Biallelic variants in WARS1 cause a highly variable neurodevelopmental syndrome and implicate a critical exon for normal auditory function
Sheng-Jia Lin and Barbara Vona should be considered joint first authors.
Abstract
Aminoacyl-tRNA synthetases (ARSs) are essential enzymes for faithful assignment of amino acids to their cognate tRNA. Variants in ARS genes are frequently associated with clinically heterogeneous phenotypes in humans and follow both autosomal dominant or recessive inheritance patterns in many instances. Variants in tryptophanyl-tRNA synthetase 1 (WARS1) cause autosomal dominantly inherited distal hereditary motor neuropathy and Charcot-Marie-Tooth disease. Presently, only one family with biallelic WARS1 variants has been described. We present three affected individuals from two families with biallelic variants (p.Met1? and p.(Asp419Asn)) in WARS1, showing varying severities of developmental delay and intellectual disability. Hearing impairment and microcephaly, as well as abnormalities of the brain, skeletal system, movement/gait, and behavior were variable features. Phenotyping of knocked down wars-1 in a Caenorhabditis elegans model showed depletion is associated with defects in germ cell development. A wars1 knockout vertebrate model recapitulates the human clinical phenotypes, confirms variant pathogenicity, and uncovers evidence implicating the p.Met1? variant as potentially impacting an exon critical for normal hearing. Together, our findings provide consolidating evidence for biallelic disruption of WARS1 as causal for an autosomal recessive neurodevelopmental syndrome and present a vertebrate model that recapitulates key phenotypes observed in patients.
1 INTRODUCTION
Aminoacyl-tRNA synthetases (ARSs) are essential enzymes that catalyze the aminoacylation of specific amino acids onto each cognate tRNA for protein synthesis. In humans, ARS genes encode 17 cytoplasmic, 17 mitochondrial, and three bifunctional enzymes. These evolutionarily conserved enzymes are required for protein synthesis involving all 20 amino acids in both the cytoplasm and mitochondria (Antonellis & Green, 2008). As protein structure and function depend on amino acid sequence, any compromise with respect to protein synthesis usually has drastic clinical consequences. The fidelity of translation largely relies on the selectivity and specificity of the recognition of both amino acid and tRNA by ARSs and are critical for maintaining protein synthesis integrity (Shen et al., 2006). Biallelic variants in cytoplasmic ARS genes cause various severe human diseases affecting a range of tissues (Meyer-Schuman & Antonellis, 2017). The increasing number of ARSs being implicated in human diseases further highlight their role in processes other than tRNA charging, such as regulation of transcription and translation, cell signaling, and immune response.
The human WARS1 gene (MIM# 191050, NM_004184.4) encodes a multi-domain cytoplasmic tryptophanyl-tRNA synthetase 1 that spans 471 amino acids. The domains encompass an N-terminal domain (amino acid residues 1-153) containing a WHEP helix-turn-helix motif (residues 8-64) that is named after the domain that is present in five ARSs: WARS1, HARS1, and EPRS1 (from which the acronym derives), GARS1 and MARS1), two catalytic domains (residues 154–362 and 453–471) and a C-terminal alpha-helical anticodon-binding domain (residues 363–452) (Shen et al., 2006). Pathogenic variants in WARS1 are most commonly associated with autosomal dominant distal hereditary motor neuropathy (dHMN) type IX (MIM# 617721) and Charcot-Marie-Tooth syndrome, with five heterozygous variants presently described in 15 patients (Li et al., 2019; Nam et al., 2022; Tsai et al., 2017; Wang et al., 2019). Recently, one family with compound heterozygous variants in WARS1 defined a novel neurogenetic syndrome comprised of microcephaly, intellectual disability, epilepsy, and delayed myelination (Okamoto et al., 2022). We present further molecular genetic evidence for biallelic WARS1 variants causing a clinically heterogeneous syndrome. To elucidate WARS1 function in vivo, we knocked down wars-1 in Caenorhabditis elegans using RNA interference (RNAi) and showed depletion in wars-1 is associated with a defect in cell division in the germline of C. elegans. Furthermore, we generated a zebrafish wars1 knockout using CRISPR/Cas9-mediated targeted mutagenesis. We showed that wars1 knockout (wars1−/−) recapitulates key clinical human phenotypes related to the brain, ear, and musculoskeletal system. In aggregate, we add to the limited data about biallelic variants in WARS1 and characterize an apparently clinically heterogeneous syndrome.
2 MATERIALS AND METHODS
2.1 Editorial policies and ethical considerations
The genetic and clinical data were obtained as part of regular clinical care and to confirm molecular genetic diagnoses. This study was approved by the Ethics Committees of the University of Würzburg (46/15) and Augusta University (624456-4). Written informed consent for genetic testing and publication of the clinical information, including clinical images, was obtained from the parents of each individual by the local clinicians/geneticists according to the Declaration of Helsinki.
2.2 Cohort
In this series, three affected individuals (Family 1 IV:3 and IV:4, Family 2 VI:1) with WARS1 biallelic variants were compared to the previously reported individuals retrieved from querying WARS1 in PubMed (https://pubmed.ncbi.nlm.nih.gov/). Phenotyping was carried out by local clinicians. All affected individuals underwent radiological examination. The proband in Family 1 (IV:3) underwent audiological assessment by pure-tone audiometry adhering to previously described recommendations (Mazzoli et al., 2003). Brain magnetic resonance imaging (MRI) and electroencephalogram (EEG) studies were performed on the probands from Families 1 and 2. Additionally, newborn hearing screening and an echocardiogram were completed in the proband from Family 2.
2.3 Exome sequencing
Genomic DNA (gDNA) from individuals in Families 1 (III:1, III:2, IV:3, IV:4) and 2 (V:1, V:2, and VI:1) was extracted from whole blood. The gDNA of the proband in Family 1 (IV:3) and a parent–child trio in Family 2 (V:1, V:2, VI:1) were exome sequenced. Exome library preparation using gDNA from the proband in Family 1 was performed with the Nextera Rapid Capture Exome (Illumina) according to the manufacturer's instructions and paired-end sequenced (2 × 76 bp) with the NextSeq 500 sequencer (Illumina). A v2 high output reagent kit (Illumina) was used, and the data were aligned to the human reference genome GRCh37 assembly. gDNA from the mother, father, and proband in Family 2 were subjected to exome sequencing using a VCRome version 2.1 capture method with paired-end sequencing (2 × 100 bp) on the HiSeq sequencer as previously described (Yang et al., 2014).
2.4 In silico variant analysis and variant prioritization
Data analysis from the proband in Family 1 was performed using Gensearch NGS software (PhenoSystems SA). The sequencing data alignment for the proband of Family 2 and variant calling were performed using the Edico Dragen BioIT Platform. Variant interpretation was performed by the Baylor Genetics analytics pipeline (Liu et al., 2019). Variant filtering was done using a minor allele frequency (MAF) <0.01 and alternate allele present at >20. Frequency data employed gnomAD (v2.1.1 and v3.1.2) (Liu et al., 2019), the Greater Middle Eastern (GME) Variome (Scott et al., 2016), and the Gene-Based Association Summary Statistics (Genebass) from the UK Biobank (Karczewski et al., 2021) resources. Pathogenicity prioritization was performed using CADD (Rentzsch et al., 2019), PolyPhen-2 (Adzhubei et al., 2010), SIFT (Ng & Henikoff, 2001), and MutationTaster (Schwarz et al., 2014). Multiple in silico splice prediction tools such as MaxEntScan (Yeo & Burge, 2004), NNSPLICE (Reese et al., 1997), Genesplicer (Pertea et al., 2001), and Human Splicing Finder (Desmet et al., 2009) were used to predict splicing effects and were determined through Alamut Visual software (Sophia Genetics). Public archives of clinically relevant variants were queried using ClinVar (Harrison et al., 2016) and the Human Gene Mutation Database (HGMD) (Stenson et al., 2020). Variants were classified according to the American College for Medical Genetics and Genomics (ACMG)/American College of Pathology (AMP) guidelines in accordance with the ClinGen Sequence Variant Interpretation recommendations (Richards et al., 2015). ACMG/AMP clinical significance was aided through using the public version of the Human Genomic Variant Search Engine (VarSome) (Kopanos et al., 2019). Mutalyzer 2.0.35 (Lefter et al., 2021) and VariantValidator (Freeman et al., 2018) were used to validate variant nomenclature according to Human Genome Variation Society (HGVS) recommendations.
2.5 Validation of the WARS1 variant and segregation testing
Validation of the WARS1 variant identified in Family 1 was carried out using Sanger sequencing from PCR-amplified gDNA from the available affected and unaffected family members using standard cycling conditions and primers (Supporting Information: Table S1) to validate the c.1A>G, p.Met1? variant. The variant in the parents and proband of Family 2 was visualized in Integrated Genomics Viewer (IGV) (https://software.broadinstitute.org/software/igv/home).
2.6 Prediction of translation initiation sites
To predict alternative in-frame translation initiation sites as a means to estimate the effect of the start loss variant (c.1A>G, p.Met1?), the tool Predict TIS (https://www.tispredictor.com/tis) was used (Gleason et al., 2021). This machine learning-based tool utilizes balanced training data with a well-suited machine learning algorithm for limited data, splits ATG and near-cognate translation initiation codon data to train separately, and reduces features to capture the most critical nucleotides for translation initiation. The wild-type messenger RNA (mRNA) sequence of WARS1 (NM_004184.4) was queried for ATG predictions with color-coded scoring using the Kozak similarity score. The Kozak similarity score code uses a scale from less than 0.5 to greater than 0.8 and is colored as follows: blue (scores <0.5), teal (≥0.5 and <0.6), green (≥0.6 and <0.7), orange (≥0.7 and <0.8), and red ≥0.8.
2.7 Visualization of single-cell RNA-seq data in the mouse
To assess transcript isoform structure and gene expression in the wild-type postnatal day (P) 15 mouse inner ear, 140 murine single-cell samples from three cochlear cell types (inner hair cells [IHCs], outer HCs [OHCs], and Deiters’ cells [DC]) were subjected to single-cell RNA-seq (scRNAseq). These data were made publicly available via the Molecular Otolaryngology and Renal Research Laboratories (MORL) scRNAseq transcript browser that was used to profile transcript structure (Ranum et al., 2019). To assess Wars1 RNA expression in the mouse cochlear epithelium at various stages (embryonic day [E] 14, and P1 and P7), publicly available single-cell RNA-seq data were visualized using the gene Expression Analysis Resource (gEAR) (Kolla et al., 2020; Orvis et al., 2021). Similarly, 10× Genomics RNA expression in the adult mouse whole cortex and hippocampus were visualized using the gene expression atlas provided by the Allen Institute for Brain Science (Harris et al., 2019; Lein et al., 2007; Oh et al., 2014; Yao et al., 2021).
2.8 C. elegans maintenance
C. elegans N2 Bristol strain was used in this study. Worms were maintained at 20°C on a Nematode Growth Medium (NGM) plate seeded with OP50 (Escherichia coli) bacteria.
2.9 Knocking down wars-1 using RNAi
The wars-1 gene was knocked down using the RNAi clone from the Ahringer C. elegans RNAi library (Source Bioscience) (Fraser et al., 2000; Kamath et al., 2003). Briefly, wars-1 and the control RNAi expressing bacteria (HT115) containing an empty L4440 vector were grown overnight in 5 ml LB broth media (L3022; Sigma-Aldrich) supplemented with 100 mg/ml ampicillin (BP1760-25; Fisher Scientific) and 10 mg/ml tetracycline (J61714, Alfa Aesar; Thermo Fisher Scientific). Bacteria were harvested with centrifugation and regrown until they reached OD (optical density) of 0.6–0.8 in 15 ml LB supplemented with 100 mg/ml ampicillin at 37°C, shaking at 210 rpm. RNAi expressing bacteria were seeded on the NGM plates containing 1mM IPTG (isopropyl-beta-d-thiogalactopyranoside, R0392; Thermo Scientific™). Larva (L1) stage worms were transferred to the corresponding RNAi plates and were analyzed after 72 h.
2.10 Progeny assay
Worms were treated with the RNAi against wars-1 from the L1 stage to adulthood. Each adult worm (P0) was transferred to a new plate every 24 h. The first transfer was counted as a Day 1 transfer, and the P0 worms were transferred to a new NGM plate until they no longer laid embryos. The total number of hatched progenies was counted for individual worms, and the data was summed by counting all the embryos from each replicate plate. The Student's t test was performed on replicates to calculate the significance.
2.11 Germline extraction and DNA staining
Adult worms were transferred to the unseeded plates, letting them crawl for 1–2 min to eliminate the bacteria attached to their bodies. Eight microliters of dissection buffer (0.2 mM Levamisole, 0.2% Tween-20 dissolved in egg buffer) were added to a 22 × 22 mm coverslip, and 8–10 worms were placed into the solution. Worms were dissected using a 23 Gauge syringe needle. Germlines were then fixed using fixation buffer (4% formaldehyde, 0.2 mM Tween-20 dissolved in egg buffer) and gently placed on a poly-l-lysine coated slide. The slide was incubated for 5 min at room temperature and then flash-frozen in liquid nitrogen for a few minutes. Slides were then removed from liquid nitrogen and freeze-cracked by removing the coverslip. 0.5 μg/ml diamidino-2-phenylindole (DAPI) stain (62248; Thermo Scientific™) was used to stain DNA. The slides were mounted using an antifade mounting media (Vectashield, H-1000; Vector Laboratories).
2.12 Microscopy and image analysis
NGM plates containing worms were screened with a progeny assay under a Leica stereomicroscope. A Leica Thunder Imager microscope was used to visualize DAPI-stained germ cells using ×40 or ×63 objectives by capturing z stacks (each ~0.3 μm) covering the whole diameter of the germline. Mitotic cells were counted using ImageJ software (NIH) as follows: The images were imported into the software, the scale was set, and the mitotic zone as the region of interest was selected. The border of the mitotic zone was determined by identifying cells entering the transition zone, in which their chromatin exhibits a crescent shape visualized by DAPI staining. Next, the image was converted to 8-bit while preserving its scale. The threshold was adjusted to get the best color in which the cell shapes were obvious. Then using the “Analyze Particles” option, the number of cells in each stack were counted. Since the diameter of the germline in worms treated with control RNAi is more than the diameter of one layer of mitotic cells, the number of cells from three independent stacks were summed up to calculate the number of mitotic cells.
2.13 Protein structure analysis
The 3D protein structure of WARS1 was initially obtained using the iTASSER (IterativeThreading ASSEmbly Refinement) server (Yang et al., 2015). The further 3D conformational similarity of human and C. elegans WARS1 protein was visualized using the PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC. (https://pymol.org/2/). The individual rainbow ribbon presentation of the human and C. elegans WARS1 was displayed by selecting the “C” (color) from the command buttons, followed by selecting “spectrum” that colors the N-terminus blue and the C-terminus red. The overlay of the two corresponding WARS1 structures was visualized by color, that is, human in cyan and C. elegans in tints. Then both structures were opened in the same PyMOL session, followed by selecting “A” (align) > “all to this” from the command section.
2.14 Ethics statement and zebrafish care
Zebrafish (Danio rerio) were raised and maintained in an AALAC accredited facility at the Oklahoma Medical Research Foundation (OMRF) under standard conditions, and all experiments were performed as per protocol 20-03 approved by the OMRF Institutional Animal Care Committee (IACUC). All zebrafish handling, embryo care, and microinjections were performed according to procedures described in the zebrafish book (Westerfield, 1993). All zebrafish experiments were performed either using wild-type zebrafish strain, NHGRI-1, or transgenic line Tg(pou4f3:GAP-GFP) (Xiao et al., 2005).
2.15 Sequence comparisons
The GenBank accession numbers of the compared proteins are listed as follows: human WARS1 (NP_776049.1), mouse WARS1 (NP_035840.3), zebrafish Wars1 (NP_957066.1), worm Wars1 (NP_507683.2), yeast Wars1 (NP_014544.1). Amino acid sequences were aligned and analyzed using Clustal W and JalView.
2.16 Whole-mount in situ hybridization
Whole-mount in situ hybridization was performed using digoxigenin-labeled antisense probes, as described earlier (Thisse & Thisse, 2008). A wars1 antisense probe was synthesized from a polymerase chain reaction (PCR) amplified template from zebrafish complementary DNA (cDNA) harboring the T7 promoter sequences at the 3′-end. The primer sequences are listed in Supporting Information: Table S1. The T7 polymerase was used to synthesize the RNA.
2.17 Zebrafish wars1 knockouts by CRISPR/Cas9 genome editing
Four target sites were identified and the guide RNAs were designed using the CRISPOR tool (Concordet & Haeussler, 2018). The single guide RNAs (sgRNAs) were synthesized as described previously (Varshney et al., 2016). A mixture containing 1 μl of 40 μM Cas9-NLS protein (UC Berkeley QB3 Macrolab), 500 ng of each sgRNA (in 3 μl), and 2 μl of 1 M potassium chloride was injected into one-cell-stage wild-type NHGRI-1 genetic background or transgenic line Tg(pou4f3:GAP-GFP) (Xiao et al., 2005) embryos as described previously (LaFave et al., 2014; Lin et al., 2021).
2.18 Site-directed mutagenesis and cloning
The full-length human cDNA clone was purchased from Horizon discovery (MHS6278-202828896), PCR amplified with zebrafish Kozak sequence at 5′, and cloned into the pCS2+ vector (a kind gift by Dr. David L. Turner, University of Michigan). Zebrafish cDNA was amplified from mRNA extracted from zebrafish larvae of different stages, and cloned into the pCS2+ vector. Patient-specific variants were generated by site-directed mutagenesis using Quick change II site-directed mutagenesis (Agilent). Primers are listed in Supporting Information: Table S1. All plasmids were sequence-verified end-to-end (Primordium Labs).
2.19 Morphological phenotyping, histology, and whole-mount immunohistochemistry
Phenotype analysis was performed at 3, 5, and 8 days postfertilization (dpf) stages. The embryos/larvae were placed either in an agarose mold or 2% methylcellulose (Sigma-Aldrich), and images were taken using an Olympus SZX12 stereomicroscope. The head and eye sizes were measured directly from scale-calibrated images using ImageJ software and the size measurements were calculated in percentage of mean value of uninjected embryos. HC stereocilia of the inner ear and muscle fibers were stained for F-actin with fluorescently labeled (FITC) phalloidin dye at 5 and 8 dpf, respectively, as described earlier (Lin et al., 2021). Whole-mount FM1-43 (Invitrogen) staining was used to investigate the function of HCs in sensory lateral line neuromasts. To visualize cartilage development, Alcian blue staining was performed in zebrafish larvae at 8 dpf according to methods described previously (Walker & Kimmel, 2007).
3 RESULTS
3.1 Clinical reports
Detailed clinical and variant summaries of all previously published families and the three affected individuals that we present are shown in Supporting Information: Tables S2 and S3, respectively. Both families described below derived from the Pakistani population.
3.2 Family 1, individuals 1 and 2
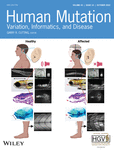
Family 1 presents two affected individuals with parental consanguinity and otherwise unremarkable family history (Figure 1a, Table 1). Both parents are physically active and mentally healthy. They do not have any auditory, speech, or mobility issues. The male proband (IV:3, individual 1, arrow) was 20 years of age at last evaluation. He had short stature (156.9 cm, −2.7 SD for age, <1st centile) and mild microcephaly with a head circumference of 52 cm (-2.1 SD for age but above 2nd centile for height). He showed a triangular face with micrognathia, abundant hair, eyebrows, and mustache. His fingers showed small, round fingernails (Figure 1c). His walking and speech development were delayed, and he is illiterate and does not know how to count. He can feed himself, is toilet trained, and can complete tasks such as cleaning and getting dressed and is sexually healthy. He has a mild loss of the sense of smell. Brain MRI and EEG were normal. X-rays showed mild medullary widening and minimal medial bowing of the mid/distal fibula shafts. Spine was normal. Pure-tone audiometry at 19 years of age revealed mild sensorineural hearing loss affecting both ears, symmetrically (Figure 1d).
| Family 1 IV:3/IV:4 | Family 2 VI:1 | |
|---|---|---|
| Molecular genetics summary | ||
| Genomic position (GRCh38) | Chr14:100369185T>C | Chr14:100335036C>T |
| Genomic position (GRCh37) | Chr14:100835522T>C | Chr14:100801373C>T |
| WARS1 c. position | c.1A>G | c.1255G>A |
| WARS1 p. position | p.Met1? | p. (Asp419Asn) |
| CADD_Phred | 0.003 | 25.4 |
| PolyPhen-2 | No entry | Benign |
| SIFT | No entry | Tolerated |
| MutationTaster | Disease causing | Disease causing |
| ACMG/AMP classification including functional study criteria at supporting level | Likely pathogenic (PVS1, PM2, PM3, PP1, PS3) | VUS, leaning likely pathogenic (PM2, PM3, PP3, PS3) |
| gnomAD v.3.1.2 | 0 | 0.00009859 |
| gnomAD v.2.1.1 | 0 | 0.0003899 |
| GME Variome | 0 | 0.0020597 |
| UK Biobank | 0 | 0.00004465 |
| Clinical summary | ||
| Developmental delay | +/+ | + |
| Microcephaly | +/+ | ? |
| Seizures | −/+ | + |
| Intellectual disability | +/+ | + |
| Spasticity | +/+ | ? |
| Hypotonia | −/+ | + |
| Deep tendon reflexes | −/+ | − |
| Walking abnormalities/abnormal gait | −/+ | + |
| Behavioral abnormalities | +/+ | − |
| Hearing loss | +/+ | − |
| Cortical visual impairment | +/+ | + |
| Abnormal brain MRI | −/+ | + |
| Abnormal X-ray | +/+ | + |
- Note: Transcript NM_004184.4, “−” means normal, “+” means abnormal, “?” means uncertain.
- Abbreviation: MRI, magnetic resonance imaging.
His sister (IV:4, individual 2) was 16 years old at last evaluation. She had a history of severe delayed developmental milestones (including delayed breast development), intellectual disability, hypotonia with contracted hands that interfere with grasping or holding objects, anosmia, short stature (135 cm, −4.2 SD, below 1st centile), microcephaly (head circumference, 46 cm, −7.8 SD, below 1st centile), large earlobes with apparent bifid tragus and micrognathia. Her hands show clinodactyly of the fifth finger, variation of her deep palmar flexion creases, and presence of bilateral hypothenar creases (Figure 1c). She cannot walk, speak, feed herself, read, or count. She has severe hearing impairment, although her level of cooperation made it impossible to quantify using pure-tone audiometry. Radiological studies showed pelvic coxa valga deformity of the hips with relative overgrowth of the lesser trochanters, with the left side more affected than the right. Most striking are her legs. On the right, there is medial bowing of the mid tibia and fibula with valgus positioning of the ankle with respect to the knee and overgrowth of the distal fibula (lateral malleolus) and, to a lesser degree, the medial malleolus. On the left, there is no significant tibial bowing but similar to the contralateral side, there is significant medial bowing of the mid-fibula, which has a long and gracile appearance and relative overgrowth of the medial and lateral malleolus. Spine was normal in anterior-posterior views. She has dental problems and other issues due to a poor environment.
Both individuals did not have any signs of neuropathy, as they were not sensing prickling sensations or numbness and tingling. Both were congenitally symptomatic and do not have high arches or hammer toes. Therefore, distal hereditary neuropathy and Charcot-Marie-Tooth syndrome were not suspected in light of absent electromyography (EMG) studies.
3.3 Family 2, individual 3
Family 2 shows evidence of multiple consanguineous marriages but an otherwise unremarkable family history apart from the proband (VI:1, individual 3, arrow) (Figure 1a, Table 1). Both parents are healthy. The proband is a 5 year, 2-month-old male with a history of global developmental delay, complex partial epilepsy, central adrenal insufficiency, cortical blindness, and multiple brain abnormalities. Ventriculomegaly and agenesis of the corpus callosum were diagnosed prenatally. He was born at 37 weeks gestation and weighed 2600 g, which was noted to be small for gestational age and was, therefore, monitored in the neonatal intensive care unit for 1 day. At discharge, he passed newborn hearing screening. At 6 days of life, he was hospitalized for hypothermia, hypoglycemia, hypernatremia, and lethargy. At 11 days old, there was a suboptimal response to adrenocorticotropic hormone (ACTH) stimulation and low random cortisol, raising concerns for central adrenal insufficiency. His adrenal insufficiency resolved after passing the ACTH stimulation test (cosyntropin) at 2 years old and he was successfully weaned off hydrocortisone. At 21 months of age, extensive metabolic work-up was unremarkable. A neonatal MRI demonstrated agenesis of the corpus callosum with hypoplastic pons, cerebellar vermis, and cerebellar hemispheres.
Due to the prenatal and neonatal history, he was followed closely for development, and early interventions were applied in infancy. At 12 months old, a formal evaluation was performed, and he was noted to be at an approximately 4-month-old skill level. At 2 years old, he gained head control, could roll to his side, sit with support for approximately 15 min, and babble. He has persistent global delays and can now bear weight on his legs, respond to his name, track, and make vocalizations. He has a history of aspiration with all consistencies and, at 2.5 years, received G-tube placement with Nissen fundoplication.
Notable for the patient is his intractable Lennox-Gastaut syndrome, with a history of prior intractable focal epilepsy and infantile spasms. For the past 4 years, his EEGs have had temporal seizures and interictal frontotemporal and temporooccipital discharges (left>right) and one EEG showed greater right occipital discharges. At 2 years 2 months, his EEG findings evolved to Lennox-Gastaut syndrome. His most recent EEG, at 4.5 years old, was extremely abnormal, as the background rhythm was disorganized and slow with very frequent epileptiform discharges. There were multifocal and generalized discharges, as well as near-continuous left posterior discharges. Several seizures were noted that had a tonic EEG signature (slowing and then decrement with fast activity) with some events noted with staring and otherwise no clear clinical correlate. None of these were patient push-button events and appeared generalized in nature. The EEG findings were consistent with epileptic encephalopathy with increased abnormality in the left posterior region. The proband's most recent brain MRI was performed at 3.5 years old and showed hypogenesis/dysgenesis of the corpus callosum. Bilateral hippocampi are small and dysplastic, without evidence of mesial temporal sclerosis. The bilateral occipital lobe is abnormal, right more than left, with likely malformation of cortical development. Despite long-term neurology follow-up, EMG studies were not performed due to a lack of neuropathy signs.
Osteopenia was noted at approximately 22 months old. He has a history of re-fracture of the femur and low bone mineral density for chronologic age. At 4.5 years old, an echocardiogram was completed and normal.
A previous dysmorphology exam at 21 months old noted microcephaly at less than the 3rd percentile. He has neutral palpebral fissures, low-set and posteriorly rotated ears with peaked helices, and notching at the peak. The pinnae had subtle creases bilaterally. A low hair line and hirsutism were observed, particularly over the sacrum and shoulders. His fingers were tapered. On neurology exam, hypotonia was noted throughout.
His most recent measurements include a weight of 27.6 kg (99th centile) and a height of 111 cm (61st centile). Head circumference has not been measured since age 3 years, when it measured 48 cm (13.6th centile), so microcephaly cannot be followed. He has normal male genitalia with bilaterally descended testes.
3.4 Molecular genetics results
Exome sequencing of the proband in Family 1 uncovered a homozygous start loss variant (c.1A>G, p.Met1?) in exon 2 of WARS1 that was classified as likely pathogenic (Table 1, Supporting Information: Table S3). This variant was absent from both versions of gnomAD, GME Variome and the UK Biobank. It was predicted to be disease causing by MutationTaster. As parental consanguinity was known, analysis of homozygosity flanking the variants confirmed the variant to reside in a roughly 8 Mb run of homozygosity (NC_000014.8:g.93,159,453-101,184,139; WARS1 variant coordinate: NC_000014.8:g.100,835,522T>C). Sanger sequencing confirmed the segregation of the homozygous variant in both affected individuals and identified their parents as carriers (Supporting Information: Figure S1).
Chromosomal microarray of the proband in Family 2 was unremarkable but showed 22.8% autozygosity. Therefore, exome sequencing was recommended. Parent-child trio exome sequencing of Family 2 was performed when the proband was 21 months old. No pathogenic variants in disease-associated genes related to the clinical phenotype were initially reported. A homozygous c.1255G>A, p.(Asp419Asn) variant was identified in WARS1, with a visual inspection of the variant in his parents showing carrier status (Supporting Information: Figure S1, Supporting Information: Table S3). This variant is present in both versions of gnomAD, GME Variome, and UK Biobank with a maximum minor allele frequency in the Middle Eastern and South Asian subpopulations, each around 0.003, but it was never observed in a homozygous state.
Additional variants that were identified in both probands are included in Supporting Information: Table S4. Candidate variants in genes associated with hearing impairment were excluded.
3.5 Prediction of translation initiation sites using Kozak similarity scoring
As Wars1 knockout mice experience preweaning lethality (International Mouse Phenotyping Consortium Warstm1b(EUCOMM)Hmgu allele) (Brown, 2021), it is hypothesized that instead of a complete loss of WARS1 expression in individuals 1 and 2, that an alternative translation initiation site is used. These isoforms could be the consequence of two possibilities: First, complete loss of the long isoform could result in exclusive use of two shorter human isoforms (NM_213645 and NM_213646) that both lack partial N-terminal and WHEP domains or, second, use of an alternative Kozak consensus sequence. By querying the mRNA sequence of human WARS1, 21 predicted Kozak consensus sequences were identified (Supporting Information: Figure S2a). The closest in-frame predicted Kozak consensus sequence to the original start codon (position 139 with a score of 0.77 in Supporting Information: Figure S2) can be seen at position 262 in exon 3 (with a score of 0.7), corresponding to c.124. If this start codon would be used, a deletion spanning all of exon 2 and part of exon 3 would result (NC_000014.8:g.100828235_100835522del, c.4_126del, p.(Pro2_Met42del)) (Supporting Information: Figure S2b) based on this prediction. This would truncate the WHEP helix-turn-helix motif that resides at amino acid positions 8 to 64 (Figure 1b). From in silico prediction and comparison with other human isoforms, this is equivalent to use of the two shorter human isoforms.
3.6 Expression of Wars1 in the mouse inner ear and brain
Single-cell RNAseq in the E14, P1, and P7 mouse cochlear epithelium showed diffuse expression across nearly all cell types (Supporting Information: Figure S3a). Similarly, nearly ubiquitious expression is observed in the mouse whole cortex and hippocampus (Supporting Information: Figure S3b). Analysis of transcript expression in the P15 mouse inner ear using MORL scRNAseq showed the abundant use of the long Wars1 isoforms (Wars-201, Wars-205) in the DCs, IHCs, and OHCs that all use the same ATG start codon and both encode proteins with 475 amino acid residues. These are the longest Wars1 isoforms in the mouse, corresponding to the longest isoform in humans (NM_004184.4, encoding 471 amino acids) (Supporting Information: Figure S4).
3.7 Defective mitotic cell cycle in C. elegans germline upon wars-1 depletion
WARS-1 is highly conserved from human to C. elegans in its protein sequence and three-dimensional (3D) structure (Supporting Information: Figure S5a-c). Using PyMol, we predicted the 3D structures of human and C. elegans WARS1 protein (Supporting Information: Figure S5a-c). The overall 3D structures of both proteins are highly similar, as seen in their overlay.
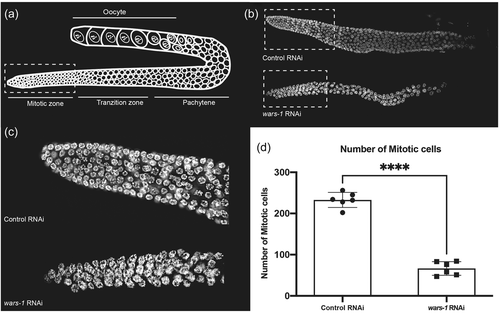
To elucidate the function of WARS-1 protein in vivo, we knocked down wars-1 using RNAi in L1 stage developing C. elegans. Knocking down wars-1 did not lead to a developmental arrest or developmental abnormalities, and affected worms reached adulthood with an aberrant gross phenotype. However, depletion of WARS-1 resulted in a 100% sterility suggesting a defect in germline proliferation (Supporting Information: Figure S5d,e). C. elegans germline is the only mitotically active tissue in an adult worm. Like many other organisms, the C. elegans germline is highly responsive to physiological and pathological changes. Germ cells are oriented from the distal to the proximal end of the U-shaped germline in C. elegans. The proliferative region (mitotic zone) is located at the distal end of the germline, where most of the proliferative cells are at the G1, S, or G2 stages of the mitotic cell cycle, with few in the M phase, undergoing active mitosis (Hubbard, 2007). In wild-type C. elegans, the mitotic zone contains ~250 germ cells. From the proliferative zone, germ cells enter the early stages of meiotic cell division (early prophase of meiosis) at the transition zone. During meiosis, germ nuclei have high nuclear and chromatin reorganization, such that chromatin and chromosome staining can be used to determine the exact stages of the cell cycle, such as pachytene, diplotene, and diakinesis that are located at the most proximal region of the germline (Figure 2a). To understand the effect of wars-1 knockdown on the sterility of C. elegans, we isolated the germline to stain the germ cells’ chromatin using DAPI. The germline of wars-1 RNAi worms was dramatically smaller than the control RNAi with an apparent reduction in the proliferative, mitotically active region (Figure 2b,c). We, therefore, counted the number of mitotic cells in the proliferative zone of the wars-1 RNAi and found that the mitotic cells were reduced to ~60 cells on average, while the germline of the control RNAi contained around 230 cells (Figure 2d). From these data, we conclude that the depletion of WARS-1 significantly reduces the proliferation of the germ cells in the mitotic region of the C. elegans germline, while it does not have an apparent effect on the proliferation of the somatic cells.
3.8 Zebrafish functional studies
Human WARS1 has a single ortholog in zebrafish, wars1. We performed whole-mount in situ hybridization to identify temporal and spatial expression patterns of wars1 mRNA at embryonic stages. wars1 was expressed ubiquitously at 24 h postfertilization (hpf) stage but was relatively enriched in the brain, eyes, otic vesicle, and muscles (Figure 3a).
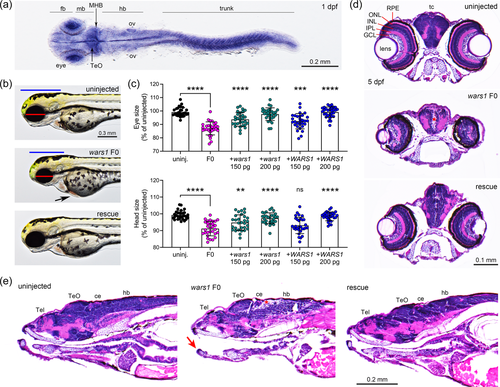
To study the function of Wars1 in zebrafish, we generated knockouts using CRISPR/Cas9 and analyzed phenotypes in injected animals (F0-founding generation, F0 mutant). Four sgRNAs were co-injected with Cas9 protein as ribonucleoprotein (RNP) complex, and the resulting phenotypes were observed every day. wars1 mutant embryos start showing morphological phenotypes such as small eyes, smaller head, and pericardial edema at 2 dpf, and by 3 dpf, phenotypes become more prominent (Figure 3b) when compared to the wild-type control. We further quantified the eye and head size, which were significantly reduced in wars1 F0 mutants compared to control animals (Figure 3c). To confirm whether the morphological phenotypes observed in wars1 F0 animals are due to loss of Wars1 function, we performed an mRNA rescue experiment by injecting either zebrafish WT wars1 or the human WARS1 mRNA. We performed coinjection of sgRNA/Cas9 RNP with variable amounts (150 and 200 pg) of mRNA encoding wars1/WARS1 into one-cell stage eggs and measured the level of phenotype rescued (Figure 3b,c). We found that 200 pg of mRNA injection was optimal to rescue morphological phenotypes, and both zebrafish and human mRNA were able to rescue phenotypes suggesting mutant phenotypes are caused by loss of Wars1 function and is evolutionarily conserved (Figure 3c).
Histological analysis of the 5 dpf larvae revealed brain abnormalities such as reduced cell density in many areas, disorganized retinal layers in the eyes, and shorter lower jaw in the mutants (Figure 3d,e). These phenotypes recapitulated some of the clinical symptoms related to brain development, visual impairment, and one individual who showed triangular face and micrognathia. These phenotypes in zebrafish mutants were rescued with coinjection of human WARS1 mRNA (Figure 3d,e). We further investigated abnormal jaw development by analyzing cartilage development using Alcian blue stain. In zebrafish embryonic development, palatoquadrate (pq) cartilage makes the upper embryonic jaw, and Meckel's (mk) cartilage makes the embryonic lower jaw (Figure 4a). In wars1 F0 mutants, Meckel's length is reduced to ~50% of the wild-type animals; coinjection of human WARS1 mRNA restored the length to the wild-type level. This phenotype is similar to micrognathia observed in individuals affected by WARS1 variants (Figure 4b). Furthermore, palatoquadrate and ceratohyal cartilages are connected at 10–20° angles in control animals, but in mutants, these are connected at 70–100° angles, and the phenotype was rescued by WARS1 mRNA injection (Figure 4c). Given that some individuals affected by WARS1 variants show movement abnormalities, we investigated whether muscle development is affected in zebrafish F0 mutants. Phalloidin staining that labels actin showed weaker staining in the myotomes, and many muscle fibers were detached from the myotendinous junction (MTJ), suggesting this phenotype might be contributing to the movement abnormalities in the affected individuals (Figure 4d). Coinjection of WARS1 mRNA restored the muscle-related abnormalities to the WT levels confirming the specificity of the phenotypes (Figure 4d).
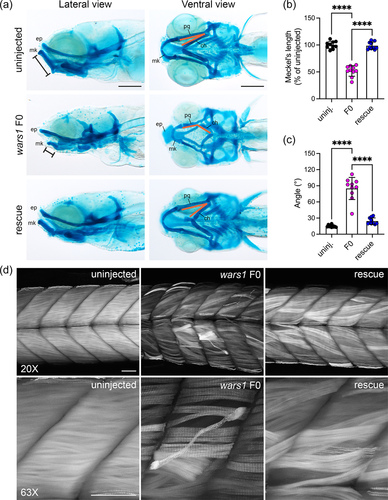
Two individuals with WARS1 variant alleles also have a hearing impairment; therefore, we investigated whether wars1 F0 mutants also show hearing abnormalities. To this end, we injected wars1 sgRNA/Cas9 RNP in a transgenic line, Tg(pou4f3:GAP-GFP) in which sensory HCs are labeled by green fluorescent protein (GFP) in the inner ear as well as in the lateral line neuromasts which are morphologically and functionally similar to the HCs of the inner ear. The wars1 F0 animals displayed smaller otic vesicles and otoliths, as well as fewer HCs in sensory epithelia including lateral crista (Figure 5a). Detailed examination of the anterior and posterior maculae did not show any difference in stereocilia length but a reduced number of HCs in the posterior macula that could be a contributing factor of reduced hearing (Figure 5b,d). We further used vital stain FM1-43, that is taken up by functional mechanotransduction channels of the HCs showing fewer HCs in the sensory lateral line as well (Figure 5e). Taken together, these data suggest that wars1 plays a critical role in HC formation, leading to hearing abnormalities in zebrafish similar to hearing loss found in some individuals affected with WARS1 variants.
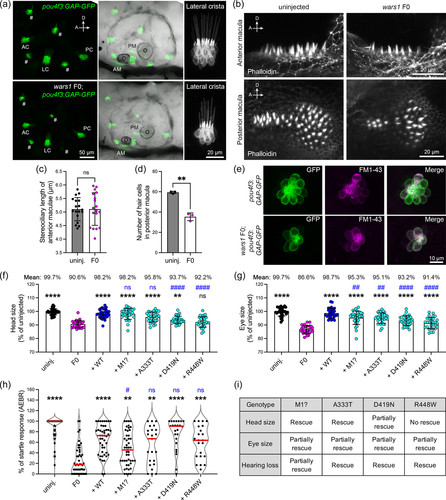
Finally, to test the pathogenicity of human WARS1 variants from our cohort, as well as previously reported in autosomal recessive cases (Okamoto et al., 2022), we introduced these variants in human WARS1 cDNA and then co-injected embryos with wars1 sgRNAs/Cas9 RNP together with WT or mutant mRNAs that encode WARS1.
We measured eye, head size, and hearing phenotype using auditory evoked behavior response (AEBR) at 6 dpf after microinjections, and the phenotypes were compared with F0 mutants and the WT rescue (Figure 5f–h). The mRNA encoding p.Met1? rescued head size but only partially rescued eye size and hearing. The rescue result is in line with the patient phenotypes as individuals affected with this variant have neurological problems—one sibling has a severe intellectual disability, visual impairment, and hearing abnormalities. The mRNA encoding the p.(Asp419Asn) variant found in Family 2 was able to rescue head and eye sizes partially, while the hearing phenotype was rescued completely. Interestingly, this individual only had neurological and visual abnormalities without any auditory symptoms (Figure 5i). We further tested two published variants, p.(Ala333Thr) and p.(Arg448Trp), found in a compound heterozygous state, and individuals affected with these variants mostly showed neurodevelopmental phenotypes without hearing loss (Figure 5i). Rescue experiments with mRNA carrying these variants rescued hearing phenotypes and partially rescue eye phenotypes. However, only the p.(Arg448Trp) variant failed to rescue the brain phenotype, suggesting this variant might be contributing to the neurological phenotypes in the affected individual (Figure 5i). Taken together, our data suggest that the zebrafish wars1 knockout recapitulates many of the human clinical phenotypes, confirms the pathogenicity of the variants, and serves as a model for functional studies.
3.9 Final variant classification
Following completion of the vertebrate rescue experiments, we applied PS3 at supporting weight in the classification of variants. The variants in Families 1 and 2 both yielded evidence of lack of rescue considering several organ systems. The c.1A>G, p.Met1? variant was classified as likely pathogenic (PVS1, PM2, PM3, PP1, PS3), while the c.1255G>A, p.(Asp419Asn) variant was classified as a variant of uncertain significance, leaning likely pathogenic (PM2, PM3, PP3, PS3). Both variants from the Okamoto et al. (2022) study that described compound heterozygous alleles (c.997G>A, p.(Ala333Thr) and c.1342C>T, p.(Arg448Trp)) were originally classified as variants of uncertain significance. Applying PS3 to both variants at supporting strength would reclassify them to likely pathogenic (PM2, PP3, PP4, PS3 for both).
4 DISCUSSION
We report two WARS1 variants in three individuals with complex neurodevelopmental disorders of varying severities (Supporting Information: Table S3). Individuals 1 and 2 in Family 1 show drastically different severities. Individual 1 reports neither seizures, hypotonia, abnormal deep tendon reflexes nor severe difficulties walking, which are all present in his younger sister (individual 2). Of note, his younger sister could not speak, walk, or be toilet trained. They both report developmental delay, intellectual disability, microcephaly, aggressive behavior, hearing, and visual impairment, as well as abnormal X-rays. Individual 3 in Family 2, with a homozygous missense variant (c.1255G>A, p.(Asp419Asn)) in the terminal exon, follows a severe clinical course since birth with severe epilepsy and shares overlap with individual 2 in that he cannot walk or speak.
Interestingly, hearing impairment was only reported in individuals 1 and 2 in Family 1 leading to the hypothesis that the loss of an essential start codon (c.1A>G. p.Met1?) may exert a tissue-specific effect. Transcript isoform studies have hinted to the first coding exon, exon 2, as a potentially crucial exon for hearing, due to expression in at least three different inner ear cell types (DCs, IHCs, and OHCs). This is further supported by rescue experiments that tested other variants (p.(Ala333Thr), p.(Asp419Asn), and p.(Arg448Trp)) in the zebrafish that exhibited a rescue of AEBRs. On the basis of preweaning lethality in the Wars1 knockout mouse and lethality observed in our zebrafish mutants, we hypothesize that usage of an in-frame Kozak consensus sequence would leave most of the protein intact and essentially deplete the long isoform in exchange for use of two possible shorter isoforms. From the perspective of the long isoform that appears to be exclusively expressed in the P15 mouse auditory system at a stage following the onset of hearing, by using the closest in-frame start codon, a predicted deletion spanning all of exon 2 and part of exon 3 (NM_004184.4:c.4_126del, p.(Pro2_Met42del)) would truncate the WHEP helix-turn-helix motif by 34 amino acids out of 56 amino acids. This is corroborated by two shorter isoforms that use the same start codon as predicted from Kozak sequence analysis. WHEP motifs are key domains in several ARS genes that are responsible for high-affinity protein-protein (tRNA) interactions (Ray et al., 2011). Because the analysis of the Kozak consensus sequence was limited to in silico prediction, this should be regarded as a possibility that warrants further investigation but would likely result in WHEP motif disruption. Additionally, further studies to dissect phenotypic differences are essential considering the limited cumulative number of families with biallelic WARS1 variants reported to date.
The c.1255G>A, p.(Asp419Asn) variant in individual 3 impacts all transcripts. Rescue experiments that assayed pathogenicity of the p.(Asp419Asn) variant showed only partial rescue with respect to head size and eye size, which would signal severe neurological effects.
When dissecting the clinical information from the three patients we present, along with a previously published patient from Japan (Okamoto et al., 2022), all patients report developmental delay and intellectual disability (Supporting Information: Table S3). Three out of four patients have confirmed microcephaly, seizures, hypotonia, absent speech, and cortical vision impairment. Larger cohort studies are needed to understand characteristic versus rare clinical phenotypes. Corroborating our postulate, autosomal recessive alleles in another ARS gene called KARS1 (lysl-tRNA synthetase 1) in two Pakistani consanguineous families with isolated hearing impairment (Santos-Cortez et al., 2013) and three rare variants of KARS1 in two patients with peripheral neuropathy have been reported (McLaughlin et al., 2010) and is exemplary of the complexity of phenotypes exhibited in ARS genes.
Heterozygous variants in WARS1 have been associated with autosomal dominant dHMN (Li et al., 2019; Tsai et al., 2017; Wang et al., 2019) and Charcot-Marie-Tooth disease (Nam et al., 2022) (Supporting Information: Table S3). Thus far, variants have clustered in the catalytic domain, with one exception being the Phe138Tyr substitution that impacts the N-terminal domain (Figure 1b, red). Presently, the recessively associated alleles are distributed across all domains (Figure 1b, black) and substitution variants impact conserved amino acids.
To further analyze the pathology of WARS1 disruption in an in vivo model system, we knocked down wars-1, the C. elegans orthologue of human WARS1, using RNAi. The ARS protein family members are highly conserved, with over 60% identity at protein level between worm and human WARS1 (data not shown). Here, we provide evidence that depletion of WARS-1 in C. elegans is associated with a defect in germ cell development, especially those in the mitotic zone. Surprisingly, knocking down wars-1 at the L1 stage in the developing C. elegans did not lead to developmental arrest or abnormality. However, RNAi-treated worms were completely sterile and did not lay any progeny, suggesting wars-1 depletion is associated with improper germline development. In an adult worm, the germline is the only mitotically active tissue, and most of the cells are born during the invariant embryonic development (Sulston et al., 1983). In essence, an adult worm contains 959 cells, out of which over 600 cells are born during embryogenesis. The postembryonic development in worms is mainly associated with increased cell size (Sulston & Horvitz, 1977). This explains the lack of somatic phenotype upon depleting wars-1 during post-embryonic development. Staining the germ cells with DAPI indicated that the germline of the wars-1 RNAi treated worms are much smaller, and germ cell numbers were dramatically reduced, especially in the mitotic zone, suggesting a defect in germ cell mitosis. This observation is in line with the previous studies that link microcephaly with loss of function variants in wars1 or other members of the aminoacyl tRNA synthases protein family (Siekierska et al., 2019; Stenson et al., 2020). Primary microcephaly is predominantly considered a cell cycle and apoptosis disorder. In C. elegans, germline apoptosis is restricted to the pachytene stage of meiosis, and germ cells at the mitotic zone are resistant to apoptotic cell death. Therefore, we conclude that the dramatic decrease in the mitotic cells of wars-1 RNAi worms is linked to defects in cell cycle progression. Further studies are required to dissect the mitotic cell cycle defect associated with WARS-1 depletion. It is of significant interest to define if such a mitotic phenotype is associated with cell cycle arrest or caused by the decline in the pool of germline stem cells.
Additionally, we used a vertebrate model, zebrafish, to investigate wars1 function in vivo. Many ARS encoding genes such as GARS1, HARS, VARS1, KARS1 have been studied in zebrafish to understand their function (Lin et al., 2021; Malissovas et al., 2016; Siekierska et al., 2019; Waldron et al., 2019). Recently, a zebrafish model for bifunctional tRNA synthetase, Kars1 has been shown to recapitulate key clinical symptoms, and kars1 knockout animals showed increased expression of genes related to p53 signaling and apoptosis that leads to downregulation of key myelin-related genes (Lin et al., 2021). Similarly, a knockout model for vars1 recapitulates microcephaly, and developmental encephalopathy caused by biallelic variants in VARS (Siekierska et al., 2019).
Here, we generated the first knockout model for wars1 that exhibited many of the key phenotypes reported in individuals affected by biallelic variants in WARS1. Zebrafish wars1 F0 mutants displayed microcephaly, hearing loss, and musculoskeletal abnormalities, and the homozygous animals do not survive past Day 10 (Brown, 2021; Lin et al., 2021; Siekierska et al., 2019). Interestingly many other ARS knockouts in zebrafish, and mouse are also embryonic/larval lethal. Microinjection of exogenous human WARS1 mRNA rescued most of these phenotypes suggesting phenotype specificity as well as a conserved function of WARS1. Whole-mount in situ expression analysis revealed ubiquitous expression at early stage but later enriched expression in the brain, eye, otic vesicle, and muscles, similar to Wars1 expression in mouse cochlea and brain, suggesting a tissue-specific conserved role across species. Many other ARSs have also shown similar expression patterns (Lin et al., 2021; Malissovas et al., 2016; Siekierska et al., 2019; Waldron et al., 2019). Individuals affected by WARS1 variants report diverse clinical symptoms such as syndromic neurodevelopmental disorders and hearing loss. We tested specific variants for their abilities to rescue phenotypes caused by loss of Wars1 function in zebrafish. All variants except p.Met1? rescued the hearing loss phenotype. The start codon is located in exon 2, suggesting a specific role of exon 2 in normal hearing function. However, the identification of additional patients presenting variants in exon 2 would be needed for a deeper understanding, as well as additional sampling of relevant tissues to support RNA studies.
In summary, we present two wars1 disease models in C. elegans and zebrafish and showed tissue-specific requirement for WARS1 function in disease pathology. Our disease models offer new tools to study specific variants for understanding their roles in WARS1-related pathologies.
AUTHOR CONTRIBUTIONS
Barbara Vona, Franz Rüschendorf, Linda Schnapp, and Pengfei Liu generated, analyzed, and validated high-throughput sequencing datasets and variant segregation results. Barbara Vona performed gene expression analysis in the mouse and analyzed transcript isoforms and initiation sites via Kozak similarity scoring. Ehsan Pourkarimi, Mahmoud Izadi, and Tayyiba Akbar Ali performed C. elegans experiments and contributed to manuscript preparation. Sheng-Jia Lin, Kevin Huang, Cassidy Petree, and Gaurav K. Varshney provided functional experiments on zebrafish. Barbara Vona, Sheng-Jia Lin, Ehsan Pourkarimi, and Gaurav K. Varshney wrote the first manuscript draft. Hillary M. Porter, Yves Lacassie, Jill A. Rosenfeld, Saadullah Khan, Pengfei Liu, and Marie-Louise Haymon edited subsequent manuscript drafts. Hillary M. Porter, Yves Lacassie, Jill A. Rosenfeld, Saadullah Khan, NazifM, Sher A. Khan, Noor Muhammad, Marie-Louise Haymon, Il-Keun Kong, Natasha Shur, Lynn Chorich, and Lawrence Layman recruited the families and/or performed clinical examinations. Barbara Vona submitted variants to LOVD v. 3.0. Barbara Vona, Thomas Haaf, Hyung-Goo Kim, and Gaurav K. Varshney conceived the study. All authors approved the final version of the manuscript.
ACKNOWLEDGMENTS
The authors thank the families for their participation in this study. This study was supported by intramural funding from the Oklahoma Medical Research Foundation, and Presbyterian Health Foundation (PHF-4411-07-04-0) (GKV), the German Research Foundation DFG VO 2138/7-1 grant 469177153 (BV), and through the Collaborative Research Center 889 and the Multiscale Bioimaging Cluster of Excellence (MBExC). IKK is funded by the National Research Foundation of Korea (NRF), a grant funded by the Korean government (MSIT; Grant No. 2020R1A2C2006614), Republic of Korea. EP was funded by Hamad Bin Khalifa University (Qatar Foundation, Qatar) and HGK was funded by Qatar Biomedical Research Institute at Hamad Khalifa University. We are thankful to the Caenorhabditis Genetics Center (CGC) which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440), for providing the worm strains. We thank Ben Fowler, OMRF Imaging Core Facility for providing histology services. Open Access funding enabled and organized by Projekt DEAL.
CONFLICTS OF INTEREST
The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing completed at Baylor Genetics Laboratories. All other authors declare no conflict of interest.
WEB RESOURCES
-
- Allen Brain Map [Mouse whole cortex and hippocampus brain atlas, Allen Institute for Brain Science
-
- https://portal.brain-map.org/atlases-and-data/rnaseq
-
- CADD
-
- https://cadd.gs.washington.edu/
-
- ClinVar
-
- https://www.ncbi.nlm.nih.gov/clinvar/
-
- CRISPOR
-
- http://crispor.tefor.net/
-
- gEAR
-
- https://umgear.org/
-
- Genesplicer
-
- https://ccb.jhu.edu/software/genesplicer/
-
- HGMD
-
- http://www.hgmd.cf.ac.uk/ac/index.php
-
- Human Splicing Finder
-
- https://www.genomnis.com/access-hsf
-
- IGV
-
- https://software.broadinstitute.org/software/igv/home
-
- I-TASSER
-
- https://zhanggroup.org/I-TASSER/
-
- LOVD v.3.0
-
- https://www.lovd.nl/3.0/home
-
- MORL scRNA-Seq
-
- https://morlscrnaseq.org/
-
- MaxEntScan
-
- http://hollywood.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html
-
- MutationTaster
-
- https://www.mutationtaster.org/
-
- MIM
-
- https://www.omim.org/clinvar
-
- PolyPhen-2
-
- http://genetics.bwh.harvard.edu/pph2/
-
- PubMed
-
- https://pubmed.ncbi.nlm.nih.gov/
-
- Predict TIS
-
- https://www.tispredictor.com/
-
- PyMOL
-
- https://pymol.org/2/
-
- SIFT
-
- https://sift.bii.a-star.edu.sg/
-
- UCSC Genome Browser
-
- https://genome-euro.ucsc.edu/index.html
-
- VarSome
-
- https://varsome.com/
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available upon request from the corresponding authors. The genetic data are not publicly available due to data privacy or ethical restrictions. WARS1 variants were submitted to LOVD v.3.0 under the variant accession identifiers 0000846763 (https://databases.lovd.nl/shared/variants/0000846763#00022706), 0000846766 (https://databases.lovd.nl/shared/variants/0000846766#00022706), 0000846767 (https://databases.lovd.nl/shared/variants/0000846767#00022706).