Genome sequencing reveals a novel genetic mechanism underlying dihydropyrimidine dehydrogenase deficiency: A novel missense variant c.1700G>A and a large intragenic inversion in DPYD spanning intron 8 to intron 12
Contract grant sponsors: BC Children's Hospital Research Institute Foundation; Genome British Columbia (SOF-195, #174DE); Canadian Institutes of Health Research (#301221); Michael Smith Foundation for Health Research Scholar Award.
Communicated by David S. Rosenblatt
Abstract
Dihydropyrimidine dehydrogenase (DPD) deficiency is associated with a variable clinical presentation. A family with three DPD-deficient patients presented with unusual clinical phenotypes including pregnancy-induced symptoms, transient visual impairment, severe developmental delay, cortical blindness, and delayed myelination in the brain. DPYD Sanger sequencing showed heterozygosity for the c.1905+1G>A mutation and a novel missense variant c.1700G>A (p.G567E). The recombinantly expressed p.G567E DPD variant showed increased temperature lability probably caused by structural rearrangements within the DPD protein. Genome sequencing of the affected son established compound heterozygosity for the c.1700G>A and an imperfect 115,731 bp inversion with breakpoints at chr1: 98,113,121 (intron 8) and chr1: 97,997,390 (intron 12) of the DPYD associated with a 4 bp deletion (chr1: 97,997,386_97,997,389del). Whole exome and mitochondrial DNA analyses for the mother and daughter did not reveal additional mutated genes of significance. Thus, an inversion in DPYD should be considered in patients with an inconclusive genotype or unusual clinical phenotype.
Dihydropyrimidine dehydrogenase (DPD) is the initial and rate-limiting enzyme of the pyrimidine degradation pathway, catalyzing the reduction of uracil and thymine to 5,6-dihydrouracil and 5,6-dihydrothymine, respectively. A DPD deficiency (MIM# 274270) is an infrequently described autosomal recessive disease characterized by thymine-uraciluria. In patients with a complete DPD deficiency, a considerable variation in the clinical presentation has been observed. Psychomotor retardation and convulsive disorders are relatively frequent manifestations, whereas growth retardation, microcephaly, dysmorphia, autism, hypotonia, and ocular abnormalities are less frequently observed (van Kuilenburg et al., 2002; van Kuilenburg et al., 1999). However, a consistent phenotype has not yet emerged (van Kuilenburg et al., 2002; van Kuilenburg et al., 1999). In addition, DPD plays an important role in the breakdown of the antineoplastic agent 5-fluorouracil (5FU). Patients with a partial or complete DPD deficiency have a strongly reduced capacity to degrade 5FU and therefore, an increased likelihood of suffering from severe and sometimes fatal multiorgan toxicity (van Kuilenburg, 2004).
Genetic variations in the gene encoding DPD (DPYD; MIM# 612779) have emerged as predictive risk factors for severe fluoropyrimidine toxicity (Meulendijks et al., 2015). To date, many mutations and polymorphisms have been described in DPYD including large genomic deletions and amplifications (van Kuilenburg et al., 2017; van Kuilenburg et al., 2009; van Kuilenburg et al., 2010). Analysis of the mutational spectrum of DPYD in pediatric DPD patients and cancer patients experiencing severe toxicity showed that the splice-site mutation c.1905+1G>A, leading to skipping of exon 14 in the process of DPD pre-mRNA splicing, is most commonly involved (van Kuilenburg et al., 1999; van Kuilenburg, Meinsma, Zoetekouw, & van Gennip, 2002).
There is, however, mounting evidence that additional rare variants may collectively explain an appreciable fraction of patients with DPD deficiency and that these variants may well be candidate risk alleles for fluoropyrimidine toxicity (Offer et al., 2014; van Kuilenburg et al., 2017). Therefore, the identification of novel genetic mechanisms underlying DPD deficiency will not only allow analysis of genotype–phenotype relationships in DPD-deficient patients but also screening of cancer patients at risk.
In this brief report, we present a family with three DPD-deficient patients with an unusual clinical presentation in whom initial sequence analysis of DPYD did not reveal a conclusive genotype. A previously healthy woman with a history of gastrointestinal dysmotility and normal development and intellect experienced significant pregnancy-induced symptoms including severe abdominal pain, increased gastrointestinal symptoms, pancreatitis, and intermittent changes in consciousness in three consecutive pregnancies. Following her third pregnancy, she developed recurrent stroke-like episodes that included symptoms of headaches, blurring to near loss of vision, and right hemiparesis. Her blood pressure remained normal through all three pregnancies. Mild right-sided spasticity persisted after delivery, as did her gastrointestinal dysmotility. Her eldest child, a girl now 19 years old, was diagnosed with congenital blindness and delay of early gross motor and social milestones. By 8 months of age, her visual impairment improved and her development normalized. In her teens, she developed gastrointestinal dysmotility. She is now intellectually normal and in college. A second pregnancy ended in miscarriage due to fetal renal agenesis. The third child, the index case, was a son born with apparent cortical blindness and subsequently exhibited severe developmental delay. At 2 years of age, he had severe spasticity and hyperreflexia felt to be secondary to an intrauterine stroke. Multiple brain MRIs over the first four years of life were consistent with a delay in myelination. He is now 18 years of age and exhibits a clinical picture of spastic paraparesis with mild intellectual disability (ID).
As part of a diagnostic evaluation for inborn errors, the pyrimidine bases uracil and thymine were analyzed in urine and plasma samples of the son (Supp. Methods). Strongly elevated levels of uracil and thymine were observed in urine and plasma, and were subsequently also seen in the mother and daughter (Table 1). The observed uracil and thymine in urine strongly suggested DPD deficiency. Subsequent analysis of the DPD activity in fibroblasts of the mother and son showed no residual activity (<0.006 nmol/mg/hr). In contrast, normal DPD activity was present in the fibroblasts of the father (Table 1). Immunoblot analysis showed that the DPD protein could not be detected in fibroblasts of the affected mother and son whereas a normal amount of DPD protein was observed in the fibroblasts of the unaffected father (Supp. Figure S1).
| Patient (age) | Genotypea | DPD activity (nmol/mg/hr) | Urine (μmol/mmol creatine) | Plasma (μM) | ||
|---|---|---|---|---|---|---|
| Uracil | Thymine | Uracil | Thymine | |||
| Mother (31 years) | g.[97915614 C>T (c.1905+1G>A)]; g.[98.113.121_97.997.390inv; 97.997.386_97.997.389del] | <0.006 | 186 | 135 | 35 | 25 |
| Son (2 years) | g.[97981322C>T (c.1700G>A)]; g.[98.113.121_97.997.390inv; 97.997.386_97.997.389del] | <0.006 | 742 | 380 | 15 | 11 |
| Daughter (3 years) | g.[97915614 C>T (c.1905+1G>A)]; g.[97981322C>T (c.1700G>A)]; | n.a. | 493 | 199 | 19 | 15 |
| Father (33 years) | g.[97981322C>T (c.1700G>A)];[ = ] | 0.7 | n.a. | n.a. | n.a. | n.a. |
| Controls (<3 years)b | 11.8 ± 9.1 (n = 104) | 0.5 ± 0.6 (n = 104) | 0.2 ± 0.4 (n = 40) | 0.05 ± 0.03 (n = 40) | ||
| Controls (>3 years) | 7.1 ± 5.5 (n = 112) | 0.1 ± 0.3 (n = 112) | 0.3 ± 1.0 (n = 57) | 0.01 ± 0.03 (n = 57) | ||
| Controlsc | 1.1 ± 1.2 | |||||
- n.a., not available. Reference sequence of DPYD: Ref Seq NM_000110.3; Ensembl ENST00000370192).
- a Nomenclature according to https://varnomen.hgvs.org/.
- b Data taken from van Kuilenburg et al., (2004).
- c Data taken from van Kuilenburg et al., (2000).
Sequence analysis of all 23 coding exons and flanking intronic regions of DPYD in three family members with a complete DPD deficiency demonstrated the presence of three intronic, one splice-site, and four missense variants (Supp. Table S1). The mother and daughter proved to be heterozygous for the known pathogenic splice-site variant g.97915614C>T [c.1905+1G>A], whereas the son and daughter were heterozygous for the novel missense variant g.97981322C>T [c.1700G>A (p. Gly567Glu)], which Sanger analysis revealed to be inherited from the father (Table 1). The c.1700G > A variant is extremely rare in the general population with an allele frequency of 4.1e−6 (https://gnomad.broadinstitute.org/variant/1-97981322-C-T). In silico analysis using the UMD predictor [https://umd-predictor.eu/index.php [(Salgado et al., 2016) version March 2018, scores of 75–100 indicate pathogenicity] and the Combined Annotation Dependent Depletion tool [CADD, v1.2, scores>15 indicate high probability of deleterious effects, (Kircher et al., 2014)] resulted in scores of 96 and 32, respectively, suggesting that the c.1700G>A variation is likely to be deleterious.
Since potential compound heterozygosity, in line with the recessive DPD deficiency, was identified in the daughter only, we performed copy number analysis on mother and the son but did not identify any deletions or amplifications within DPYD. Quantitative PCR analysis of mRNA coding for DPD in fibroblasts of the mother and her affected son showed that DPD mRNA levels were reduced to 31% and 43%, respectively, whereas the DPD mRNA levels in the father were unaffected (138%) compared with control fibroblasts. Surprisingly, cDNA analysis of the mother and her affected son revealed a discordant genotype. Both the mother and son were heterozygous for the c.85T>C variant in DPYD and the mother was heterozygous for c.1905+1G>A. cDNA analysis using forward and reverse primers located in exons 1 and 8, respectively, showed that the mother and son were heterozygous for the c.85T>C variant (Supp. Table S2). In contrast, only the wild-type sequence was observed using forward and reverse primers located in exons 1 and 9 (Supp. Table S2), respectively, suggesting that an intragenic rearrangement occurred in cis with c.85T>C after exon 8. Furthermore, in cDNA analysis of the mother using forward and reverse primers located in exons 1 and 16 only a smaller sized PCR product was produced, indicating homozygosity for the c.[1741_1905del] mutation (p.581_635del) caused by c.1905+1G>A (Supp. Table S2). Therefore, these results suggested that both the mother and affected son carried an allele containing a DPYD gene with intragenic rearrangements occurring after exon 8.
To search for and identify the second variant in the DPYD that would explain the observed DPD deficiency in this family, we used singleton-WGS (son only) and state-of-the art bioinformatics (i.e., tailored semi-automated bioinformatics approach (Tarailo-Graovac et al., 2016)) (Supp. Methods). Using this approach, we have identified 11 genes (Supp. Table S3) affected by rare impactful variants corresponding to recessive mode of inheritance: (1) homozygous (TNXB and WDR36), (2) compound heterozygous (DNAH11, DPYD, KBTBD12, SCN10A, SNTA1, SRRM2, and ZNF180), and (3) hemizygous (SRPX and VCX2). WGS analysis demonstrated a rare heterozygous DPYD variant in the son: the Illumina paired-end reads from introns 8 and 12 of the DPYD partially mapped to both introns 8 and 12, suggesting the presence of an inversion (Figure 1A) with a base-pair resolution. The WGS results were confirmed using PCR analyses combined with Sanger sequencing and showed that both the mother and the son were heterozygous for the 115,731 bp inversion with the breakpoints at chr1: 98,113,121 (intron 8) and chr1: 97,997,390 (intron 12), as well as a small deletion of four nucleotides (GAAA) crh1: 97,997,386_97,997,389del (Figure 1).
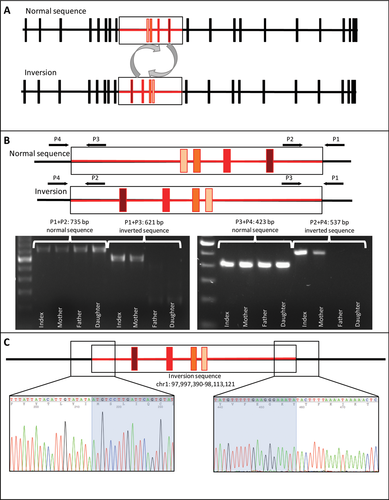
To perform deeper genomic analysis for potential variants that may be contributing to the ID observed in the son, pancreatitis observed in the mother, as well as the visual impairment observed in both the son (cortical blindness) and the affected daughter (resolved with now normal vision) but which are atypical for DPD deficiency, the duo-WES analysis (affected daughter–unaffected mother) was performed. These analyses of WES data and mtDNA did not reveal pancreatitis pre-disposing variants and explanations for the transient vision loss.
The missense variant c.1700G>A was investigated in more detail by functional expression of the DPD mutant in mammalian cells and subsequent analysis of the DPD activity and protein expression levels. The protein expression and DPD activity of the recombinant DPD mutant was comparable to that observed for the wild-type DPD enzyme (Supp. Figure S2). To investigate whether the p.Gly567Glu mutation would be associated with an increased temperature lability of the DPD enzyme, the activity of DPD was assayed at different temperatures. A gradual increase in activity was observed for both the wild-type and mutant DPD enzyme between 37°C and 45°C, followed by a drop at 50°C (Supp. Figure S2). At 45°C, the DPD activity of the mutant enzyme was approximately 11% lower than that of the wild-type enzyme. Analysis of the DPD activity in fibroblasts of the affected son, a carrier of the c.1700G>A variant, showed that the DPD activity (<0.006 nmol/mg/hr, n = 3) was undetectable for fibroblasts cultured at 37°C but a very low DPD activity (0.024 ± 0.002 nmol/mg/hr, n = 3) could be detected for fibroblasts grown at 30°C.
To analyze potential structural effects of the p.G567E mutation, we generated a three-dimensional model of the human enzyme based on the crystal structure of recombinant pig DPD. From the 1025 amino acids per subunit, 92.8% are strictly conserved between both enzymes, and the majority of the nonidentical amino acids is conservatively exchanged or located on the protein surface. It can therefore be expected that the molecular structures of human and pig DPD are highly similar and that the analysis of the three-dimensional context of the mutation site in the homology model allows confident predictions about structural effects of the resulting amino acid exchange. G567 is strictly conserved in available DPD sequences, most likely because it serves as the C-capping residue of the first α8β8-barrel helix of the FMN-binding domain (Supp. Figure S3), and has a backbone dihedral angle (Φ/Ψ) combination disallowed for nonglycine residues. It is located ca. 20–25 Å from the FMN and pyrimidine binding site, and thus direct effects of the p.G567 mutation on substrate binding or active site architecture can be excluded. Its main chain oxygen forms a hydrogen bond with the carboxamide-nitrogen atom of p.N544, which in turn is hydrogen-bonded to the backbone amide of p.F546 from the first barrel-strand via its carboxamide-oxygen. Since this site is solvent-exposed the replacement of p.G567 by the larger (and charged) glutamate does not cause steric clashes. Preservation of both the C-capping interactions with p.A563 and the hydrogen bond to p.N544 will however be energetically less favorable for a glutamate due to the disallowed Φ/Ψ-angle combination, and structural adjustments, possibly accompanied by increased flexibility, of this region involved in dimerization can be expected. Surprisingly, the unaffected enzymatic activity of the mutant DPD indicates that this is not detrimental to enzyme function, at least not in vitro under the assay conditions. Detection of low residual DPD activity at a fibroblast cultivation temperature of 30°C, and of no residual DPD activity at 37°C, is in line with the predicted slightly impaired dimerization of the enzyme.
Although analysis of the crystal structure of DPD suggested that the p.Gly567Glu mutation might affect the dimerization of DPD and thus the activity of the enzyme a clear pathogenic role of this novel variant could not be shown and the c.1700G>A still remains a variant of unclear significance. Of the three additional missense variants, only the c.496A>G (p.Met166Val) was in cis with the c.1700G>A variant (Supp. Table S1). The two mutation sites are about 26 Å distant from each other and there is no apparent direct interaction between the two variants. For the c.496A>G (p.M166V) variant it has been shown that the mutant protein has a mildly reduced DPD activity compared with that observed for the wild-type enzyme (van Kuilenburg et al., 2016). Thus, the presence of this variant, in addition to the c.1700G>A (p.Gly567Glu) variant might affect the degree of residual DPD activity in the son. However, we cannot exclude the possibility that additional genetic mechanisms, undetectable by WGS analysis, underlie the observed DPD deficiency in the son and affected daughter. In this respect, it is worthwhile to note that the bioavailability of DPD is affected not only by pathogenic mutations in DPYD but also by epigenetic and posttranslational processes such as the regulation by two microRNAs miR-27a and miR-27b (Offer et al., 2014). However, the affected son was heterozygous, whereas the affected daughter was homozygous wild-type (A/A) for a common A>G polymorphism in MIR27A, the gene encoding miR-27a, associated with increased expression of miR-27a and a reduced DPD activity.
Although considerable phenotypic variability has been reported in patients with complete DPD deficiency, the majority presents with neurological features (van Kuilenburg et al., 2002; van Kuilenburg et al., 1999). The most conspicuous clinical abnormalities encountered in our patients were pregnancy-induced symptoms (mother and daughter), visual impairment (cortical blindness in both children), and gastrointestinal dysmotility (mother and daughter). Severe intellectual developmental disorder with delayed myelination in the brain was present in the son, but his neurologic status was complicated by a presumed in utero stroke. Cognitive impairment is one of the most frequently encountered clinical presentations of DPD-deficient patients, but both mother and daughter in this family were of normal intelligence with college educations (van Kuilenburg et al., 1999). Brain malformations on neuroimaging have only occasionally been reported in DPD-deficient patients (Brussel, van Kuilenburg, & Janssens, 2006; Chen et al., 2014; Enns et al., 2004). Delayed myelination is not a malformation and overall an unspecific feature. The transient cortical blindness is more difficult to explain. An appreciable number of DPD patients have presented with ocular abnormalities, but to our knowledge no transient visual impairment has been reported (van Kuilenburg et al., 1999). Cortical blindness is a central nervous system phenomenon, and in our cases possibly secondary to the myelination abnormalities. Alternatively, in utero exposure to high pyrimidine metabolite concentration due to maternal DPD deficiency may have contributed to the visual symptoms in both children.
Whole genome sequencing of the affected son identified 10 additional genes containing rare and impactful variants corresponding to a recessive mode of inheritance. However, these genes have not been associated with the clinical presentation as reported for the three DPD-deficient patients. Furthermore, WES and mtDNA analysis of the mother and daughter did not reveal pancreatitis predisposing variants and explanations for the transient vision loss. Extensive genome analysis did not identify other candidate genes, apart from DPYD, which could be reasonably assumed to explain the observed phenotypic features of the DPD-deficient patients. The observation that some patients with a complete DPD deficiency do not present with any clinical abnormalities suggests that other factors are involved in determining the clinical outcome (van Kuilenburg et al., 1999). Although whole genome sequencing allows detection of variants in the non-protein-coding regions of the genome, currently there are limited possibilities to interpret pathogenic potential of the variants affecting non-protein-coding regions that may affect temporal and spatial protein expression.
In the present study, unambiguous evidence for a large (∼116 kb) inversion of DPYD was demonstrated in two out of three family members. Polymorphic inversions are a type of structural genomic variants which are difficult to analyze owing to their balanced nature and the location of breakpoints within complex repeated regions. To date, only a handful of inversions have been studied in humans and their possible functional effects are still largely unknown (Puig, Casillas, Villatoro, & Caceres, 2015). Inversions are often generated by nonallelic homologous recombination between inverted repeats or by double-strand break repair mechanisms (Puig et al., 2015). In this respect, it is interesting to note that the inversion, encompassing intron 8–12, was located in the common fragile site FRA1E which extends from intron 8 to 18 within DPYD (Hormozian, Schmitt, Sagulenko, Schwab, & Savelyeva, 2007). Common fragile sites represent chromosomal structures that are particularly prone to breakage under replication stress and the genomic instability can give rise to deletions, amplifications, and translocations (Hormozian et al., 2007). Several molecular mechanisms have been proposed by which inversions can lead to phenotypic consequences including the suppression of recombination within the inverted sequence in heterozygotes; direct disruption of the coding sequence of a gene and altering gene expression of adjacent genes by separating regulatory elements from the corresponding coding sequences (Puig et al., 2015). In our case, the deleterious effect of the inversion encompassing introns 8–12 of DPYD is most likely due to the disruption/truncation of the coding sequence of the DPYD gene.
An interesting observation is the fact that inversions may lead to an increased predisposition to acquire other rearrangements in the same genomic region in particular deletions (Puig et al., 2015). Previously, we showed that genomic deletions affecting DPYD occurred in 7% of pediatric patients with a complete DPD deficiency (van Kuilenburg et al., 2009). Genomic rearrangements in DPYD have also been observed in patients with ID (Willemsen et al., 2011), autism-spectrum disorder (Carter et al., 2011; Prasad et al., 2012), syndromic obesity (D'Angelo, Moller Dos Santos, Alonso, & Koiffmann, 2015), and amyotrophic lateral sclerosis (Uyan et al., 2013). To date, no clear genotype–phenotype correlation has been established in patients with a DPD deficiency but it is noteworthy that patients with gross deletions in DPYD presented with a severe phenotype when compared with that observed in other DPD patients (van Kuilenburg et al., 2009).
DPD deficiency has been shown to be a major determinant of fluoropyrimidine-associated toxicity (Meulendijks et al., 2015). Patients with a partial or complete DPD deficiency have a strongly reduced capacity to degrade 5FU and are, therefore, at risk to develop severe and sometimes lethal toxicity (van Kuilenburg, 2004). Thus, there is an increased awareness that genotyping of DPYD will enable the identification of patients at risk prior to the start of the therapy with fluoropyrimidines and allow the adjustment of the initial fluoropyrimidine dose (Swen et al., 2011). However, in a significant number of cancer patients with a reduced DPD activity, no mutations could be identified in the coding part of DPYD (van Kuilenburg et al., 2000). Therefore, screening for the presence of the novel inversion in DPYD should be considered in case initial sequence analysis does not reveal pathogenic mutations.
ACKNOWLEDGMENTS
The authors thank the patient and family for participating in this research study, and acknowledge Ms. A. Ghani for research coordination, Ms. X. Han and Ms. M. Higginson for sample handling and Sanger analysis, Ms. E. Lomba for research administration. This research did not receive any specific grant from funding agencies in the commercial sector.
DISCLOSURE STATEMENT
The authors declare no conflict of interest.