A CTGF-RUNX2-RANKL Axis in Breast and Prostate Cancer Cells Promotes Tumor Progression in Bone
ABSTRACT
Metastasis to bone is a frequent occurrence in patients with breast and prostate cancers and inevitably threatens the patient's quality of life and survival. Identification of cancer-derived mediators of bone metastasis and osteolysis may lead to novel therapeutic strategies. In this study, we established highly bone-metastatic PC3 prostate and MDA-MB-231 (MDA) breast cancer cell sublines by in vivo selection in mice. In bone-metastatic cancer cells, the expression and secretion of connective tissue growth factor (CTGF) were highly upregulated. CTGF knockdown in bone-metastatic cells decreased invasion activity and MMP expression. RUNX2 overexpression in the CTGF knockdown cells restored the invasion activity and MMP expression. In addition, CTGF increased RUNX2 protein stability by inducing its acetylation via p300 acetyl transferase. The integrin αvβ3 receptor mediated these effects of CTGF. Furthermore, CTGF promoted RUNX2 recruitment to the RANKL promoter, resulting in increased RANKL production from the tumor cells and subsequent stimulation of osteoclastogenesis from precursor cells. In addition, animal model with injection of CTGF knocked-down prostate cancer cells into 6-week old BALB/c male mice showed reduced osteolytic lesions. More importantly, the expression levels of CTGF and RANKL showed a strong positive correlation in human primary breast tumor tissues and were higher in bone metastases than in other site metastases. These findings indicate that CTGF plays crucial roles for osteolytic bone metastasis both by enhancing invasiveness of tumor cells and by producing RANKL for osteoclastogenesis. Targeting CTGF may lead to the development of effective preventive and therapeutic strategies for osteolytic metastasis. © 2019 American Society for Bone and Mineral Research.
Introduction
Breast and prostate cancers have the common feature of a high frequency of metastasis to the skeleton, which is accompanied by pathologic bone resorption.1 Molecular insights into the mechanisms mediating the interplay between tumor and bone cells that lead to alterations in bone remodeling in tumor metastasis to bone have been the focus of recent studies. Once they reach the metastatic loci of bone the tumor cells cause modifications of the bone microenvironment in favor of their growth, including increased osteoclastogenesis and subsequent bone resorption. Secretion by tumor cells of soluble factors such as PTHrP that stimulate osteoblasts to upregulate expression of RANKL, the osteoclast differentiation factor, is one of the mechanisms that mediate the tumor-bone interplay.2 In turn, by degrading bone matrix, osteoclasts induce the release of latent growth factors like TGF-β to promote tumor growth, resulting in further secretion of tumor-derived factors. This vicious cycle between tumor cells and bone resorption3 has recently been proposed as a characteristic of the overt metastasis stage.1
Connective tissue growth factor (CTGF), also called CCN2, is a small secreted matrix protein that regulates various cellular functions including proliferation, adhesion, migration, and invasion.4, 5 The physiological roles of CTGF include skeletogenesis, angiogenesis, and injury repair.4, 6-9 Pathologically, CTGF has been linked to fibrotic diseases and malignancies.5 Elevated CTGF expression was detected in some types of cancers including breast and pancreatic cancers.5 In breast cancer, higher CTGF expression was reported to be associated with poor prognosis, lymph node involvement, and resistance to taxol treatment.10, 11 In a prostate carcinoma xenograft model, stromal overexpression of CTGF led to increased microvessel density and tumor growth.12 Of note, CTGF is known to be barely expressed in normal adult tissues except in the developmental stages.13 Although these studies suggest that CTGF might have important functions in modulating the bone microenvironment as well as tumor stroma, its roles in osteolysis and bone metastasis have not been investigated.
In this study, we found that CTGF was upregulated in highly bone-metastatic sublines of prostate and breast cancer cells. CTGF increased the invasion activity of these cancer cells via induction of matrix metalloproteinases (MMPs) and stimulated osteoclastogenesis by increasing RANKL secretion. Consistently, CTGF promoted osteolytic bone metastasis and tumor growth in vivo. In addition, there was a strong positive correlation between CTGF and RANKL in specimens from patients with invasive breast carcinoma. Our results indicate that CTGF is a critical mediator of osteolytic bone metastasis.
Materials and Methods
The Supplemental Methods provide more experimental methods in detail.
Establishment of bone-metastatic tumor cell lines
Highly bone-metastatic PC3 (mtPC3) and MDA-MB-231 (mtMDA) cell lines were established as described.14 A total of 1 × 105 cells of PC3 or MDA-MB-231 were injected into left ventricles of 8-week-old BALB/c-nude male or female mice. After 8 weeks, metastasized cells in femurs and tibias were flushed out from the bone marrow and expanded in culture dishes for 2 months. Cultured cancer cells were validated by detecting human leukocyte antigens by FACS analyses. The cultured cancers were re-injected into ventricles of mice for another round of selection. The bone-metastatic cancer cells collected after the second round of selection were named mtPC3 and mtMDA.
In vivo tumor inoculation experiment
Six-week old male BALB/c-nude mice (21 ± 1 g/mouse) were purchased from OrientBio (Sungnam, Korea) and randomly assigned to PBS (10 mice), mtPC3-C-sh (13 mice), and mtPC3-CTGF-sh (13 mice) groups. A total of 5 × 105 cells in 5 μL or equal volume of PBS was injected into left tibias of mice. To minimize physical bone destruction by needles, thin needles (27G) were used. Left tibias were collected 4 weeks after injection and subjected to micro–computed tomography (μCT) or decalcification for histologic analysis.
Human breast tumor sample analysis
The primary tumor tissue samples used for experiment in Fig. 7 A were collected from 18 breast carcinoma patients during surgery. Matched cancer tissue and adjacent normal tissue were isolated from the same patients. Total RNA prepared from the tissues was reverse transcribed into cDNA for real-time PCR analyses. For the experiments shown in Fig. 7 C, cDNA arrays prepared from pathologist-verified breast cancer tissues were purchased from OriGene (Rockville, MD, USA). qPCR plates were coated with cDNA and analyzed by real-time PCR using human CTGF primers.
Statistical analysis
The unpaired Student's t test was used to determine statistical differences between two groups. The statistical dependence between two variables in Fig. 7 A was assessed by a nonparametric Spearman's rank correlation coefficient method. Error bars represent SD of mean values. All p values are two-sided. A p value <0.05 was considered statistically significant.
Study approval
Mice were maintained under specific pathogen-free conditions and all animal experiments were approved by the Committee on the Care and Use of Animals in Research at Seoul National University. The study with human tumor tissue samples collected during surgery from breast carcinoma patients with informed consent was approved by the Institutional Review Board of College of Medicine, Seoul National University.
Results
Upregulation of CTGF in bone-metastatic breast and prostate cancer cells
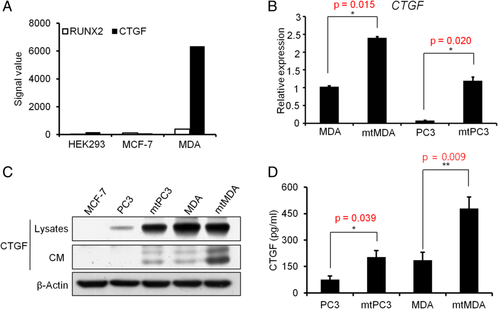
By cDNA microarray analysis we found that the CTGF gene expression signal was much higher in metastatic MDA-MB-231 (MDA) than in nonmetastatic MCF7 human breast cancer cell lines. The signal intensity of CTGF was 74.3-fold higher in MDA than in MCF7, and the RUNX2 signal was 3.45-fold higher (Fig. 1 A). To examine CTGF expression in bone metastases, we generated highly bone-metastatic breast (MDA) and prostate (PC3) cancer sublines (named mtMDA and mtPC3, respectively) by in vivo selection of bone-metastasized cells after intracardiac injection of parental cells. The mtMDA and mtPC3 cells expressed even higher levels of CTGF compared to the respective parental cells at both mRNA and protein levels (Fig. 1 B,C). ELISA with culture supernatants showed more CTGF secretion from mtPC3 and mtMDA than from the parental cell lines (Fig. 1 D). These results suggest that bone-metastatic breast and prostate cancers upregulate CTGF expression.
Role of CTGF in matrix invasion of bone-metastatic cancer cells
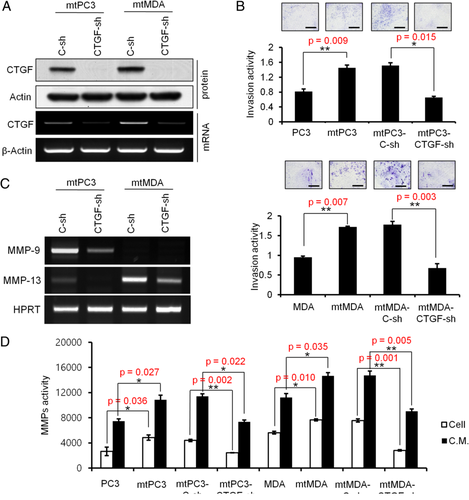
We next evaluated whether CTGF upregulation is associated with proliferation and invasion properties of cancer cells. CTGF-targeted shRNA (CTGF-sh) was used to reduce CTGF expression. Efficient reduction of CTGF expression at both mRNA and protein levels was achieved in both mtPC3 and mtMDA (Fig. 2 A). In addition, ELISA confirmed decreased secretion of CTGF from CTGF-sh cells in comparison with control cells (C-sh) (Supplemental Fig. 1A). To determine whether CTGF knockdown influences cell proliferation, we performed a BrdU assay. The proliferation rate was not significantly different between CTGF-sh and C-sh cells in both mtPC3 and mtMDA groups (Supplemental Fig. 1B). In matrix invasion assays, mtPC3 and mtMDA cells showed significantly stronger invasion activity than the parental cells (Fig. 2 B). Furthermore, introduction of CTGF-sh RNA reduced the invasion potency of both mtPC3 and mtMDA cells (Fig. 2 B). To gain insights into the molecular mechanism by which CTGF regulates matrix invasion, we examined the expression and activity of MMPs. High levels of MMP-9 mRNA were detected in mtPC3 cells and high levels of MMP13 mRNA were detected in mtMDA; expression of both MMPs was suppressed by CTGF knockdown (Fig. 2 C). In subsequent analyses of MMP activity, mtPC3 and mtMDA showed higher MMP activities than PC3 and MDA, respectively, in both cell lysates and CM (Fig. 2 D). More importantly, CTGF knockdown in mtPC3 and mtMDA reduced MMP activities (Fig. 2D). Collectively, these data show that increased MMPs expression could be one of the mechanisms for enhanced matrix invasion activity by CTGF in bone metastatic cancer cells.
Involvement of RUNX2 in CTGF regulation of matrix invasion
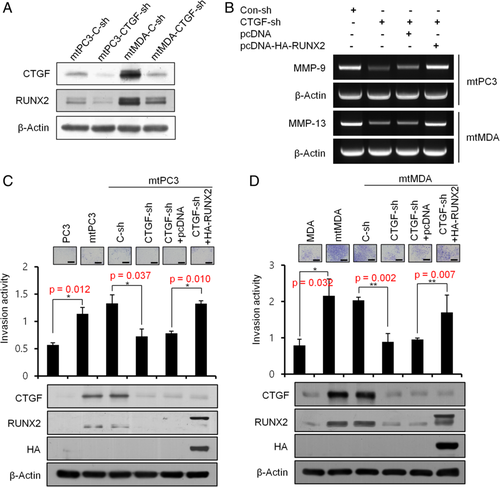
As the RUNX2 transcription factor has been suggested to play a role in the invasive properties of bone-metastatic breast and prostate cancers through regulation of MMP expression,15-18 we next investigated whether the CTGF-dependent augmentation of invasiveness of mtPC3 and mtMDA is mediated by RUNX2. First, we evaluated the effect of CTGF knockdown on RUNX2 expression. Real-time PCR analyses revealed no difference in RUNX2 mRNA levels between CTGF knockdown and control knockdown groups for both mtPC3 and mtMDA (Supplemental Fig. 2). In contrast, the level of RUNX2 protein was evidently decreased by CTGF knockdown (Fig. 3 A), indicating posttranscriptional regulation of RUNX2 by CTGF. Importantly, RUNX2 overexpression restored the MMP-9 and MMP-13 levels that were reduced by CTGF knockdown in mtPC3 cells and mtMDA cells, respectively (Fig. 3 B). Furthermore, forced expression of RUNX2 negated the reduction in invasion activity induced by CTGF knockdown in both mtPC3 and mtMDA cells (Fig. 3 C,D). These results indicate that CTGF increases matrix invasion activity of bone-metastatic PC3 and MDA cells by upregulation of RUNX2 protein levels followed by an increase in MMP activities.
Regulation of RUNX2 acetylation and stability by CTGF
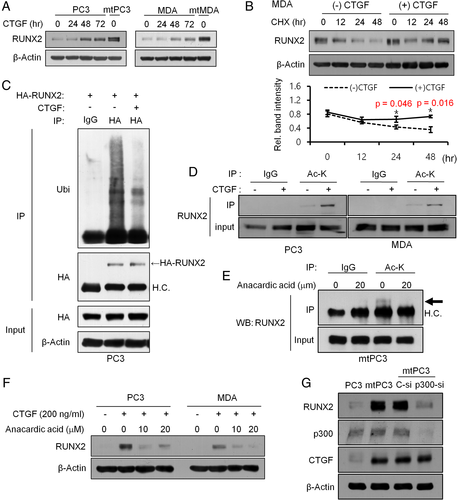
The decreased RUNX2 protein levels in CTGF-silenced mtPC3 and mtMDA cells (Fig. 3) led us to investigate the response of the parental cells to recombinant CTGF (rCTGF) treatment. RUNX2 protein levels increased after rCTGF treatment in both PC3 and MDA cells (Fig. 4 A). In agreement with the result of CTGF-sh experiments (Supplemental Fig. 2), RUNX2 mRNA levels did not change following rCTGF treatment (Supplemental Fig. 3A). Next, we assessed the effect of cycloheximide, a protein synthesis inhibitor, on RUNX2 protein levels. Cycloheximide decreased RUNX2 protein levels in a time-dependent manner in both PC3 and MDA in the absence of CTGF. The addition of rCTGF markedly attenuated this effect of cycloheximide on RUNX2 (Fig. 4 B and Supplemental Fig. 3B). These data suggest RUNX2 protein degradation in PC3 and MDA cells that could be blocked by CTGF. Previous studies in osteoblasts reported that RUNX2 was degraded after ubiquitination by Smurf119 and this degradation was inhibited by p300-dependent acetylation.20 Therefore, we examined the effect of rCTGF treatment on RUNX2 ubiquitination and acetylation in cancer cells. As shown in Fig. 4 C, rCTGF decreased the level of ubiquitinated RUNX2 in PC3 cells. In addition, rCTGF increased RUNX2 acetylation in PC3 and MDA cells (Fig. 4 D). Expression levels of p300 acetyl transferase were similar in these cancer cells and their mt sublines (Supplemental Fig. 4A). Subsequent co-immunoprecipitation analyses revealed an interaction between p300 and RUNX2 in PC3 and MDA cells, and this interaction remained unchanged by rCTGF treatment (Supplemental Fig. 4B). However, treatment with anacardic acid, a p300-specific inhibitor, reduced RUNX2 acetylation in mtPC3 cells (Fig. 4 E) and blocked CTGF-mediated RUNX2 stabilization in PC3 and MDA cells (Fig. 4 F). In addition, the increase in RUNX2 protein levels in mtPC3 was nullified by p300 knockdown (Fig. 4 G). Together, these data suggest that CTGF increases RUNX2 protein stability at least in part by promoting RUNX2 acetylation by p300.
Role of integrin αvβ3 in the regulation of RUNX2 by CTGF
Integrin αvβ3 was reported to function as one of the major receptors for CTGF in mediating migration and adhesion of endothelial and hepatic stellate cells.21-23 In addition, increased expression of integrin αvβ3 has been associated with the bone-invasive phenotype of cancer.24 Therefore, we investigated whether αvβ3 was involved in the CTGF regulation of RUNX2 in PC3 and MDA cells. We first examined integrin αvβ3 levels and showed that mRNA levels of both integrin αv and β3 were elevated in mtPC3 compared to PC3 (Fig. 5 A). The mRNA level of integrin β3 was also increased in mtMDA compared to MDA whereas αv mRNA levels were not different between mtMDA and MDA (Fig. 5 B). FACS analyses also indicated a higher surface expression of αvβ3 in mtMDA than in MDA cells (Supplemental Fig. 5). Next, we used an integrin αvβ3 neutralizing antibody (Ab) to block the receptor-ligand interaction. Neutralization of αvβ3 attenuated the CTGF-induced acetylation of RUNX2 in MDA and PC3 (Fig. 5 C,D). In addition, the enhancement of RUNX2 protein stability by CTGF was abrogated by treatment with anti-αvβ3 antibody (Fig. 5 E,F). These results suggest that integrin αvβ3 mediates CTGF-stimulated acetylation and stabilization of RUNX2 in highly bone-metastatic cancer cells.
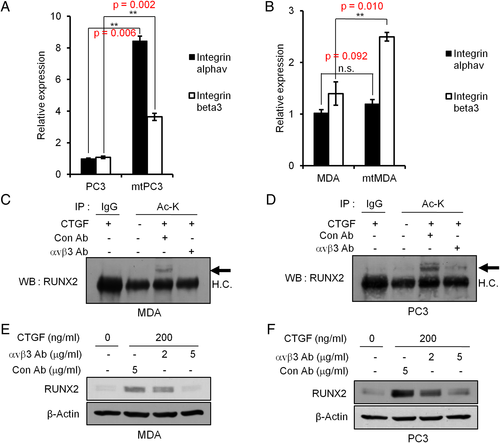
CTGF regulation of RANKL expression via RUNX2
As RUNX2 has been implicated in expression of the osteoclast differentiation factor RANKL in prostate cancer, vascular smooth muscle, and osteoblastic cells,25, 26 we next investigated whether CTGF could regulate RANKL expression in mtMDA and mtPC3 cells by increasing RUNX2 protein levels. As shown in Fig. 6 A, mRNA expression of RANKL was significantly higher in mtMDA and mtPC3 than in their parental cells. RANKL expression is also regulated by several transcription factors such as CREB or β-catenin.27 However, phosphorylation levels of CREB (Supplemental Fig. 6A) and nuclear localizations of beta-catenin (Supplemental Fig. 6B,C) were not altered by rCTGF. Next, to functionally validate the increased expression of RANKL, we obtained conditioned medium (CM) from cancer cells and examined its effects on osteoclastogenesis. Consistent with the increase in RANKL mRNA, CM from mtMDA and mtPC3 increased the number of osteoclasts generated from mouse preosteoclasts more potently than CM from MDA and PC3 cells (Fig. 6 B). The osteoclasts appeared bigger and more spread in cultures treated with mtMDA-CM and mtPC3-CM. We next searched for potential RUNX2 binding sites in the human RANKL promoter by in silico analyses. Based on the resulting information, we performed chromatin immunoprecipitation assays and found that recruitment of RUNX2 to the RANKL promoter was much higher in mtPC3 and mtMDA than in PC3 and MDA, respectively (Fig. 6 C). More important, CTGF knockdown decreased the accumulation of RUNX2 on the RANKL promoter in mtPC3 and mtMDA (Fig. 6 D). Furthermore, a reduction in CTGF led to a decrease in RANKL mRNA in these cells (Fig. 6 E). In line with this result, CM collected from CTGF-sh cells showed a reduced ability to promote osteoclastogenesis compared with CM from control C-sh cells (Fig. 6 F). Moreover, forced expression of RUNX2 in CTGF-sh mtMDA cells partially restored the osteoclastogenic potential (Fig. 6 G). Additionally, p300 knockdown or anacardic acid treatment decreased RANKL mRNA expression in mtMDA (Fig. 6 H). Taken together, these results suggest that CTGF induces RANKL expression in bone-metastatic cancer cells via p300-dependent stabilization of RUNX2.
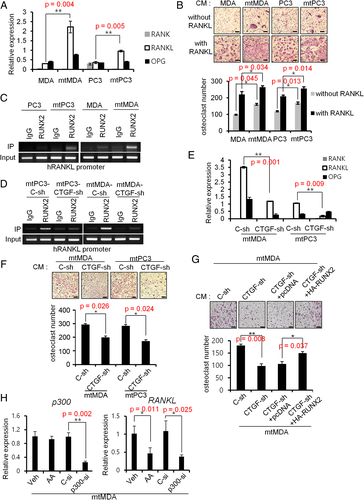
Role of CTGF in osteolysis and tumor growth in vivo
Next, we performed in vivo experiment by directly injecting mtPC3-C-sh cells into tibia. At 4 weeks after injection, tibias were analyzed. In μCT analyses, tibias with mtPC3-C-sh showed a mixed phenotype with osteosclerotic trabecular bone and osteolytic lesions in cortical area (Fig. 7 A,B,G, and Supplemental Fig. 7). These changes were barely observed in tibias with mtPC3-CTGF-sh. H&E staining of tibia sections showed less tumor area in mtPC3-CTGF-sh than in mtPC3-C-sh group (Fig. 7 C,D). Tumor-induced osteoclast numbers were also decreased by CTGF-sh (Fig. 7 E,F). Although tibias with mtPC3-CTGF-sh showed no mixed lesion, only three of 13 samples showed mild osteosclerosis (Fig. 7 A,C,G). Taken together, these results suggest that CTGF contributes to the stimulation of bone remodeling, bone lesion progression, and tumor growth of bone-metastasized prostate cancer cells.
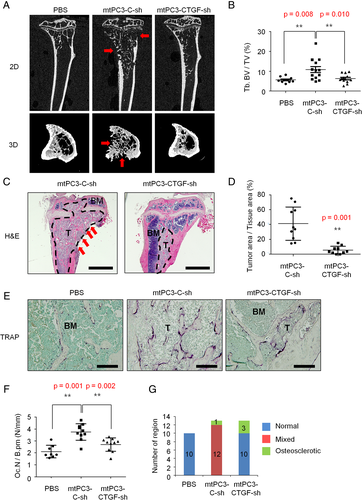
Correlation between CTGF and RANKL expression levels in human breast cancer tissues
To obtain more clinically relevant evidence for the role of CTGF in RANKL upregulation, we next evaluated the relationship between CTGF and RANKL expression levels in human invasive breast cancer tissues. Matched cancer tissues and adjacent normal tissues were obtained from 18 breast cancer patients of various stages (from 0 to IIIC) with estrogen receptor positive (ER+; n = 7) and ER− (n = 11). In real-time PCR analyses, normal tissues showed a moderate positive correlation between CTGF and RANKL (Spearman correlation coefficient = 0.540) whereas cancer tissues exhibited a very strong positive correlation (Spearman correlation coefficient = 0.814) (Fig. 8 A). Moreover, analyses of a public microarray data set (GSE14020) obtained from tissues of 65 breast cancer patients revealed significantly higher expressions of CTGF and RANKL in tumors that metastasized to bone than in those that metastasized to other sites (Fig. 8 B). Notably, RUNX2 mRNA expression signals were comparable among the different groups of metastasis sites (Fig. 8 B). In addition, quantitative PCR analyses of cDNA from a commercial source of breast cancer tissues with verified stages showed a trend for association of CTGF mRNA levels with cancer progression up to stage IIIA (Fig. 8 C). Taken together, these results indicate that CTGF may play a crucial role in bone metastasis and osteolysis in human breast cancer patients through induction of RANKL.
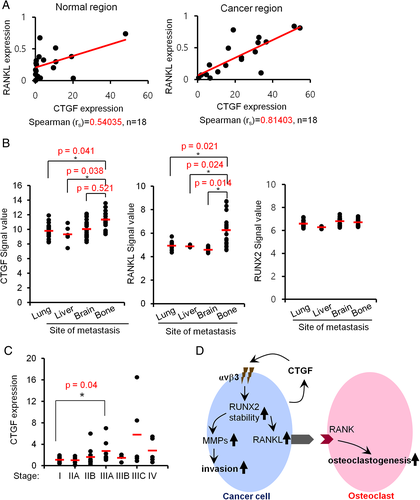
Discussion
In this study, we found that CTGF induces RANKL and MMPs via RUNX2 stabilization in bone-metastatic breast and prostate cancer cells (Fig. 8 D). The expression of CTGF was closely associated with increased matrix invasion and augmented osteoclastogenesis of these cancer cells. These invasion-promoting and osteoclastogenic effects implicate CTGF as one of the key players in making the microenvironment more susceptible to bone metastasis. Indeed, our findings suggest a close association of CTGF expression with bone metastasis of breast and potentially prostate cancers. We found that CTGF levels were elevated in highly bone metastatic mtMDA and mtPC3 cells (Fig. 1). The higher CTGF expression in metastasized tumor specimens from breast cancer patients with bone metastasis compared to those from patients with metastasis to other sites (Fig. 8 B) provides clinical relevance of CTGF expression for bone metastasis. In line with this notion, inhibition of bone metastasis of MDA cells by BMP9 was reported to be linked to CTGF downregulation.28 Another group also showed that osteolytic lesions elicited by intracardiac injection of MDA cells could be reduced by administration of a CTGF-neutralizing Ab.29 Nevertheless, there are few studies on the precise mechanisms of CTGF-mediated bone metastasis.
In this study, we for the first time showed that CTGF induces RANKL expression in bone-metastatic cancer cells (Fig. 6). In addition, we showed that RANKL and CTGF mRNA levels were more strongly positively correlated in tumor tissues than in non-tumor tissues of human breast cancer patients (Fig. 8 A). These results not only indicate the effect of CTGF on RANKL upregulation but also imply the presence of a currently undisclosed mechanism in tumor tissue that facilitates enhanced RANKL expression in the tumor context. Because RANKL induction is essential for osteolysis and tumor growth in the bone microenvironment, suppressing RANKL expression by restraining CTGF induction in cancer cells may lead to reduced metastatic bone lesions. Consistent with this hypothesis, knockdown of CTGF suppressed osteoclast numbers and tumor area in vivo (Fig. 7). However, CTGF also has direct impact on osteoclastogenesis by interacting with dendrocyte expressed seven transmembrane protein (DC-stamp).30, 31 Thus, the results of our in vivo study with CTGF knockdown may be attributed to two mechanisms at least, direct effect of CTGF on osteoclasts and indirect effect via promoting RANKL expression. However, our animal model imitated the post bone metastasis. Thus precise role of CTGF on cancer cell migration into bone needs to be elucidated. As suggested,32, 33 CTGF may also contribute to tumor metastasis by stimulating angiogenesis. However, our results provide strong evidence that bone-metastatic cancer cells can directly stimulate osteoclastogenesis by expressing RANKL in response to autocrine CTGF leading to promotion of tumor growth in bone.
We identified RUNX2 as a crucial mediator of RANKL induction by CTGF in mtMDA and mtPC3 cells. To the best of our knowledge, the regulation of RUNX2 by CTGF has not been previously reported. Our results indicate that CTGF stimulates RUNX2 acetylation and reduces ubiquitination-dependent degradation of RUNX2 in cancer cells. Enhanced RUNX2 stability leads to upregulated expression of MMPs and RANKL, which may allow invasive cancers to dominate the bone microenvironment. A previous study has shown that CTGF increased expression of MMPs by regulating of microRNA miR-519d.34 In this regard, it would be intriguing if RUNX2 stability is linked to miR-519d levels in CTGF-stimulated cells. In osteoblastic cells, the E3 ubiquitin ligase smurf1 was reported to mediate ubiquitination for RUNX2 degradation19 whereas p300 prevented smurf1-dependent degradation of RUNX2 by acetylation.20 Similar posttranslational regulation of RUNX2 in cancer cells and osteoblasts might be another molecular mechanism for the osteomimicry of bone metastases. In the present study, posttranslational upregulation of RUNX2 by CTGF was evident in both biochemical experiments (Fig. 4 B) and human cDNA microarray data (Fig. 8 B). In addition to upregulation of RANKL, RUNX2 also mediated the upregulation of MMP-9 and MMP-13 by CTGF (Fig. 3). The controlled degradation of the extracellular matrix by MMPs comprises a process that is critical for metastasis. In our human breast cancer tissue array analysis, there was a noticeable increase in CTGF expression from stage I to stage IIIA progression (Fig. 8 C). According to the definition of the TNM staging system, stage I delineates non-spread/small tumor size status and stage IIIA indicates the status of a tumor larger than 50 mm with internal lymph node metastasis.35 Therefore, it is probable that increased CTGF during the early stage of tumor progression promotes local invasion and metastasis by inducing MMP-9/13 expression via RUNX2 stabilization.
CCN family proteins, including CTGF, are thought to exert their biological activities through integrin.36 Recently, integrin αvβ3 upregulation in bone metastatic cancer was reported26 and, more important, exogenous expression of integrin αvβ3 was shown to be sufficient to promote bone metastasis.37 In our study, greatly increased expression of integrin αvβ3 in mtMDA and mtPC3 cells was responsible for RUNX2 acetylation and its subsequent stabilization by CTGF (Fig. 5). Analogous to our results, Tan and colleagues38 reported that CTGF-integrin αvβ3 signaling increased MMP-13 expression in human chondrosarcoma cells. Even though we showed that neutralizing antibody against integrin αvβ3 inhibited CTGF-induced Runx2 acetylation (Fig. 5 C,D), the possibility of additional mechanisms could not be ruled out. Several studies have reported that CTGF interacts with various integrins such as α2β1, α5β1, αvβ1, and αvβ6 in various normal cells.39-41 The interaction between CTGF and these integrins in cancer pathology needs to be elucidated. The CTGF-induced RUNX2 acetylation was due to stimulation of p300 activity rather than an increase in p300 association (Fig. 4 F and Supplemental Fig. 4B). Therefore, integrin αvβ3 signaling triggered by CTGF binding is transduced to regulation of p300 activity via a pathway that remains to be revealed. Of note, under the stimulation of thyroid hormone, internalization of integrin αvβ3 and subsequent complex formation between αv monomer and p300 has been reported.42 Although the detailed molecular mechanisms must be further elucidated, our findings delineate the relationship of integrin αvβ3, RUNX2, and p300 with tumor progression and metastatic bone diseases, and position CTGF as the tumor-derived element that triggers the αvβ3-p300-RUNX2 axis for RANKL induction in bone metastases.
In conclusion, our data indicate that bone-metastatic breast and prostate cancers express elevated levels of CTGF, which enhances RUNX2 stabilization via p300-mediated acetylation resulting in the induction of MMPs and RANKL. Subsequent matrix modulation and osteolysis may render the microenvironment favorable to cancer bone metastases. Therefore, we suggest CTGF as a promising target in the development of new therapeutics for bone-metastatic cancers.
Disclosures
All the authors declare that they have no conflict of interest concerning this paper.
Acknowledgments
This work was supported by grants from the National Research Foundation of Korea (NRF-2017R1A2A1A17069648 and NRF-2018R1A5A2024418 [to HHK] and NRF-2019R1A2C4070083 [to HJK]).
Authors' roles: HJK, AM, and HHK perceived the experiments. BK, HK, SJ, and HJK performed the experiments. HJK, HK, ZHL, and HHK analyzed the data. AM and ZHL provided expertise and feedback. DYN procured human breast tumor specimens. BK, HK, HJK, and HHK wrote the manuscript.




