. Furthermore, actin localization in the Golgi apparatus is regulated by ARF1, Rac, and Cdc42 [Fucini et al., 2002]. Thus after actin structure disruption by ARF6, degraded actin is drawn into the trans-Golgi apparatus by actin-binding proteins which are connecting to the trans-Golgi apparatus. As described above, GFP-PLD1 and 2 were co-localized with actin in the dense structure. Further it is known that PLD binds to actin [Lee et al., 2001]. Thus it is possible that PLD was also drawn into the trans-Golgi along with actin. As noted above, cytochalasin D dispersed actin, in contrast to DA-ARF6, which concentrated it. Cytochalasin D is an inhibitor of actin polymerization, which causes the degradation of cell wide actin filaments. On the other hand, ARF6 destroys peripheral actin structures and induces membrane ruffling. Thus this fact suggests that actin remaining near the trans-Golgi can induce PLD to localize there but completely destroyed actin structures cannot.
The overlapping localization of (WT)-ARF6 and (DA)-ARF6 with PLD1 and PLD2 led us to examine the effects of these ARF6 forms on PLD1 and PLD2 activity in vivo. This revealed a striking difference between the ability of (DA)-ARF6 to activate the PLD isozymes, i.e., (DA)-ARF6 stimulated PLD2 only. In contrast (DA)-ARF1 and (DA)-Rac1 activated both isozymes. Because (DA)-ARF6 caused collapse of actin fibers into a dense structure containing PLD1 and PLD2, we tested to see the effects of cytochalasin D, which is well known to disrupt the actin cytoskeleton. However, this compound did not induce the appearance of the dense structure seen with (DA)-ARF6. Neither did it affect the activation of PLD2 by (DA)-ARF6 or phorbol ester.
The present studies extend the observations that, although PLD2 cannot apparently be activated by PKC or members of the ARF and Rho families of GTPases in vitro [Colley et al., 1997; Kodaki and Yamashita, 1997; Lopez et al., 1998; Sung et al., 1999] this isozyme can be stimulated by phorbol ester or PKC isozymes in vivo [Siddiqi et al., 2000; Han et al., 2002; Xie et al., 2002; Mitchell et al., 2003] and by (DA)-Rac1, (DA)-ARF1, and (DA)-ARF6 in vivo (Fig. 4). These results suggest that crucial components required for activation of PLD2 in vivo are lacking in the in vitro studies or that the assay conditions used in the in vitro studies are suboptimal for revealing the effects. Santy and Casanova [2001] have observed that ARNO a guanine nucleotide exchange factor for ARF6 can increase PLD activity in MDCK cells, but the specific PLD isozyme involved was not defined. Mitchell et al. [2003] obtained evidence that PLD could be regulated by ARF6 in COS7 cells, but were unable to determine if PLD1 or PLD2 was involved. Honda et al. [1999] noted that ARF6 and PLD2 co-localized in membrane ruffles in HeLa cells treated with epidermal growth factor, but no measurements of PLD activity were made. In NIH3T3 cells, transformation with H-Ras led to the activation of PLD, which could be blocked by (DN)-ARF6, and the expression of both activated Ral1 and ARF6 increased PLD activity [Xu et al., 2003]. The PLD isozyme involved was not determined. In summary, several studies indicate that ARF6 can increase PLD activity in vivo and the present findings point to PLD2 as the selective PLD target of ARF6 in vivo. In contrast, ARF1 and Rac1 are not so selective.
Further, we carried out cell fractionation after methyl-β-cyclodextrin extraction to determine the extent to which PLD isoforms and ARF6 mutants exist in cholesterol-containing microdomains. van Deenen and colleagues [Demel et al., 1968] established that the biological properties of cell membranes were dependent on their free cholesterol content. Free cholesterol is not randomly distributed in synthetic lipid monolayers [Darke et al., 1971] or in biological membranes [Schroeder et al., 1991]. It forms local, free cholesterol/sphingolipid-rich patches whose properties differ from those of adjacent regions. Purified preparations of these patches from cell membranes are enriched in signaling proteins [Anderson, 1993]. The surfaces of most living cells contain at least two distinct classes of free cholesterol/sphingolipid-rich microdomains (lipid rafts and caveolae) that differ in their structure, stability and content of associated proteins [Schnitzer et al., 1995; Iwabuchi et al., 1998; Abrami et al., 2001; Sowa et al., 2001]. Caveolae, which are flask-shaped membrane invaginations, were first detected on the surface of endothelial cells. It is known that a considerable proportion of PLD1 activity in HL-60 cells is located in a detergent-insoluble fraction of the cell membrane that is unlikely to be a caveolae-like domain, but is probably cytoskeletal [Hodgkin et al., 1999]. PLD2 is preferentially targeted to microdomains that contain the caveolae marker protein caveolin-1 in human keratinocytes and human umbilical vein endothelial cells [Czarny et al., 1999; Yoon et al., 2003; Cho et al., 2004]. Further it is reported that PLD1 also exists in caveolae [Kim et al., 1999a; Xu et al., 2003]. There is disagreement regarding to PLD isoforms location in caveolae. It is possible that the difference comes from the approaches used. The previous study showed the colocalization of PLD and caveolae as detected by fractionation using sucrose gradients or detergent, but in the present study we used a combination of fractionation and extraction by methyl-β-cyclodextrin. Our finding is consistent with reports that ARF6 exists in caveolae, for example, ARF6 colocalizes with Ral1 in caveolin-enriched light membrane fractions, which together lead to elevated PLD activity [Xu et al., 2003].
In the in vivo PLD assay, actin filament disruption did not inhibit PLD activity. That is not consistent with a previous report in vivo, but is in accord with in vivo results using latrunculin B to directly sequester free G-actin monomers [Kusner et al., 2002]. Figure 5 shows that PLD1 and PLD2 overlap partially with actin-dots labeled with phalloidin, which recognizes F-actin.
The domain in PLD1 that is the target of ARF1 remains undefined, although there is evidence that it is not in the N-terminal 319 amino acids [Park et al., 1998; Sung et al., 1999]. Concerning the interaction site for PLD1 on ARF1, this has been localized to the N-terminus [Zhang et al., 1995; Jones et al., 1999a], but the amino acid residues involved have not been defined. Therefore, the basis for the selectivity of ARF6 for PLD2 remains to be discovered. Another question requiring much further work is to explore the role of PLD2 in the physiological actions of ARF6.




