Rare germline variants in PALB2 and BRCA2 in familial and sporadic chordoma
Abstract
Chordoma is a rare bone tumor with genetic risk factors largely unknown. We conducted a whole-exome sequencing (WES) analysis of germline DNA from 19 familial chordoma cases in five pedigrees and 137 sporadic chordoma patients and identified 17 rare germline variants in PALB2 and BRCA2, whose products play essential roles in homologous recombination (HR) and tumor suppression. One PALB2 variant showed disease cosegregation in a family with four affected people or obligate gene carrier. Chordoma cases had a significantly increased burden of rare variants in both genes when compared to population-based controls. Four of the six PALB2 variants identified from chordoma patients modestly affected HR function and three of the 11 BRCA2 variants caused loss of function in experimental assays. These results, together with previous reports of abnormal morphology and Brachyury expression of the notochord in Palb2 knockout mouse embryos and genomic signatures associated with HR defect and HR gene mutations in advanced chordomas, suggest that germline mutations in PALB2 and BRCA2 may increase chordoma susceptibility. Our data shed light on the etiology of chordoma and support the previous finding that PARP-1 inhibitors may be a potential therapy for some chordoma patients.
1 INTRODUCTION
Chordoma is a rare bone tumor, with an age-adjusted incidence rate of less than 0.1 per 100,000 in the United States (US) and Europe (Bakker et al., 2018). Chordoma is believed to originate from notochordal remnants and occurs in bones of skull base and vertebra. The tumor occurs more frequently in males than females and is diagnosed at a median age of 57 years with a range from infancy to over 98 (Bakker et al., 2018; Parry et al., 2020; Zuckerman et al., 2018). Skull-base patients are associated with earlier age onset and longer survival compared with patients with sacral tumors (Zuckerman et al., 2018).
Although chordoma is typically sporadic, more than 10 families with multiple relatives affected with chordoma have been described worldwide (Parry et al., 2020), suggesting that hereditary factors exist for this rare tumor. We previously identified germline TBXT duplication as a major susceptibility mechanism in several chordoma families (Yang et al., 2009). The TBXT gene encodes brachyury, a tissue-specific transcription factor that is expressed in notochord cells and is essential for the formation and maintenance of the notochord (Kispert et al., 1995). Subsequent studies reported that common and rare germline variants in TBXT were associated with increased chordoma risk in familial as well as sporadic chordoma patients (Kelley et al., 2014; Pillay et al., 2012), further highlighting the importance of TBXT in chordoma susceptibility. However, genetic causes in the majority of sporadic cases and chordoma families remain largely unknown.
PALB2 (Partner and Localizer of BRCA2) codes for a key binding partner of BRCA2, a major breast cancer susceptibility protein (Xia et al., 2006). The two proteins form a tight complex and play an essential role in homologous recombination (HR)-based repair of DNA double-strand breaks, largely by promoting the loading of the recombination enzyme RAD51 to damaged DNA ends (Prakash et al., 2015). PALB2 also directly interacts with BRCA1, the other major breast cancer suppressor protein, serving as a linker between BRCA1 and BRCA2 in HR and the DNA damage response (Sy et al., 2009; Zhang et al., 2009). Like BRCA1 and BRCA2, germline mutations in PALB2 have been linked to breast cancer, ovarian cancer, and pancreatic cancer susceptibility (Nepomuceno et al., 2017; Nepomuceno et al., 2021). Interestingly, a study by Rantakari et al. showed that Palb2(−/−) mouse embryos revealed a notochord with diffuse and discontinuous morphology (Rantakari et al., 2010), implicating PALB2 in chordoma development. Moreover, a Brca1(−/−) mouse model demonstrated that abnormalities in Brca1-deficient embryos were most evident in the neural tube (Gowen et al., 1996), the development of which is induced by notochord-derived sonic hedgehog (Shh) signaling (Kahane & Kalcheim, 2020). In line with this, a recent genomic study of 11 chordoma patients demonstrated that SBS3, a mutational signature associated with HR deficiency (HRD) and BRCA1/2 germline mutations, was significantly enriched in advanced chordoma samples (Gröschel et al., 2019), further supporting the potential importance of PALB2/BRCA2-mediated HR repair in chordoma development.
In this study, we aimed to identify additional chordoma susceptibility genes in 19 familial patients or obligate mutation carriers in TBXT duplication negative families and 137 sporadic chordoma patients using whole-exome sequencing (WES). We found that rare germline variants in PALB2 and BRCA2 occurred with higher frequency in chordoma patients compared with population-based controls that were sequenced using the same sequencing platform and bioinformatic analysis pipeline. Experimental work demonstrated that some of the rare PALB2 and BRCA2 variants observed in chordoma patients might have functional relevance in affecting protein binding and HR, suggesting that rare germline variants in these genes may play a role in chordoma susceptibility.
2 MATERIALS AND METHODS
2.1 Study population
The details of the family and sporadic chordoma studies have been previously described (Kelley et al., 2014; Parry et al., 2020). In this analysis, we included five chordoma families with ≥2 chordoma cases/obligate gene carriers (n = 19) and 137 sporadic chordoma cases identified from the United States and Canada. The average age at diagnosis among sporadic patients was 46.9 years (range 7–78), and the vast majority of patients (97%) had classic chordoma histology. The chordoma site distribution was 55.7% skull-base, 23% spinal, and 20.3% sacral. All diagnoses of chordoma were confirmed by reviewing pathologic slides or reports, medical records, or death certificates. All study subjects were Europeans and negative for TBXT duplications. Both the family and sporadic chordoma studies were approved by institutional review boards at the National Institutes of Health (approval number 78C0039 for the family study and 10CN188 for the sporadic study) and all participants provided written informed consent.
We also used WES data from 598 healthy Caucasian cancer-free unrelated individuals, from the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO) and Cancer Prevention Study (CPS) as reference/controls for conducting rare variant burden tests. These controls were sequenced and analyzed using the same sequencing platform and Ensemble variant calling pipeline as used for chordoma cases.
2.2 WES and bioinformatics analysis
WES of germline DNA bioinformatic analyses were performed at the Cancer Genomics Research Laboratory (CGR), NCI, as previously described in detail (Goldstein et al., 2017). We only included variants for further evaluation if they (1) passed the quality control (QC) filter in the in-house bioinformatics pipeline; (2) were called by at least two of the three variant callers; (3) had a minor allele frequency (MAF) <0.1% in public databases such as the 1000 Genomes Project, Exome Sequencing Project (ESP6500), and Exome Aggregation Consortium (ExAC); (4) were present in ≤2 families from an in-house database of ~2000 exomes in ~1000 cancer-prone families (excluding chordoma families); and (5) were classified as nonsynonymous (NS) including frameshift, stopgain, inframe deletion or insertion, or NS substitutions (missense).
Rare variants in PALB2 and BRCA2 identified in chordoma patients were technically validated using Sanger sequencing at CGR. Variants were annotated according to the Human Genome Variation Society (HGVS) guidelines and interpreted using RefSeq NM_024675.3 (PALB2) and NM_000059.4 (BRCA2). Sequence variant descriptions were verified by Mutalyzer online tool (https://mutalyzer.nl/; accessed on June 9, 2022).
2.3 Cell lines and cultures
U2OS/DR-GFP HR reporter cells were previously described (Xia et al., 2006). HEK293T cells were purchased from American Type Culture Collection (ATCC). These cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 1× penicillin-streptomycin at 37°C in a humidified incubator with 5% CO2. PL2F7 mouse embryonic stem (mES) cells were previously described (Kuznetsov et al., 2008) and cultured on SNL feeder cells in Knock-out DMEM media supplemented with 15% fetal bovine serum, GPS (glutamine-penicillin-streptomycin), 0.1 mM β-mercaptoethanol at 37°C in a humidified incubator with 5% CO2.
2.4 HR repair assay
The assay was conducted using U2OS/DR-GFP cells as previously described (Anantha et al., 2017; Foo et al., 2017). Briefly, U2OS/DR-GFP cells were first depleted of the endogenous PALB2 using a small interfering RNA (siRNA) in 10 cm plates and then split/seeded into six-well plates. Cells were then cotransfected with a plasmid expressing I-SceI (pCBASce) and a PALB2 expression vector (pOZC-PALB2) in which the siRNA target sequence had been mutated (Xia et al., 2006). Cells were harvested approximately 54–68 h post the second transfection, and GFP-positive cells were counted by flow cytometry. The sequence of the PALB2 siRNA is 5′-UCAUUUGGAUGUCAAGAAAdTdT-3′. Variant sequences were introduced into the pOZC-PALB2 expression vector by site-directed mutagenesis following the QuikChange protocol (Agilent) and verified by Sanger sequencing. Two independent clones of each variant were used for the assay.
2.5 Mammalian two-hybrid assay
Fusion constructs GAL4DBD-BRCA2 N-terminus (aa 1–60), VP16AD-PALB2 C-terminus (aa 859–1186), and fusions containing control variants (PALB2 p.Leu21Ala and p.Ala1025Arg) were previously described (Rodrigue et al., 2019). Chordoma-associated PALB2 missense variants were generated using the QuikChange II Site-Directed Mutagenesis Kit (Agilent) and confirmed by Sanger sequencing. Mammalian two-hybrid assays were conducted as previously described (Rodrigue et al., 2019). Briefly, HEK293FT cells were cotransfected with GAL4DBD-BRCA2 N-terminus and VP16AD-PALB2 C-terminus (wild type or variants) plasmids together with pG5luc luciferase reporter vector and pGR-TK internal luciferase control. Expression of luciferases was quantified 24 h posttransfection using the Dual-Luciferase Reporter Assay System (Promega) according to manufacturer's instructions.
2.6 Generation of mES cells expressing BRCA2 variants and cell viability assay
Variants were generated in BAC clone containing full-length BRCA2 as described previously (Biswas, Das, et al., 2012; Biswas, Stauffer, et al., 2012). Twenty micrograms of BAC DNA was electroporated into 1.0 × 107 PL2F7 mES cells and selected in the presence of G418 (Invitrogen). Colonies expressing human BRCA2 variants were selected either by reverse-transcription polymerase chain reaction or western blot as described previously (Biswas, Das, et al., 2012; Biswas, Stauffer, et al., 2012). Two independent clones expressing each variant were used for cell viability assay. Twenty micrograms of Pgk-Cre plasmid was electroporated to delete the conditional allele of Brca2 in mES cells expressing BRCA2 variants as described previously (Kuznetsov et al., 2008). After electroporation, 1 × 106 cells were plated in triplicate in 100 mm dishes and cultured in the HAT media (Gibco) to select recombinant colonies. Another set of 3 × 100 mm dishes were used as plating efficiency control. ES cell colonies were stained with methylene blue and counted as described previously (Biswas et al., 2020).
2.7 Immunoprecipitation (IP) and western blot analysis
To assess the complex formation between variant PALB2 proteins with BRCA2, RAD51, and BRCA1, HEK293T cells were seeded into six-well plates at 6 × 105 cells per well and cotransfected with 1 µg pOZC-PALB2 (wt and variants) and 1 µg pcDNA3-Myc-BRCA1 (Chen et al., 1998) using 5 µl of X-tremeGENE HP (Roche). Approximately 30 h after transfection, cells were collected and lysed in 350 µl of NETNG-250 (20 mM Tris-HCl [pH 7.4], 1 mM EDTA, 5 mM NaF, 0.5% NP-40, and 10% glycerol), of which 300 µl was used for IP and the remaining as inputs. The FLAG-HA-tagged PALB2 proteins were IPed with 10 µl 1:1 slurry of Anti-HA Agarose (A2095; Sigma) at 4°C for 3 h. Samples were resolved on 4%–12% Tris-Glycine sodium dodecyl sulphate (SDS) polyacrylamide gels (Invitrogen) and transferred to nitrocellulose membrane at 4°C overnight. Blots were probed with anti-PALB2 (aa601-880) (Xia et al., 2006), anti-BRCA2 (OP95; Millipore), anti-RAD51 (H92; Santa Cruz), and anti-BRCA1 (07-434; Millipore). Secondary antibodies used were horseradish peroxidase (HRP)-conjugated sheep anti-mouse IgG (NA931V; GE Healthcare), and donkey anti-rabbit IgG (NA9340V; GE Healthcare). Immobilon Western Chemiluminescent HRP Substrate (Millipore) was used to develop the blots.
2.8 Statistical analysis
Rare variant burden test for PALB2 and BRCA2 in 137 sporadic cases and 598 population-based controls was conducted using the SKAT statistic, which aggregates individual score test statistics of SNPs in a SNP set and efficiently computes gene-level p values (Wu et al., 2011). All exonic rare variants with MAF < 0.1% in ESP and 1000 Genomes were included, regardless of their predicted functions. HR assays were conducted in at least eight independent experiments for each variant, mostly using two independent clones for each variant and two clones of the wt construct in each experiment. Results were normalized against the average of the wt clones in each experiment. Two-tailed unpaired t test was used to test the differences between the activities of each variant and the wt protein. Final data are presented as mean ± SD (standard deviation). Mammalian two-hybrid assays were conducted in three independent experiments, each performed in four technical replicates, and the results were normalized against the luciferase activity of the wt construct. Final data are expressed as mean ± SEM (standard error of the mean). Statistical significance was accessed by one-way ANOVA followed by Dunnett's post hoc analysis for the burden test and unpaired Student's t test for the functional assays. All p values were two-sided, with p < 0.05 considered as statistically significant.
3 RESULTS
3.1 Cosegregation of a PALB2 variant in a chordoma family
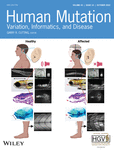
To identify genetic variants that may predispose to chordoma, we first analyzed WES data in a family with two confirmed chordoma cases, one obligate gene carrier, and one case with juvenile pilocytic astrocytoma (Figure 1). Both chordoma patients had skull-base tumors and were diagnosed at young ages, 46 and 11 years, respectively. The patient with juvenile pilocytic astrocytoma was diagnosed at 16 years old. WES data analysis identified only three rare NS variants that showed disease cosegregation in this family. Among them, a missense NS variant in PALB2 (NM_024675.3: c.1042C>A, p.Gln348Lys) was of particular interest because of the well-established important role of PALB2 in cancer susceptibility and the previous report of abnormal notochord morphology in Palb2 knockout mouse embryos (Rantakari et al., 2010). This variant was present in both chordoma cases, the obligate gene carrier, and the patient with juvenile pilocytic astrocytoma, as confirmed by targeted Sanger sequencing. Among eight unaffected family members we sequenced in this family, only one had the variant (Figure 1), but this subject was only 14 years old at the examination, and therefore it is possible that the clinical manifestation was not yet present. This variant was reported with minor allele frequency (MAF) of 0.000039 in the Genome Aggregation Database (gnomAD, v2.1.1) and was not seen in our in-house exome database including 598 population-based healthy controls and ~2000 exomes in ~1000 cancer-prone families (excluding chordoma patients). The variant is listed in ClinVar (accession VCV000141588) with conflicting interpretations of pathogenicity (Table 1).
| Variants | AA change | MAF | ClinVar | In silico prediction | Age | Tumor site | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| gnomAD | In-house | Polyphen2 HDIV |
Sift | Mutation Taster (simple model) |
Mutation Assessor |
Provean | |||||
| c.398G>C | Ser133Thr | 0 | 0 | VUS | B (0.225) | B (0.28) | B (1) | L (1.355) | B (−0.59) | 27 | Skull |
| c.1042C>A | Gln348Lys | 3.9 × 10−5 | 0 | Conflict | B (0.176) | B (0.418) | B (0.999) | L (1.87) | B (−1.57) | 11, 46 | Skull |
| c.1600T>G | Ser534Ala | 4.0 × 10−6 | 0 | VUS | B (0.021) | B (0.403) | B (1) | L (1.39) | B (−1.09) | 66 | Vertebral |
| c.2755G>A | Val919Ile | 1.6 × 10−5 | 0 | VUS | D (0.353) | D (0.062) | D (0.99) | M (2.36) | B (−0.74) | 44 | Skull |
| c.3103A>G | Ile1035Val | 8.0 × 10−6 | 0 | Conflict | B (0.033) | B (0.305) | B (0.91) | L (0.86) | B (−0.47) | 47 | Skull |
| c.3494C>T | Ser1165Leu | 4.0 × 10−6 | 0 | VUS | D (1) | D (0.002) | D (0.997) | M (2.24) | D (−4.65) | 10 | Skull |
- Note: All variants were interpreted using NM_024675.3 (GRCH37/HG19). ClinVar: https://www.ncbi.nlm.nih.gov/clinvar/; VUS: variant of unknown significance. PolyPhen-2 (Polymorphism Phenotyping v2) HDIV: identifies human damaging mutations by assuming differences between human proteins and their closely related mammalian homologs as nondamaging. The PolyPhen score represents the probability that a substitution is damaging, so values nearer 1 are more confidently predicted to be deleterious; SIFT (Sorting Intolerant From Tolerant); Provean (Protein Variation Effect Analyzer). A SIFT score is a normalized probability of observing the new amino acid at that position, and ranges from 0 to 1. A value of between 0 and 0.05 is predicted to affect protein function; The MutationTaster score is the probability that the prediction is true; Mutation Assessor creates a conservation score which is combined with a specificity score to determine a functional impact score. Variants classed as “neutral” (N) or “low” (L) are predicted to not impact protein function, whereas variants classed as “medium” (M) or “high” (H) are predicted to result in altered function. Prediction: B: Benign; D: Deleterious; Scores for each algorithm are presented in parentheses.
- Abbreviations: AA, amino acid; MAF, minor allele frequency; gnomAD, The Genome Aggregation Database.
- In silico prediction was based on algorithms included in Emsembl (https://useast.ensembl.org/info/genome/variation/prediction/protein_function.html).
3.2 Rare germline PALB2 variants in other familial and sporadic chordoma patients
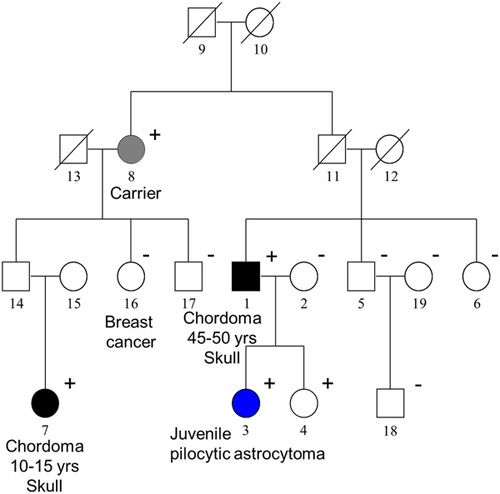
We then screened for PALB2 variants in all remaining chordoma families and sporadic chordoma patients with WES data and identified five additional PALB2 rare NS variants, each in a sporadic patient (Table 1). All variants were technically validated by targeted Sanger sequencing. Four of these patients had skull-base chordoma and were diagnosed before age 50, including a pediatric case who was diagnosed at 10 years old. The remaining patient had vertebral chordoma and was diagnosed at age 66 years. All five variants were missense changes, were extremely rare in public and in-house databases, and were listed as variants of unknown significance (VUS) in ClinVar (Table 1). Three of the variants are located in the carboxy-terminal WD-40 domain (Figure 2a), which is responsible for its association with BRCA2 and RAD51 (Nepomuceno et al., 2021; Park et al., 2014). Among them, the variant carried by the pediatric case (NM_024675.3: c.3494C>T, p.Ser1165Leu) was predicted as deleterious by most in silico prediction algorithms.
To assess the overall genetic burden due to these rare exonic variants in PALB2, we conducted a rare variant burden test by comparing 137 sporadic cases to 598 population-based controls sequenced and analyzed at the Cancer Genomics Research Laboratory (CGR), NCI, using similar approaches as used for chordoma cases (see Methods). Sporadic chordoma cases showed significantly higher frequency of PALB2 rare variants when compared to controls (3.6% vs. 0.2%, p = 0.028).
3.3 Functional characterization of the PALB2 variants
To determine the functional impact of the PALB2 variants identified, we first assessed their interaction with BRCA2, RAD51, and BRCA1 by co-IP (co-IP). All six variants were found to co-IP with all three proteins, and the affinity of the associations appeared to be similar to that of the wild-type (wt) protein (Figure 2b), indicating that the sequence alterations did not affect stable associations of PALB2 with the three critical interaction partners. Next, we assessed their intercellular localization and HR repair (HRR) activity using a “protein replacement” strategy, wherein the endogenous PALB2 protein was depleted in the U2OS/DR-GFP HR reporter cells using an siRNA, and the variants were introduced by transient transfection of complementary DNAs that were resistant to the siRNA used (Anantha et al., 2017; Foo et al., 2017). PALB2-Ala1025Arg, a previously reported artificial mutant with severely reduced ability to interact with BRCA2 (Oliver et al., 2009), was used as a reference. All six variants were found to be expressed at levels comparable to that of the wt protein, localized in the nucleus and partially colocalized with BRCA1 in nuclear foci after DNA damage (Supporting Information: Figure S1). However, four of the six variants, namely p.Ser133Thr, p.Gln348Klys, p.Val919Ile, and p.Ser1165Leu, showed modestly reduced HRR activity (Figure 2c), suggesting that these variants may be partially defective in a certain aspect(s) of PALB2's HR-related functions.
As PALB2 interacts via its C-terminal WD40 repeats with BRCA2, we also used a mammalian two-hybrid assay to assess the effect of the three C-terminal WD40 variants (p.Val919Ile, p.Ile1035Val, p.Ser1165Leu) on their interactions with BRCA2 (Figure 2d). Compared to wt PALB2, all three variants showed significantly reduced interactions, although the differences were less significant than that of Ala1025Arg (Figure 2e). Considering that similar amount of BRCA2 was found to associate with these three variants in the co-IP assay, which measures stable associations, these mammalian two-hybrid results suggest that there may be a transient or unstable component of the PALB2-BRCA2 interaction, which is affected by these C terminal variants.
3.4 Rare germline BRCA2 variants in familial and sporadic chordoma patients
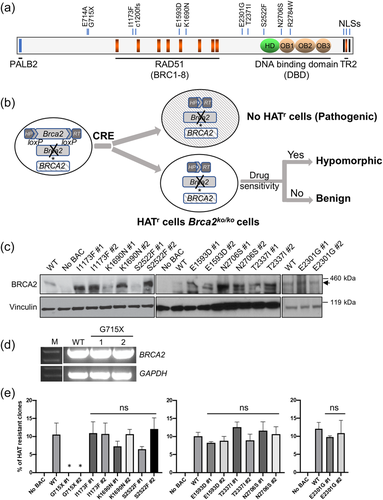
Given the close functional connection of BRCA2 with PALB2, we also analyzed BRCA2 rare NS variants in all familial and sporadic chordoma patients with WES data. We identified 11 rare NS BRCA2 variants in 10 sporadic chordoma patients (Table 2 and Figure 3a), all of which were technically validated by targeted Sanger sequencing. Two BRCA2 variants (NM_000059.4: p.Glu714Ala and p.Gly715Ter) were seen in the same patient, who was diagnosed with vertebral chordoma at age 51 years. The remaining variants were each seen in a single patient. Among the 11 variants, the majority result in a single amino acid change (Table 2). Eight of these variants are listed in ClinVar; p.Ile1173Phe and p.Thr2337Ile are considered VUS, p.Glu1593Asp, p.Lys1693Asp, p.Ser2522Phe and p.Asn2706Ser are benign or likely benign, and p.Cys1200fs and p.Arg2784Trp are pathogenic or likely pathogenic. The other three (p.Glu714Ala, p.Gly715Ter and p.Glu2301Gly) are novel and not listed in ClinVar. p.Gly715Ter is likely to be pathogenic given its protein-truncating nature and the fact that two other truncating variants (c.2137C>T, p.Gln713Ter and c.2147del, p.Gln716fs) in its vicinity are reported to be pathogenic in ClinVar. Similar to PALB2 variants, we also observed a higher genetic burden of BRCA2 rare variants in chordoma patients compared with controls (8% vs. 3%, p = 0.025).
| Variants | AA Change | gnomAD | ClinVar | HGMD | LOVD | Patient Age |
Tumor site | ES cell rescue | Drug sensitivity |
Functional impact |
|---|---|---|---|---|---|---|---|---|---|---|
| c.2141A>C | Glu714Ala | 0 | 51 | Vertebral | ND | |||||
| c.2143G>T | Gly715* | 0 | 51 | Vertebral | No | P | ||||
| c.3517A>T | Ile1173Phe | 0 | VUS | 19 | Skull | Yes | No | B | ||
| c.3599_3600delGT | Cys1200* | 0 | P | DM | 56 | Pelvic | ND | P | ||
| c.4779A>C | Glu1593Asp | 0.00037 | B/LB | B | 52 | Skull | Yes | No | B | |
| c.5070A>C | Lys1690Asn | 0.00022 | B/LB | DM? | B | 29 | Skull | Yes | No | B |
| c.6902A>G | Glu2301Gly | 0 | 70 | Multi-site | Yes | No | B | |||
| c.7010C>T | Thr2337Ile | 2.5 × 10−5 | VUS | 53 | Skull | Yes | No | B | ||
| c.7565C>T | Ser2522Phe | 7.1 × 10−6 | Conflict | DM | P | 53 | Pelvic | Yes | No | B |
| c.8117A>G | Asn2706Ser | 6.7 × 10−5 | B/LB/VUS | DM? | B | 42 | Skull | Yes | No | B |
| c.8350C>T | Arg2784Trp | 8.2 × 10−6 | P/LP | B/B/VUS | 28 | Pelvic | ND | P |
- Note: All variants were interpreted using NM_000059.4 (GRCH37/HG19).
- Abbreviations: AA, amino acid; B, benign; Conflict, conflicting evidence for pathogenicity; gnomAD, The Genome Aggregation Database; LB, likely benign; LP/DM?, likely pathogenic; MAF: minor allele frequency; ND, not determined; P/DM, pathogenic; VUS, variant of unknow significance.
- ClinVar: https://www.ncbi.nlm.nih.gov/clinvar/;
- HGMD (The Human Gene Mutation Database): http://www.hgmd.cf.ac.uk/ac/index.php;
- LOVD (Leiden Open Variation Database): https://www.lovd.nl/.
3.5 Functional characterization of BRCA2 variants
We used a mouse ES cell-based assay for functional evaluations of the BRCA2 variants. Two variants, p.Cys1200fs and p.Arg2784Trp, were not further evaluated because they were reported to be pathogenic in ClinVar or in our previous studies (Biswas et al., 2020). p.Glu714Ala was not evaluated because it was seen in the patient with p.Gly715Ter that is more likely to be pathogenic. The remaining eight variants were introduced into a bacterial artificial chromosome (BAC) construct containing full-length human BRCA2 by recombineering-based gene editing, and their impact on cell survival and sensitivity to DNA damaging agents was examined in Brca2 knockout (KO) ES cells (Figure 3b). Two independent clones that express the protein at levels at least equivalent to the clone expressing wt BAC were selected (Figure 3c,d) and analyzed for colony formation upon deletion of the conditional mouse Brca2 allele. As functional BRCA2 is required for the viability of mouse ES cells, variants that fail to rescue the lethality of Brca2KO/KO ES cells are considered pathogenic. Expression of p.Gly715Ter did not result in viable ES cells, confirming that it is indeed a pathogenic variant, while the other seven full-length variant BRCA2 proteins tested were all able to support viability and colony formation (Figure 3e and Supporting Information: Figure S2). The BAC-reconstituted, viable Brca2KO/KO ES cells were subsequently analyzed for their sensitivity to DNA damaging agents (camptothecin, cisplatin, mitomycin c [MMC], and olaparib) to distinguish between benign and hypomorphic variants. For all seven variants, the sensitivity of their reconstituted cells was comparable to cells expressing wt BRCA2 (Supporting Information: Figure S3), suggesting that these are fully functional and likely neutral variants.
3.6 Somatic copy number alterations of PALB2 and BRCA2 among germline variant carriers
Among chordoma patients included in our germline WES analysis, Formalin-fixed, paraffin-embedded (FFPE) tumor blocks or sections were available for 52 patients, including six patients carrying the germline rare variants in PALB2 (n = 2) or BRCA2 (n = 4). Copy number analysis of these tumors based on SNP array data showed that the deletion of chromosome 13q, where BRCA2 is located, occurred in 19 of 52 tumors (36.5%) and was one of the most frequent copy number events (Supporting Information: Figure S4A). Somatic mutations in chromatin remodeling genes (PBRM1, SETD2, and SMARB1) or CDKN2A homozygous deletions, which are the most common driver events in chordoma (Bai et al., 2021; Tarpey et al., 2017), were not found in the six patients carrying germline PALB2/BRCA2 variants. Tumors in two (p.Glu2301Gly and p.Ser2522Phe) out of four BRCA2 germline variant carriers with SNP array data available showed chromosome 13q deletions (Supporting Information: Figures S4B,S4C). The tumor in a PALB2 variant (p.Ser1165Leu) carrier, who was diagnosed with chordoma at 10 years old, contained a complete loss of chromosome 16, where PALB2 resides, as well as a copy number neutral loss of heterozygosity (LOH) of chromosome 13 (Supporting Information: Figure S4B,S4D). Interestingly, one tumor (the carrier of p.Glu2301Gly) had somatic deletions of regions containing all three genes, BRCA1, BRCA2, and PALB2. Although we were unable to determine whether the wt or the variant containing allele was lost, the somatic copy number data are generally consistent with the notion that PALB2 and BRCA2 genes may play an important role in chordoma development.
4 DISCUSSION
In this WES analysis of 19 familial chordoma cases from five pedigrees and 137 sporadic chordoma patients, we identified rare germline variants in PALB2 and BRCA2. To the best of our knowledge, our study is the first and largest WES analysis of germline DNA from familial and sporadic chordoma patients. In addition, chordoma cases had an increased burden of rare germline variants in both genes as compared to population-based controls that were analyzed using the same sequencing platforms and bioinformatic pipelines as for the chordoma cases. Further, somatic deletions of chromosome regions containing PALB2 and BRCA2 were found in chordoma tumors from patients carrying germline rare variants of the two genes. Functionally, four of the six PALB2 variants modestly affected its HR function and three of the BRCA2 variants caused loss of function. Overall, our results are in line with the previously reported findings of diffuse and discontinuous morphology and improper Brachyury expression of the notochord of Palb2 knockout mouse embryo (Rantakari et al., 2010). Our results are also consistent with recent findings of genomic signatures associated with HRD and mutations in HR genes (BRCA2 and NBN) in a small series of advanced chordoma (Gröschel et al., 2019). Taken together, these lines of evidence suggest that germline mutations in PALB2 and BRCA2 may contribute to chordoma susceptibility. Our data also support the use of PARP inhibitors to treat some chordoma patients, that is, those whose tumors have HR gene mutations and HRD, as has been shown with one tumor with BRCA2 mutations in the above-noted study (Gröschel et al., 2019).
Considering that the identified PABL2 variants showed either modest or no effect on HR, it appears that the genetic mechanism of any PALB2-associated chordoma development may differ from that of breast cancer, for which only variants that strongly abrogate HR function are considered pathogenic. As such, these PALB2 variants may contribute to chordoma development either in conjunction with other, yet to be identified, genetic events or even through an HR-independent mechanism. It is also possible that they are disease-modifying rather than disease-causing. As for BRCA2, three of the 11 variants identified can be considered classical pathogenic mutations; regrettably, however, tumor materials from carriers of these pathogenic mutations were not available to determine the LOH status. Also, the information on breast cancer occurrence in families of these carriers were not collected. Further investigations are required to determine whether those classical pathogenic mutations were indeed the major cause of chordoma in patients carrying them, and whether those variants that appear to have normal or nearly normal HR function may also contribute to chordoma development through HR-independent mechanisms.
Limitations of our study included small number and size of pedigrees for new gene discovery. Therefore, the prioritization of candidate genes from WES heavily relied on existing knowledge of the biological relevance of the candidate gene(s) in chordoma or cancer development. In addition, the lack of appropriate tumor materials in patients carrying PALB2/BRCA2 variants prohibited us from further evaluating the allele-specific LOH and mutational signatures to compliment germline findings. Although results from our somatic copy number analysis are consistent with previous studies showing that chromosome 13q deletions are very common in chordoma tumors (Bai et al., 2021; Wang et al., 2016), thereby supporting our germline findings, these deletions often involve large chromosome regions which makes it challenging to pinpoint specific genes. In addition, results from several chordoma somatic mutation landscape studies do not support HRD as a common defect in chordoma, at least less common than alterations in chromatin remodeling genes and chromosome 9 deletions (Bai et al., 2021; Tarpey et al., 2017; Wang et al., 2016). However, the overall mutational burden in chordoma tumors is very low and most tumors have very quiet genome (Bai et al., 2021); therefore, it is possible that epigenetics may play a key role in these quiet tumors, possibly through inactivating tumor suppressor genes such as PALB2 and BRCA2. The lack of multiple omics data available for this rare disease and small number of patients in each individual study have limited our ability to integrate germline and somatic data for a comprehensive evaluation of the role of these genes in chordoma susceptibility. Nevertheless, together with the previous report on Palb2 knockout mice and the effectiveness of PARP-1 inhibitor in treating metastatic chordoma patients (Gröschel et al., 2019; Rantakari et al., 2010), our germline and somatic findings on PALB2/BRCA2 variants in chordoma patients warrant future investigations of these genes and involved pathways in independent chordoma patient datasets.
Finally, our findings raise the question why there have been no reports of chordoma in families and individuals known to carry BRCA2 and PALB2 pathogenic mutations or VUS. Since chordoma is very rare and the majority of chordoma patients are asymptomatic or present nonspecific back pain at early stage (Xu et al., 2022), it is possible that there are undiagnosed chordoma patients in these families. In addition, it is still unclear whether the loss-of-function mutations, which are typically seen in PALB2/BRCA2 breast cancer families/individuals, in the two genes indeed increase chordoma risk, given that most rare variants we observed in chordoma patients were missense with only modest or no effect on HR. To address these questions, we hereby recommend a closer examination of possible chordoma incidence in BRCA2 and PALB2 families, which may yield valuable information that would allow us to further understand the etiology and improve the clinical management of this rare disease.
ACKNOWLEDGMENTS
We are indebted to the participating families, whose generosity and cooperation have made this study possible. We thank the Cancer Sequencing Working Group and Cancer Genomics Research Laboratory of the Division of Cancer Epidemiology & Genetics (DCEG) at the National Cancer Institute (NCI). This study was funded by Intramural Research Program of DCEG and Center for Cancer Research at NCI, National Institutes of Health, USA; and by NCI grants R01CA138804 and R01CA262227 to BX. This workstudy was also supported by Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro, Conselho Nacional de Desenvolvimento Científico e Tecnológico, Fundação do Câncer (Programa de Oncobiologia) (MAC), and Moffitt foundation (AM). TKF and ZK were recipients of New Jersey Commission on Cancer Research (NJCCR) Postdoctoral Fellowships.
CONFLICT OF INTEREST
The authors declare no conflict of interest.