Cross-talk Between Histone and DNA Methylation Mediates Bone Loss in Hind Limb Unloading
ABSTRACT
Bone loss induced by mechanical unloading is a common skeletal disease, but the precise mechanism remains unclear. The current study investigated the role of histone methylation, a key epigenetic marker, and its cross-talk with DNA methylation in bone loss induced by mechanical unloading. The expression of G9a, ubiquitin-like with PHD and ring finger domains 1 (UHRF1), and DNA methylation transferase 1 (DNMT1) were increased in hind limb unloading (HLU) rats. This was accompanied by an increased level of histone H3 lysine 9 (H3K9) di-/tri-methylation at lncH19 promoter. Then, alteration of G9a, DNMT1, or UHRF1 expression significantly affected lncH19 level and osteogenic activity in UMR106 cells. Osteogenic gene expression and matrix mineralization were robustly promoted after simultaneous knockdown of G9a, DNMT1, and UHRF1. Furthermore, physical interactions of lncH19 promoter with G9a and DNMT1, as well as direct interactions among DNMT1, G9a, and UHRF1 were detected. Importantly, overexpression of DNMT1, G9a, or UHRF1, respectively, resulted in enrichment of H3K9me2/me3 and 5-methylcytosine at lncH19 promoter. Finally, in vivo rescue experiments indicated that knockdown of DNMT1, G9a, or UHRF1 significantly relieved bone loss in HLU rats. In conclusion, our research demonstrated the critical role of H3K9 methylation and its cross-talk with DNA methylation in regulating lncH19 expression and bone loss in HLU rats. Combined targeting of DNMT1, G9a, and UHRF1 could be a promising strategy for the treatment of bone loss induced by mechanical unloading. © 2021 American Society for Bone and Mineral Research (ASBMR).
Introduction
Disuse osteoporosis (DOP) is caused by mechanical unloading or reduced mechanical stimulation, which leads to low-energy fractures, deterioration of body function, increased morbidity, and impaired quality of life.(1-3) Although an imbalance between bone formation and resorption is the key pathological change in response to mechanical unloading, the underlying mechanisms remain unclear.
The main mechanisms of epigenetic regulation involve DNA methylation, histone modification, and non-coding RNAs, which result in an external stimulus being imprinted on DNA/chromatin, influencing gene expression.(4) There is accumulating evidence indicating that abnormal gene expression and cellular processes that occur during the development of DOP are strongly linked to dysregulated epigenetic modifications.(5, 6) Hind limb unloading (HLU)-induced upregulation of DNA methylation transferase 1 (DNMT1) leads to DNA hypermethylation at lncH19 promoter, thus inhibiting its expression and resulting in decreased osteogenic activity.(7, 8) However, gene expression levels do not completely recover after the administration of a DNMT1 inhibitor, indicating that mechanisms other than DNMT1-mediated DNA methylation contribute to lncH19 silencing.(8)
Histone methylation, which is catalyzed by histone methyltransferase, predominantly occurs in lysine residues of histone H3 and H4. Methylation at H3K9, H3K27, and H4K20 is generally linked to transcriptional repression.(9, 10) Mono-(me), di-(me2), and tri-(me3) methylation of lysine residue 9 of histone H3 (H3K9) catalyzed by G9a are primarily found to be associated with silent genes and are involved in numerous physiological and pathological processes, including bone metabolism-associated signaling pathways and cellular differentiation.(11-13) Loss of H3K9 methylation at the Zfp423 gene promoter leads to increased Zfp423 expression and adipocyte commitment during bone morphogenic protein-induced mesenchymal precursor differentiation.(14) In addition, the levels of H3K9 me/me2/me3 in RunX2-binding regions of Bglap2 and Ibsp genes decrease during the differentiation of MC3T3-E1 cells.(15) The above results indicate the potential role of G9a-mediated H3K9 methylation in the regulation of osteoblast differentiation and bone metabolism. However, less attention has been given to its association with bone disorders.
Epigenetic regulation is a complex process involving various mechanisms, in which DNA and histone methylation coordinate with each other for chromatin remodeling and gene expression control.(16, 17) The functional interaction of G9a with DNMT1 through ubiquitin-like with PHD and ring finger domains 1 (UHRF1), an adaptor in the cross-talk between DNA and histone methylation, has been identified in vitro(18, 19) and in disease models.(20-22) In view of the crucial role of H3K9 methylation and its cross-talk with DNA methylation, we speculated that it may be involved in transcriptional repression of lncH19 in response to mechanical unloading. The purpose of the current study is to investigate the role of histone methylation and its cross-talk with DNA methylation in bone loss induced by mechanical unloading. Further understanding of these complex epigenetic regulations will facilitate the identification of novel therapeutic targets.
Materials and Methods
Animals and interventions
Animal care and experimental design were manipulated as previously described.(7, 8) The animal procedures were approved by the Ethical Committee of Tianjin Hospital (2019012). Briefly, 12-week-old male Sprague Dawley (SD) rats were randomly assigned into different experimental groups of 12 rats each and kept under the same standard conditions; water and food were available ad libitum. Functional disuse was realized by hind limb unloading as in our previous study.(8) Rats in HLU groups were euthanized at 1, 2, 3, and 4 weeks after the beginning of tail suspension, and normal controls were euthanized after 4 weeks of breeding. In another set of experiments, siRNA transfection in vivo was conducted according to the manufacturer's instructions. SiRNAs targeting DNMT1, G9a, and UHRF1 were mixed with Entranster in vivo transfection reagent (Engreen, Beijing, China) for 15 minutes at room temperature. The siRNA-Entranster complex was then injected into the medullary cavity using a 1-mL syringe after 1 and 3 weeks of HLU.(8, 23) Euthanasia was performed at week 4 and the femurs were collected for subsequent experiments.
Micro-computed tomography (μCT) scan
The left femur was scanned using μCT (Skyscan 1276, Bruker microCT, Kontich, Belgium) to obtain high-quality images of the trabecular and cortical bone. The scanning parameters were set as follows: 17.5 μm spatial resolution, 70 kV, 200 uA, and 498 ms of integration time. The length of region of interest (ROI) is 1977 μm, starting from 262 μm proximal to the distal epiphyseal plate of the femur. Global thresholding, which were selected visually by having the same operator compare raw images to thresholded images, was applied to avoid bias in determining μCT outcome variables. Trabecular microarchitecture was quantified by using the following values: bone volume fraction (bone volume/total volume [BV/TV]), trabecular thickness (Tb.Th), trabecular number (Tb.N), and trabecular separation (Tb.Sp). For the analysis of cortical bone thickness, the volume of interest was defined as a cylindrical area covering the distal femur.(24)
RNA isolation, quantitative real-time PCR (qRT-PCR), and RNA sequencing
RNA isolation was performed according to our previously procedures.(7, 8, 25) For bone tissues, the right femur was ground in liquid nitrogen after removing the attached soft tissues. Then the total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. After isolation, RNA sequencing was performed as described previously.(7) Alternatively, a First-Strand cDNA Synthesis Kit (Tiangen, Beijing, China) was used to reverse-transcribe the isolated RNA. Then the RNA expression was evaluated by SYBR green-based real-time PCR with standard protocols (Roche, Mannheim, Germany). Then β-actin or Gapdh was used as housekeeping gene to calculate the relative expression levels of target genes. The primer sequences are listed in Supplemental Table S1.
Protein isolation and Western blot
The procedures of protein extraction from both cell and bone tissue were conducted as previously described.(26) Thirty micrograms of total protein was electrophoresed and electrotransferred to polyvinylidene difluoride (PVDF) membrane. TBST containing 5% milk was used to block the PVDF membranes, which then were incubated at 4°C overnight with the following primary antibodies: anti-β-actin (1:5000, ProteinTech, Rosemont, IL, USA), anti-DNMT1 (1:1000, Abcam, Cambridge, MA, USA), anti-G9a (1:1000, Abcam), anti-UHRF1 (1:1000, Abcam), anti-H3K9me2 (1:1000, Active Motif, Carlsbad, CA, USA), anti-H3K9me3 (1:1000, Active Motif), anti-H3K27me2 (1:1000, Active Motif), anti-H3K27me3 (1:1000, Active Motif). Immunoreactive bands, which were visualized by an enhanced chemiluminescence detection system (MilliporeSigma, Burlington, MA, USA), were detected after 1 hour incubation with HRP-conjugated secondary antibodies (1:2000, ProteinTech).
Chromatin immunoprecipitation (ChIP)
The ChIP assay was performed using EZ-ChIP (MilliporeSigma) following the manufacturer's instruction. Briefly, samples were cross-linked with 1% formaldehyde at room temperature for 10 minutes and then sonicated to shear DNA. Genomic fragments of proper length were immunoprecipitated with IgG, anti-H3K9me3 antibody, anti-H3K9me2 antibody, anti-H3K27me3 antibody, and anti-H3K27me2 antibody overnight at 4°C. Precipitation of the complexes were performed with protein A/G magnetic beads (Thermo Fisher Scientific, Waltham, MA, USA) and then the DNA was eluted with elution buffer. The DNA was purified using a PCR purification kit (Sangon Biotech, Shanghai, China). The purified DNA was amplified by PCR and the products were analyzed by electrophoresis on an agarose gel. The binding capacity of required chromatins to the lncH19 promoters was evaluated by quantitative PCR with the shear DNA sample serving as an input control. Primer sequences in ChIP-qPCR are listed in Supplemental Table S2.
Cell culture
Cell cultures were carried out as previously described.(7, 8, 23, 25) Briefly, UMR106 cells and HEK293T cells obtained from Cell Bank of Chinese Academy of Sciences were cultured in Dulbecco's modified Eagle's medium (Gibco, Thermo Fisher Scientific) with 10% fetal bovine serum (Gibco) at 37°C under a humidified atmosphere of 5% CO2 in air. Primary rat mesenchymal stem cells (MSCs) were isolated by flushing the bone marrow of tibias and femurs. The cells were then cultured in α-MEM (Gibco) supplemented with 10% fetal bovine serum. For osteogenic induction, cells were treated with osteogenic medium containing 50 μg/mL ascorbic acid, 5 mM β-glycerophosphate, and 100 nM dexamethasone (all from Sigma, St. Louis, MO, USA).
Plasmid construction and transfection
The CDS sequence encoding DNMT1 was synthesized and cloned into pcDNA3.1-FLAG and pCDH vector (CD510B, System Biosciences, Palo Alto, CA, USA). G9a CDS sequence was synthesized and cloned into pcDNA3.1-Myc and pCDH vector. UHRF1 CDS sequence was synthesized and cloned into pCDH vector. The shRNA targeting DNMT1, G9a, and UHRF1 were synthesized and cloned into pKLO.1 vector (Addgene, Watertown, MA, USA). The shRNA targeting sequence is listed in Supplemental Table S3. HEK293T cells were transfected with Lipofectamin 2000 according to manufacturer's protocol.
Lentivirus preparation and infection
In brief, pCDH or pKLO.1, pMDLg/pRRE (12251, Addgene), pRSV-Rev (12253, Addgene), and pMD2.G (12259, Addgene) plasmids were transfected into HEK293T cells with lipofectamin 2000. Lentiviral particles were then harvested 48 hours after transfection. Targeted genes were overexpressed or knocked down by infection of UMR106 cells with recombinant lentiviral units in the presence of 10 μg/mL polybrene (Sigma) and subsequent puromycin (2 μg/mL, Sigma) selection.
Alizarin red staining
Matrix mineralization was assessed for samples using Alizarin red staining. After osteogenic induction, the cells were fixed in 4% paraformaldehyde and stained by 2% Alizarin red for 10 minutes (pH 7.2, Sigma). Samples were then washed and imaged.
DNA pull-down and liquid chromatography-mass spectrometry
A biotinylated lncH19 promoter probe (2 kb) was synthesized by GenScript Biotech Corporation (Piscataway, NJ, USA). The promoter sequence is listed in Supplemental Table S4. The biotinylated DNA was incubated with nuclear proteins and then lncH19 promoter-interacting proteins were pulled down using streptavidin-conjugated magnetic beads according to the manufacturer's instructions (Thermo Fisher Scientific). The bond between streptavidin and biotin was disrupted by resuspending the pull-down mixtures in distilled water at 70°C for 3 minutes. Retrieved proteins were resolved by gradient gel electrophoresis, which was followed by regular liquid chromatography-mass spectrometry (LC-MS) analysis by Bio-tech & Consult Co., Ltd. (Shanghai, China).
Dual luciferase reporter assay
An lncH19 promoter (2 kb) was synthesized and cloned into pMirGLO vector (Promega, Madison, WI, USA) to replace PGK promoter. The lncH19 promoter reporter and DNMT1, G9a, or UHRF1-expression plasmids were co-transfected into cells using lipofectamine 2000. Seventy-two hours after transfection, cells were harvested and a dual-luciferase reporter assay kit (Promega) was used to evaluate the function of lncH19 promoter, in which the normalized luminescence was calculated by the ratio of firefly to renilla luciferase activity.
Co-immunoprecipitation assay
Cells were rinsed three times with ice-cold PBS and then were lysed by lysis buffer (9803, Cell Signaling Technology, Danvers, MA, USA). Cells were scraped off the plates and sonicated on ice three times for 3 seconds each. Cell lysates were then centrifuged and the supernatant was used for the following experiments. Anti-Flag or anti-Myc magnetic beads (Thermo Fisher Scientific) were incubated with the cell lysate overnight at 4°C. After magnetization, the immunoprecipitation beads were washed with lysis buffer and resuspended in 1× SDS sample buffer, then heated at 95°C for 5 minutes. Subsequently, Western blotting was performed as described above.
Methylation-specific PCR (MSP)
DNA was extracted from samples using the Genomic DNA Kit (DP304, Tiangen) as previously described.(8) Bisulfite-treated DNA were amplified by PCR using two sets of primers, methylated DNA (M-MSP) and unmethylated DNA (U-MSP). The primer sequences of M-MSP are as follows: forward primer, AGTTTA CGTGTGATGGTTAGTTTTC; reverse primer, ATCTCTACGCTCTTAAAAAATCGAT. The forward and reverse primer sequences of U-MSP are TTTATGTGTGATGGTTAGTTTTTGG and ATCTCTACACTCTTAAAAAATCAAT. Two percent agarose gels stained with ethidium bromide were used to visualize PCR products.(8)
Histomorphology and immunohistochemistry stain
Femurs in different groups were fixed in 10% neutral buffered formalin solution for 48 hours. Decalcification was performed in 0.3 M EDTA until the femurs could be penetrated by a needle. Femurs were then dehydrated with gradient increasing concentration of ethanol, embedded in paraffin, sectioned (at 5 μm thickness), and cleared by 100% xylene.
Immunohistochemistry staining was performed as previous reported.(27) Briefly, samples were rehydrated, peroxidase inactivated (3% H2O2), permeabilized (0.5% Triton-X 100), blocked with species-appropriate serum for 30 minutes at room temperature, and incubated overnight at 4°C with primary antibodies: anti-DNMT1 (1:100, Abcam), anti-G9a (1:100, Abcam), anti-UHRF1 (1:100, Abcam). On the subsequent day, sections were incubated with secondary antibody, and peroxidase development was performed with an enzyme substrate kit (ZSGB-BIO, Beijing, China). The negative control is listed at the end of the Supplemental Figures. Alternatively, the samples were stained with hematoxylin and eosin (H&E). Then, the adipocytes per tissue area in the distal marrow and vessel number were calculated.
Toluidine blue and TRAP staining were performed in undecalcificated sections as previously reported.(28) Osteoblast surface/bone surface (Ob.s/bs) and osteoclast surface/bone surface (OC.s/bs) were analyzed subsequently with Osteo-measure software (OsteoMetrics, Decatur, GA, USA).
Serum bone turnover
Serum osteocalcin (OC) and carboxy-terminal collagen cross-links (CTX) were examined by Rat Osteocalcin ELISA kit (CUSABIO, Wuhan, China) and Rat CTX ELISA Kit (CUSABIO) according to the manufacturer's protocol.
Statistical analysis
All data were analyzed using the Prism 8.0 (GraphPad, La Jolla, CA, USA). Descriptive statistics are presented as mean ± SD or ratios. Statistical significance was assessed with Student's t test or analysis of variance (ANOVA) with Tukey's post hoc test. The number of independent experiments is indicated in the figure legends. A p value <0.05 was considered to be statistically significant.
Results
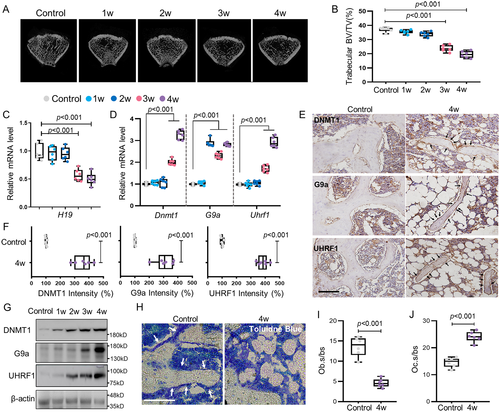
HLU leaded to increased expression of DNMT1, G9a, and UHRF1 in the distal femur of rats
Trabecular bone in the distal femur progressively decreased with time after the application of HLU (Fig. 1A, B; Supplemental Fig. S1A). This was accompanied by a decrease in the expression of lncH19 (Fig. 1C). Meanwhile, the mRNA level of Dnmt1, G9a, and Uhrf1 were increased in HLU rats (Fig. 1D), whereas no significant change was found in histone deacetylase 1 (Hdac1), Hdac3, or histone acetyltransferase 1 (Hat1) (Supplemental Fig. S1B). Immunohistochemistry staining showed that DNMT1, G9a, and UHRF1 were primarily increased in osteoblasts on trabecular bone surfaces (Fig. 1E, F). Consistently, Western blot analysis indicated that the protein level of DNMT1, G9a, and UHRF1 were upregulated (Fig. 1G). Toluidine blue staining showed significantly decreased Ob.s/bs after 4 weeks of HLU (Fig. 1H, I), whereas TRAP staining indicated increased Oc.s/bs, suggesting both osteoblasts and osteoclasts were involved in unloading-induced bone loss (Fig. 1J; Supplemental Fig. S1C). RNA-sequencing analysis was then conducted to further verify the changes of related genes. The results showed that the expression of Dnmt1, G9a, and Uhrf1 were increased (Supplemental Fig. S2A), which was consistent with previous results, whereas H3K9 demethylases (Jmjd1/a and Phf8) were not changed (Supplemental Fig. S2A). Additionally, the expression of osteogenic Alp, RunX2, and Opg were significantly decreased (Supplemental Fig. S2B). DNMT1, G9a, and UHRF1 play critical roles in both DNA and histone methylation, so these results indicated that both of them may contribute to the development of bone loss in HLU.
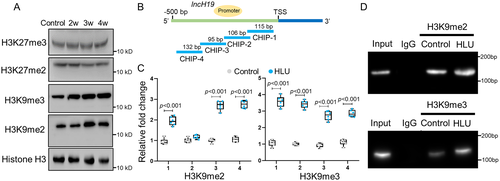
HLU resulted in increased level of DNA methylation and H3K9me2/3 at lncH19 promoter
Western blot analysis revealed increasing level of H3K9me2 and H3K9me3 after application of HLU in rats, whereas no significant change was observed in the level of H3K27me2 or H3K27me3 compared with normal controls (Fig. 2A). ChIP assays were then performed to assess the methylation status of H3K9 at the promoter region of lncH19. Significantly increased level of H3K9me2 and H3K9me3 were observed in HLU rats (Fig. 2B–D). We have found that DNA methylation level of lncH19 promoter was increased in HLU rats.(8) Taken together, these results suggested that decreased expression of lncH19 in response to mechanical unloading is closely associated with increased methylation of H3K9 and cytosine at the promoter region.
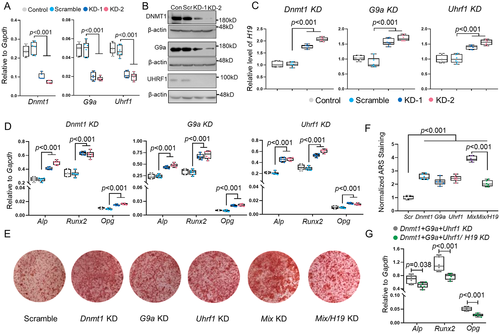
Knockdown of DNMT1, G9a, or UHRF1 increased lncH19 expression and enhanced osteogenic activity in UMR106 and primary osteoblast cells
Our previous work identified that DNMT1 plays a critical role in osteoblasts to affect HLU-induced bone loss.(8) Therefore, we first explored the function of DNA and H3K9 methylation enzymes in osteoblasts. Lentivirus-mediated transfection efficiently knocked down DNMT1, G9a, and UHRF1 at both mRNA and protein levels (Fig. 3A, B), resulting in significantly increased expression of lncH19, Alp, Runx2, and Opg (Fig. 3C, D). Importantly, simultaneous knockdown of DNMT1, G9a, and UHRF1 resulted in a greater change in lncH19, Alp, RunX2, and Opg expression (Supplemental Fig. S3A), whereas it exhibited minimal and often insignificant effect on methylation status in the promoter regions of Alp, Runx2, and Opg (Supplemental Fig. S3B–D). High osteogenic activity was further confirmed by Alizarin red staining, which showed a significant increase in matrix mineralization after knockdown of the three genes simultaneously (Fig. 3E). Importantly, knockdown of lncH19 can cancel the effects on osteogenic gene expression and mineralization ability in UMR106 cells (Fig. 3F, G), which was further verified in primary rat osteoblasts (Supplemental Fig. S4). These results indicated that Dnmt1, G9a, and Uhrf1 participate together in manipulating osteoblast fate through lncH19.
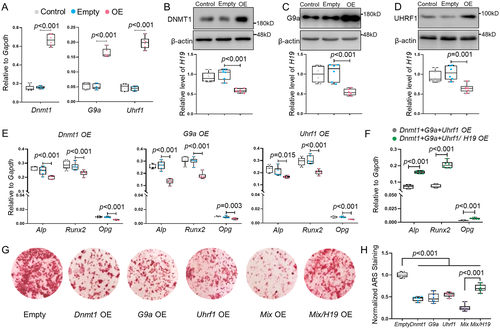
Overexpression of DNMT1, G9a, or UHRF1 decreased lncH19 expression and impaired osteogenic activity in UMR106 cells
DNMT1, G9a, and UHRF1 were significantly overexpressed at both mRNA and protein levels by using lentivirus (Fig. 4A–D). As expected, overexpression of the targeted genes resulted in a significant decreased expression of lncH19 (Fig. 4B–D). Meanwhile, the expression of osteogenic genes were also downregulated (Fig. 4E). Simultaneously overexpression of DNMT1, G9a, and UHRF1 exhibited the most significant inhibition on lncH19, Alp, RunX2, and Opg expression (Supplemental Fig. S5). Furthermore, overexpression of lncH19 can rescue the effects on osteogenic gene expression and mineralization ability (Fig. 4F–H). These results further confirmed that the effects of DNMT1, G9a, and UHRF1 on osteoblast were mediated by lncH19.
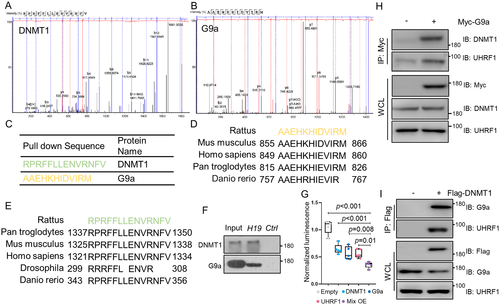
DNMT1, G9a, and UHRF1 physically interacted with each other and binded to lncH19 promoter to regulate its expression
The synthesized biotin-labeled lncH19 promoter was incubated with nuclear proteins extracted from cells. After incubation, lncH19 promoter-interacting proteins were pulled down using streptavidin-conjugated magnetic beads. Then, the beads were washed, and the eluted proteins were separated using SDS-PAGE. Each gel lane was divided into 10 bands and digested with trypsin for downstream LC-MS analyses. Both DNMT1 and G9a sequences were identified in lncH19 promoter-interacting proteins (Fig. 5A–C). Importantly, the pull-down amino fragments of DNMT1 and G9a were conserved among different species, which suggested these lncRNA promoter-protein interactions could play important functions in biological process (Fig. 5D, E). To further validate the interacting proteins identified by the LC-MS, DNA pull-down and immunoblotting assay were performed. We observed that both DNMT1 and G9a could bind with lncH19 promoter (Fig. 5F). These results suggest that lncH19 promoter is physically associated with DNMT1 and G9a. Next, we sought to determine whether the association between lncH19 promoter and DNMT1/G9a had functional significance. Because both DNMT1 and G9a are epigenetic modifiers, we investigated whether these interactions could regulate the function of lncH19 promoter. Luciferase assays showed that co-expression of lncH19 promoter reporter with DNMT1, G9a, and UHRF1 significantly decreased its activity (Fig. 5G). These results suggested that DNMT1, G9a, and UHRF1 regulate lncH19 expression through binding to its promoter region. Then, co-immunoprecipitation assays were conducted to further explore the relationship among DNMT1, G9a, and UHRF1. The results showed that both DNMT1 and UHRF1 proteins were co-immunoprecipitated with exogenous Myc-tagged G9a (Fig. 5H). Similarly, both G9a and UHRF1 proteins were co-immunoprecipitated with Flag-tagged DNMT1 (Fig. 5I). These results indicated that DNMT1, G9a, and UHRF1 physically interact with each other and the complex could be recruited to lncH19 promoter to regulate its expression.
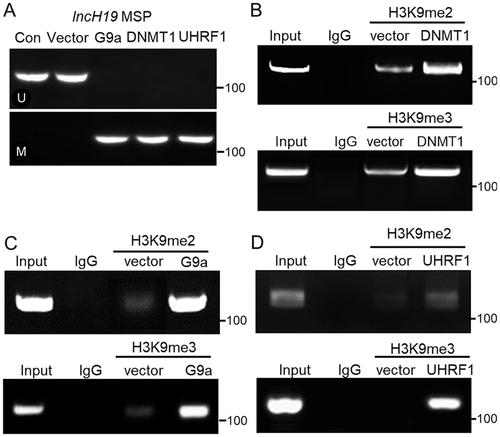
Overexpression of DNMT1, G9a, or UHRF1 resulted in DNA hypermethylation and increased level of H3K9me2/3 at lncH19 promoter in UMR106 cells
Methylated and unmethylated sequences were amplified to investigate the methylation state of CpG islands proximal to the transcriptional start site of lncH19. MSP showed that the amount of 5-methylcytosine at lncH19 promoter increased after individual overexpression of DNMT1, G9a, or UHRF1 in UMR106 cells (Fig. 6A). Moreover, ChIP assays showed that a significantly greater amount of chromosomal DNA containing lncH19 promoter was immunoprecipitated by anti-H3K9me2 and anti-H3K9me3 antibodies in response to DNMT1, G9a, and UHRF1 overexpression (Fig. 6B–D). These data indicated that DNMT1, G9a, and UHRF1 exhibited specific epigenetic effects on lncH19. However, whether DNA and histone methylation alterations induced by these genes occurred genomewide needs to be further explored.
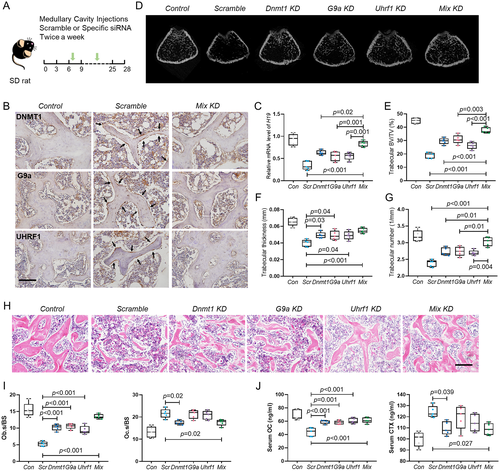
In vivo knockdown of DNMT1, G9a, and UHRF1 increased lncH19 expression and rescued bone loss in HLU rats
To further explore the critical role of DNA and histone methylation in HLU-induced bone loss, siRNA transfection in vivo was performed to knock down DNMT1, G9a, and UHRF1 (Fig. 7A, B; Supplemental Fig. S6). Significantly, the expression of lncH19 was upregulated after knockdown of three genes, especially in the mix knockdown group (Fig. 7C). Consistent with this result, siRNA treatment decreased the bone loss in HLU rats (Fig. 7D). The trabecular bone impairment was rescued most significantly in the mix knockdown group, with BV/TV, Tb.Th, and Tb.N levels increasing by approximately 2-fold, 1.4-fold, and 1.3-fold, respectively, compared with those in the HLU group (Fig. 7E–G). Meanwhile, the trabecular separation was significantly decreased (Supplemental Fig. S7A). Hematoxylin and eosin staining of the distal femur further confirmed that there was less trabecular bone loss in the rats treated with siRNA, with the greatest repair after knockdown of three genes simultaneously (Fig. 7H). Toluidine blue staining showed that Ob.s/bs was increased by siRNA treatment, which indicated that the osteogenic activity was rescued (Fig. 7I; Supplemental Fig. S7B). Consistently, serum osteocalcin was also upregulated (Fig. 7J). On the other hand, the Oc.s/bs and serum CTX was also changed after knockdown of DNMT1, but without significant change by G9a and UHRF1 (Fig. 7I, J; Supplemental Fig. S7B).These results indicated that DNMT1, G9a, and UHRF1-mediated lncH19 signaling primarily plays a function in osteoblasts. The change of osteoclasts could be due to the direct effect of DNMT1 on osteoclasts or secondary to the osteoblast alteration, which needs to be further explored in the future work. Interestingly, cortical thickness of femur was also rescued by knockdown of DNMT1 and G9a (Supplemental Fig. S8A), indicating the critical role of cross-talk between DNA and histone methylation for both trabecular and cortical bone. Last, we found that siRNA treatment in situ affects adipose tissue formation significantly but not blood vessel density (Supplemental Fig. S8B, C). This may be because knockdown of these genes promotes the differentiation of bone marrow mesenchymal stem cells into osteoblasts, leading to the reduction of adipogenic differentiation. But the blood vessel may not be the main target of DNMT1, G9a, and UHRF1.
Discussion
Our research revealed the crucial role of a cross-talk between H3K9 and DNA methylation in the development of bone loss in HLU rats through the regulation of lncH19. We found that mechanical unloading led to the upregulation of DNMT1, G9a, and UHRF1 and inhibition of lncH19, which were accompanied by increased level of H3K9me2/me3 at lncH19 promoter. In vitro experiments showed that individual alteration in DNMT1, G9a, or UHRF1 resulted in lncH19 expression and osteogenic activity changes. Importantly, knockdown or overexpression of lncH19 can cancel or rescue the effects on osteogenic gene expression and mineralization ability. We also identified that lncH19 promoter can directly interact with G9a and DNMT1. Overexpression of G9a, UHRF1, or DNMT1 in UMR106 cells, respectively, can result in increased level of H3K9me2/me3 and 5-methylcytosine at lncH19 promoter. Finally, in vivo experiments showed that bone loss induced by HLU could be significantly alleviated by inhibition of DNMT1, G9a, and UHRF1, indicating a promising approach for bone loss treatment.
As a key regulator of osteoblast differentiation and osteogenic activity, lncH19 has a major role in the development of bone loss in HLU.(7, 8, 29) DNMT1-induced hypermethylation at lncH19 promoter is one of the main reasons for dysregulation of lncH19 expression in HLU osteoporosis.(8) In addition to DNA methylation, histone methylation is also a well-documented epigenetic marker,(9) especially the methylation of H3K27 and H3K9, which are mainly catalyzed by EZH2 and G9a, respectively.(30-32) Although several studies have reported an association between H3K27 methylation and osteogenic activity,(33, 34) our results showed no significant change in H3K27me2/3 levels in mechanical unloading rats. This may be because different disease models were employed.
Compared with H3K27, there have been few studies that have focused on the association between H3K9 methylation and osteogenic activity. It has been reported that removal of H3K9me3 at DLX promoter promotes osteogenic differentiation of human bone marrow mesenchymal stem cells, which indicates a potential role of H3K9me3 in the pathogenesis of skeletal diseases.(35) In this study, we found a significantly increased level of G9a and H3K9me2/3, and decreased expression of lncH19 in HLU rats, suggesting that G9a-H3K9 methylation-mediated lncH19 inhibition promoted bone loss induced by HLU. This was further confirmed by the experiments showing that alteration of G9a led to significant change in the expression of lncH19 and osteogenic activity. But, it has been previously reported that G9a could increase osteogenesis and intramembranous ossification.(36) The reason for the different effects of G9a on osteogenic activity remains obscure. Depletion of G9a in breast cancer cells could result in both positive and negative effects on steroid hormone-regulated gene expression.(37) Therefore, the mechanism of effects of G9a on osteogenic activity needs to be further explored in the future.
UHRF1 is a nuclear protein with multiple functions, which possesses both hemi-mCpG and methylated histone-specific binding activity.(38, 39) It has been reported that UHRF1 can bind to DNMT1 and H3K9 methylation, demonstrating that UHRF1 functions as a bridge between DNA and histone methylation.(40, 41) In the present study, we observed upregulation of UHRF1 in HLU rats, indicating the potential functional cross-talk between DNA and histone methylation in bone loss induced by mechanical unloading. Our results also indicated that overexpression of DNMT1, G9a, or UHRF1, respectively, resulted in increased level of H3K9 methylation and 5-methylcytosine at lncH19 promoter. Importantly, DNMT1/G9a/UHRF1 complex was identified as novel lncH19 promoter-interacting proteins. Knockdown or overexpression of lncH19 can cancel or rescue the effects of DNMT1, G9a, and UHRF1 on osteogenic gene expression and mineralization ability. Collectively, these results demonstrated for the first time that DNMT1-UHRF1-G9a-mediated functional cross-talk between H3K9 and DNA methylation plays a vital role in the bone loss induced by mechanical unloading through regulating lncH19.
Because H3K9 and DNA methylation functionally cooperate in the pathogenesis of bone loss induced by HLU, they could be promising therapeutic targets. Actually, modifying of DNMTs and/or histone methylation are widely used therapeutic approaches for a variety of human diseases.(22, 42, 43) Several studies have reported beneficial effects of histone deacetylase inhibitors on osteoporosis, by interfering with osteoblast/osteoclast-associated signaling pathways.(44, 45) Our experiments demonstrated that simultaneous inhibition of G9a, DNMT1, and UHRF1 is an effective way to resist bone loss induced by mechanical unloading. Such a multi-targeting strategy is more efficient and flexible compared with single targeting.(46, 47) However, combination interference of multiple genes requires precise targeting; otherwise, it may result in off-target synergistic cytotoxicity.(48) Therefore, further research should focus on the development of safer and more efficient gene therapy methods.
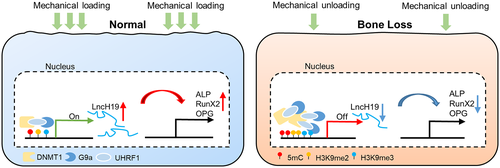
In summary, the current study demonstrated that H3K9 and DNA methylation cross-talk-mediated lncH19 expression plays a critical role in the bone loss induced by HLU (Fig. 8). Mechanical unloading led to upregulation of G9a, DNMT1, and UHRF1, which resulted in increased level of H3K9 and DNA methylation and robust functional cross-talk. Ultimately, the expression of lncH19 and osteogenic activity were inhibited in osteoblasts. Combined targeting of G9a, DNMT1, and UHRF1 presented potent therapeutic efficacy against bone loss induced by HLU, indicating a promising new approach for the treatment of mechanical unloading-associated osteoporosis.
Disclosures
All authors state that they have no conflicts of interest.
Acknowledgments
This work was supported by Natural Science Foundation of Tianjin (17JCQNJC11800, 20JCQNJC01170); Foundation of Tianjin Health Commission (ZC20192); Foundation of the Tianjin Health Bureau (14KG123); Tianjin Biomedical Engineering Technology Key Project (18ZXSGSY00010); and National Natural Science Foundation of China (81871777, 81871782).
Authors’ roles: Study design: LJ, WY, and MX. Study conduct: LB, ZJ, ZC, WW, LS, and LG. Data collection: MJ, ZY, and XG. Data analysis: LB, CW, and ZJ. Drafting manuscript: LB, MJ, and ZJ. Revising manuscript content: MJ and LB. Approving final version of the manuscript: all. LJ, WY, and MX are responsible for the integrity of the data analysis.
Author contributions: LB: Conceptualization; methodology; writing-original draft. ZJ: Conceptualization; methodology; writing-original draft. MJ: Software. CW: Investigation; methodology; project administration. ZC: Project administration. WW: Visualization. LS: Project administration. LG: Methodology; software; visualization. XG: Software. ZY: Resources. LJ: Conceptualization; data curation; formal analysis; funding acquisition. WY: Conceptualization; data curation; formal analysis; funding acquisition. MX: Conceptualization; data curation; formal analysis; funding acquisition.




