Revisiting genotype–phenotype overlap in neurogenetics: Triplet-repeat expansions mimicking spastic paraplegias†
For the Databases in Neurogenetics Special Issue
Abstract
Hereditary spastic paraplegias (HSPs) constitute a heterogeneous group of neurological disorders, characterized primarily by progressive spasticity and weakness of the lower limbs. HSPs are caused by mutations in multiple genes (at least 48 loci and 28 causative genes). The clinical spectrum of HSPs is wide and important differences have been reported between patients with distinct mutations in the same gene, or even between different family members bearing the same mutation. Many patients with HSP present clinical deficits related to the involvement of neuronal systems other than corticospinal tracts, namely, peripheral nerves, sensory, or cerebellar pathways. These cases may be difficult to differentiate from other neurological diseases (e.g., hereditary ataxias), also genetically and clinically heterogeneous. As an illustration of how overlapping this genotype–phenotype relationship is, and the difficulties that it brings upon the development of neurogenetic algorithms and databases, we review the main clinical and genetic features of HSPs, and summarize reports on cases of triplet-repeat spinocerebellar ataxias that can mimic HSP phenotypes. This complex scenario makes the necessity of high-quality, curated mutation databases even more urgent, in order to develop adequate diagnostic guidelines, correct interpretation of genetic testing, and appropriate genetic counseling. Hum Mutat 33:1315–1323, 2012. © 2012 Wiley Periodicals, Inc.
Introduction
Hereditary spastic paraplegias (HSPs), or familial spastic paraparesis, are a group of clinically and genetically heterogeneous forms of neurodegenerative disorders mainly affecting the motor neurons. HSPs are considered rare worldwide (1.3–9.6/100,000 [Erichsen et al., 2009]), and no curative treatment is currently available for these disorders. The understanding of their etiology is essential to provide adequate diagnosis and genetic counseling. Although there are two subtypes, SPG4 and SPG3A, which represent the most frequent mutations [Depienne et al., 2007], there is an increasing number of loci/genes underlying a broad spectrum of HSP phenotypes (Table 1). This situation of both clinical and genetic heterogeneity, and ever-growing new data, places an enormous challenge for the development of a rational diagnostic algorithm for this group of neurogenetic disorders [Gasser et al., 2010]. Thus, diagnostic paradigms are continuously changing, and are difficult to translate into useful guidelines for the clinical community. In this scenario, the availability of thorough, high-quality, curated databases of genetic variants identified in patients becomes even more crucial.
| Locus | Inheritance/type | Location | Gene | Protein | OMIM ID # |
|---|---|---|---|---|---|
| SPG1 | X-linked/complicated | Xq28 | L1CAM | Neuronal cell adhesion molecule L1 | 303350 |
| SPG2 | X-linked/complicated | Xq22 | PLP1 | Proteolipid protein 1 | 312920 |
| SPG3A | AD/pure and complicated | 14q22.1 | ATL1 | Atlastin-1 | 182600 |
| SPG4 | AD/pure and complicated | 2p24-p21 | SPAST | Spastin | 182601 |
| SPG5A | AR/pure | 8q21.3 | CYP7B1 | 25-Hydroxycholesterol 7-alpha-hydroxylase | 270800 |
| SPG6 | AD/pure | 15q11.2 | NIPA1 | Magnesium transporter NIPA1 | 600363 |
| SPG7 | AR/pure and complicated | 16q24.3 | SPG7 | Paraplegin | 607259 |
| SPG8 | AD/pure | 8q24.13 | KIAA0196 | WASH complex subunit strumpellin | 603563 |
| SPG9 | AD/complicated | 10q23.3-q24.2 | – | – | 601162 |
| SPG10 | AD/pure and complicated | 12q13.13 | KIF5A | Kinesin heavy chain isoform 5A | 604187 |
| SPG11 | AR/pure and complicated | 15q13-q15 | SPG11 | Spatacsin | 604360 |
| SPG12 | AD/pure | 19q13 | RTN2 | Reticulon-2 | 604805 |
| SPG13 | AD/pure | 2q33.1 | HSPD1 | 60 kDa heat shock protein, mitochondrial | 605280 |
| SPG14 | AR/complicated | 3q27-q28 | – | – | 605229 |
| SPG15 | AR/complicated | 14q23.3 | ZFYVE26 | Zinc finger FYVE domain-containing protein 26 (spastizin) | 270700 |
| SPG16 | X-linked/pure and complicated | Xq11.2 | – | – | 300266 |
| SPG17 | AD/complicated | 11q13 | BSCL2 | Seipin | 270685 |
| SPG18 | AR/complicated | 8p11.2 | ERLIN2 | Erlin-2 | 611225 |
| SPG19 | AD/pure | 9q33-q34 | – | – | 607152 |
| SPG20 | AR/complicated | 13q13.1 | SPG20 | Spartin | 275900 |
| SPG21 | AR/complicated | 15q21-q22 | SPG21 | Maspardin | 248900 |
| SPG22 | X-linked/complicated | Xq13.2 | SLC16A2 | Monocarboxylate transporter 8 | 300523 |
| SPG23 | AR/complicated | 1q24-q32 | – | – | 270750 |
| SPG24 | AR/pure | 13q14 | – | – | 607584 |
| SPG25 | AR/complicated | 6q23-q24.1 | – | – | 608220 |
| SPG26 | AR/complicated | 12p11.1-q14 | – | – | 609195 |
| SPG27 | AR/pure and complicated | 10q22.1-q24.1 | – | – | 609041 |
| SPG28 | AR/pure | 14q21.3-q22.3 | – | – | 609340 |
| SPG29 | AD/complicated | 1p31.1-p21.1 | – | – | 609727 |
| SPG30 | AR/complicated | 2q37.3 | KIF1A | Kinesin-like protein KIF1A | 610357 |
| SPG31 | AD/pure and complicated | 2p11.2 | REEP1 | Receptor expression-enhancing protein 1 | 610250 |
| SPG32 | AR/complicated | 14q12-q21 | – | – | 611252 |
| SPG33 | AD/pure | 10q24.2 | ZFYVE27 | Protrudin | 610244 |
| SPG34 | X-linked/pure | Xq24-q25 | – | – | 300750 |
| SPG35 | AR/complicated | 16q23 | FA2H | Fatty acid 2-hydroxylase | 612319 |
| SPG36 | AD/complicated | 12q23-q24 | – | – | 613096 |
| SPG37 | AD/pure | 8p21.1-q13.3 | – | – | 611945 |
| SPG38 | AD/complicated | 4p16-p15 | – | – | 612335 |
| SPG39 | AR/complicated | 19p13.2 | PNPLA6 | Neuropathy target esterase | 612020 |
| SPG40 | AD/pure | – | – | – | –a |
| SPG41 | AD/pure | 11p14.1-p11.2 | – | – | 613364 |
| SPG42 | AD/pure | 3q24-q26 | SLC33A1 | Acetyl-coenzyme a transporter 1 | 612539 |
| SPG43 | AR/complicated | 19q12 | C19orf12 | Uncharacterized protein C19orf12 | 35198b |
| SPG44 | AR/complicated | 1q41-q42 | GJC2 | Gap junction gamma-2 protein | 613206 |
| SPG45 | AR/complicated | 10q24.3-q25.1 | – | – | 613162 |
| SPG46 | AR/complicated | 9p21.2-q21.12 | – | – | 614409 |
| SPG47 | AR/complicated | 1p13.2 | AP4B1 | AP-4 complex subunit beta-1 | 37654b |
| SPG48 | AR/unclear | 7p22.1 | KIAA0415 | Protein KIAA0415 | 613647 |
- aThis locus does not have an OMIM or HGNC code, it was suggested by Subramony et al. [Subramony et al., 2009].
- bThese loci were not found at OMIM, the reference shown corresponds to HUGO Gene Nomenclature Committee (HGNC) ID number (available at: http://www.genenames.org/).
- AD, autosomal dominant; AR, autosomal recessive; OMIM, online Mendelian inheritance in man (available at: http://www.ncbi.nlm.nih.gov/omim).
In the present review, the authors summarize the clinical spectrum of HSPs and of the loci/genes that have been identified for HSPs. Although it is well recognized that several groups of neurogenetic disorders present more or less degree of clinical and/or genetic overlap with HSPs (including amyotrophic lateral sclerosis, spastic ataxias, peripheral neuropathies, neurometabolic diseases), there is less awareness on the overlap between HSPs and spinocerebellar ataxias (SCAs) caused by triplet-repeat expansions, which can occasionally mimic HSPs, particularly at the disease onset. To illustrate the evolving complexity of adapting diagnostic strategies to the rapid accumulation of data and new genomic technologies, we compare two possible diagnostic algorithms for HSPs. The authors did not intend to propose here an exhaustive diagnostic guideline for HSPs—which, in our view, should be endorsed by a commission or expert-group—but to illustrate two approaches that can be used and that may coexist in the current time of transition to a new genomic medicine: a “classical genetics” algorithm and another, more “innovative” strategy that would make use of the novel high-throughput genomic technologies combined with information available in both curated and noncurated databases.
Clinical Features and Classification of HSP
HSP is characterized by progressive spasticity and weakness of the lower limbs due to corticospinal tract dysfunction. It may also result in brisk reflexes, extensor plantar responses, muscle weakness, and urinary urgency [Depienne et al., 2007; Fink, 2003; Salinas et al., 2008]. The clinical diagnosis of HSP depends on family history, disease-course, neurological signs upon examination, and exclusion of other causes of spasticity. Cerebral and spinal MRI help to rule out other frequent neurological diseases, such as myelopathies, multiple sclerosis, leukodystrophies, or spinal tumors [Depienne et al., 2007]. In addition, these clinical studies are important to detect specific abnormalities that can sometimes be found in HSP, such as white matter changes or corpus callosum atrophy [Depienne et al., 2007].
Several classifications have been proposed for HSPs based on the mode of inheritance, presence of additional clinical features, and age at onset. Clinically, they have been classified as pure (or uncomplicated) and complicated (or complex) forms [Harding, 1983]. Pure subtypes present bilateral lower extremity spasticity and weakness in variable degree, hyperreflexia, extensor plantar responses, and mildly impaired distal vibratory sensation, comprising patients whose manifestations are limited to corticospinal signs until later stages of the disease [Coutinho et al., 1999; Fink, 2006; Harding, 1983]. Complicated forms may include additional neurological and/or extra-neurological signs, such as mental retardation, ichthyosis, pigmentary retinal degeneration, optic atrophy, amyotrophy, extrapyramidal features, sensory neuropathy, ataxia, dysarthria, and epilepsy, among others [Coutinho et al., 1999; Harding, 1983]. A revised classification of complicated HSPs has been proposed by Reid [Reid, 1997; Reid, 1999].
Age at onset is variable, from early childhood through the seventh decade of age or even later [Harding, 1983; Salinas et al., 2008]. Pure HSP forms can be further subdivided according to the age at onset. Early onset forms (typically, before 35 years) show a slow and variable evolution while later onset cases progress faster, usually with more marked muscle weakness, urinary symptoms, and sensory loss [Espinos and Palau, 2009; Fink, 2006; McDermott et al., 2000]. Age at onset, rate of progression, and degree of disability are variable not only between different genetic subtypes of HSP, but also within individual families in which all subjects present the same mutation [Fink and Hedera, 1999], possibly indicating additional influence of modifier genes and/or different exposures to environmental factors.
Genetic Classification of HSP
Due to the high clinical heterogeneity within HSP subtypes and clinical overlap between subtypes, a molecular classification seems more comprehensive, especially when individual families often cannot fit the criteria of a single clinical subtype. Genetically defined HSPs are assigned with the symbol SPG (“spastic gait”) followed by a number. To date, at least 48 distinct SPG loci have been mapped, and 28 causative genes have been reported in the literature, but this list is rapidly growing. Autosomal dominant (AD) transmission accounts for the majority of cases, but autosomal recessive (AR) and X-linked forms have also been described (Table 1). Several explanations may account for the apparently sporadic occurrence encountered frequently in clinical practice: (1) reduced or age-dependent penetrance; (2) de novo mutations; (3) premature death of the transmitting parent or undiagnosed symptoms; (4) AR or X-linked inheritance in small kindreds [Depienne et al., 2007].
Until recently, AD-HSP forms were thought to be mostly “pure” in clinical terms and to be associated with a later onset, whereas AR-HSP forms tended to be more complex and associated with an earlier onset [Depienne et al., 2007; Espinos and Palau, 2009; Stevanin et al., 2008]. However, with the increasing number of reports in the literature, the clinical spectrum of each genetic HSP subtype has widened, and differences among distinct genetic subtypes are not so clear-cut. For example, complex forms have been linked to AD-HSP loci, namely, SPG4, as well as SPG10, among others [Goizet et al., 2009b; Heinzlef et al., 1998; Mead et al., 2001; Nielsen et al., 2004; Ribai et al., 2008; Stevanin et al., 2008], and pure forms have been associated with AR-HSP loci, such as SPG5, and others [Goizet et al., 2009a; Stevanin et al., 2008].
Moreover, HSP phenotypes may be mimicked by mutations in loci associated with other groups of neurological disorders. In addition to infectious, inflammatory and other noninherited diseases, genetic neurometabolic disorders—such as lysosomal storage diseases, vitamin E deficiency, disorders of lipid metabolism, and others—may have pyramidal signs and spastic gait as primary manifestation. More recently, presenilin1 mutations have been shown in patients with spastic paraplegia [Jimenez Caballero et al., 2008]. In addition, several subtypes of hereditary ataxias, which will be further detailed in the subsequent section, can also show clinical manifestations overlapping with those of HSPs.
Clinical Overlap Between Hereditary Ataxias and Hereditary Spastic Paraplegias
Hereditary ataxias (HAs) also comprise a clinically and genetically heterogeneous group of rare neurological disorders, globally affecting ∼6/100,000 individuals [Schoenberg, 1978; Tallaksen, 2008]. HAs are characterized by progressive degeneration of the cerebellum and spinocerebellar tracts, associated with signs of the central and peripheral nervous system, in variable combinations. They are typically characterized by poor balance with falls, imprecise coordination, postural or kinetic tremor, dysarthria and dysphagia, sometimes also associated with vertigo, diplopia, and other manifestations [Fogel and Perlman, 2007]. Different types of mutations and molecular mechanisms have been associated with HAs [Schols et al., 2004]. Including Friedreich's ataxia, caused by a GAA expansion in the first intron of the frataxin gene [Campuzano et al., 1996], and polyglutamine-coding CAG tracts causing several autosomal dominant SCAs, triplet-repeat expansions in specific genes represent the most frequent genetic cause of ataxia [Durr, 2010; Koeppen, 2011]. Herein, with the purpose of illustrating some examples of the clinical overlap between HSPs and HAs, we will make a brief overview of the well-known causes of spastic ataxias, but we will also give a special emphasis to the less common reports of pyramidal syndrome as the major or first presentation of HAs caused by triplet-repeat expansions (Table 2).
| Disease/gene | Inheritance | Locus | Repeat motif | Normal range | Intermediate/premutation range | Pathologic range | OMIM ID # | Mean onset (range) | Main clinical features |
|---|---|---|---|---|---|---|---|---|---|
| Unstable repeats in coding regions (polyglutamine tracts) | |||||||||
| DRPLA/ATN1 | AD | 12p13.31 | (CAG)n | 6–35 | – | 48–93 | 125370 | 31 years (1–67) | Combination of myoclonus, epilepsy, ataxia, choreoathetosis, and dementia in the elderly or mental retardation in children. |
| SCA1/ATXN1 | AD | 6p22.3 | (CAG)n | 6–38 | 39–44 | 45–83 | 164400 | 37 years (15–65) | Progressive cerebellar ataxia, dysarthria, and eventual deterioration of bulbar functions. Patients may also present with pyramidal signs and peripheral neuropathy. |
| SCA2/ATXN2 | AD | 12q24.12 | (CAG)n | 14–31 | 32–35 | 36–500 | 183090 | 33 years (2–68) | Ataxia, dysarthria, slow saccades, hyporreflexia, titubation, dementia, and rarely parkinsonism. |
| MJD (SCA3)/ATXN3 | AD | 14q32.12 | (CAG)n | 12–44 | 45–51 | 52–87 | 109150 | 40 years (4–70) | Progressive cerebellar ataxia and pyramidal signs, and a complex clinical picture extending from extrapyramidal signs to peripheral amyotrophy. Minor but more specific features include external progressive ophthalmoplegia, dystonia, intention fasciculation-like movements of facial and lingual muscles, and bulging eyes. |
| SCA7/ATXN7 | AD | 3p14.1 | (CAG)n | 4–33 | 34–36 | 37–460 | 164500 | 33 years (1–72) | Progressive cerebellar ataxia, dysarthria, dysphagia, and cone-rod and retinal dystrophy with progressive central visual loss resulting in blindness. |
| Unstable repeats in noncoding regions | |||||||||
| FRDA/FXN | AR | 9q21.11 | (GAA)na | 5–33 | 34–65 | 66–1700 | 229300 | 10–15 years (2–75) | Progressive gait and limb ataxia, dysarthria, absent or retained deep tendon reflexes, sensory loss, and pyramidal signs. Cardiomyopathy, axonal sensory neuropathy, distal wasting, pes cavus, scoliosis, sensorineural deafness, optic atrophy, and diabetes are also frequent. |
- aOnly 2% of FRDA patients are compound heterozygous, presenting a point mutation in one of the alleles and a GAA expansion in the other.
- DRPLA, dentatorubral-pallidoluysian atrophy; SCA1, spinocerebellar ataxia type 1; SCA2, spinocerebellar ataxia type 2; MJD (SCA3), Machado–Joseph disease (spinocerebellar ataxia type 3); SCA7, spinocerebellar atacia type 7; FRDA, Friedreich ataxia; AD, autosomal dominant; AR, autosomal recessive; OMIM, online Mendelian inheritance in man (available at: http://www.ncbi.nlm.nih.gov/omim).
Spastic Ataxias
The group of spastic ataxias includes a wide range of neurological syndromes in which spinocerebellar and pyramidal pathways are affected. At least six loci have been mapped specifically for diseases called as spastic ataxias: SPAX1 (12p13; MIM# 108600); SPAX2 (17p13; MIM# 611302); SPAX3 (2q33-q34; MIM# 611390); SPAX4 (10p11.23; MIM# 613672); SPAX5 (18p11.21; MIM# 614487); and SPAX6 (13q11; MIM# 270550), usually known as Autosomal Recessive Spastic Ataxia of Charlevoix-Saguenay (ARSACS). Up to date, only MTPAP, AFG3L2, and SACS genes have been identified as the causative genes for SPAX4, SPAX5, and ARSACS, respectively. A mutation in the MTPAP gene, found only in an Amish family, was reported as the cause of SPAX4 [Crosby et al., 2010]. It is interesting to note that the AFG3L2 gene, previously described as the causative gene of SCA28 (AD) [Di Bella et al., 2010], was recently described to be also responsible for SPAX5 (AR) in a Hispanic family [Pierson et al., 2011]. ARSACS is an early-onset spastic ataxia, highly prevalent in the Charlevoix–Saguenay-Lac-Saint-Jean (carrier frequency 1/22) region of Quebec [Engert et al., 2000], and a frequent cause of early-onset cerebellar ataxia in European countries [Vermeer et al., 2008]. This disease is characterized by spasticity, dysarthria, nystagmus, distal muscle wasting, finger and foot deformities, mixed sensorimotor neuropathy, and retinal hypermyelination [Breckpot et al., 2008; Pedroso et al., 2011]. Spasticity in the lower limbs often occurs when there is still little sign of cerebellar dysfunction [Breckpot et al., 2008], evidencing the clinical overlap of ARSACS with HSPs.
Another disease associated with a phenotype that may be classified as a spastic ataxia is SPG7, which can present as a complicated HSP phenotype, often including cerebellar signs [Brugman et al., 2008; Elleuch et al., 2006; Warnecke et al., 2007; Wilkinson et al., 2004], further exemplifying the phenotypic overlap between HSPs and HAs.
HSP-Like Phenotypes Caused by Expansions in Triplet-Repeat Ataxia Loci
In addition to the spastic ataxias mentioned above, there is less awareness of the cases where triplet-repeat ataxia mutations have been found to cause a phenotype that mimics HSP (Table 2). We herein review some of these cases from the published literature, and describe additional examples from the personal experience of the authors.
Dentatorubral-pallidoluysian atrophy
Kurohara and colleagues [Kurohara et al., 1997] reported a consanguineous family, clinically diagnosed with an autosomal recessive form of spastic paraplegia, in which both patients were homozygous for intermediate size alleles (41–40 CAG repeats) in the dentatorubral-pallidoluysian atrophy (DRPLA) locus. Although the involvement of the corticospinal tracts has been reported in other patients with DRPLA, spastic paraplegia had not been noted previously as a predominant feature of this disease. Furthermore, DRPLA is transmitted in an autosomal dominant manner, whereas this atypical phenotype seemed to follow an autosomal recessive pattern of inheritance because two alleles of intermediate size were apparently necessary to cause the disease and the heterozygous parents of these patients were not affected. These authors suggested that trinucleotide expansions should also be screened in patients with neurodegenerative diseases of recessive inheritance [Kurohara et al., 1997].
Spinocerebellar ataxia type 1
Although pyramidal signs, and specifically spasticity, are thought to be frequent clinical features in spinocerebellar ataxia type 1 (SCA1) [Abele et al., 1997; Bauer et al., 2005; Burk et al., 1996; Durr, 2010; Ranum et al., 1994; Zhou et al., 2001; Zuhlke et al., 2002], we have not detected reports in the literature specifically describing SCA1 cases with pyramidal syndrome as the major or the first presentation. We screened the ATXN1 expansion, by fragment analysis, as well as triplet-repeat primed polymerase chain reaction (PCR) in a series of 161 Spanish patients, half of whom show complicated forms of HSP, including 23 with cerebellar signs. However, no expanded ATXN1 alleles were detected (detailed methods available upon request).
Spinocerebellar ataxia type 2
Miyaji and colleagues [Miyaji et al., 2010] described a female patient diagnosed with a spastic paraplegia of unknown cause, who was found to have an expanded ATXN2 allele (38 CAG repeats), but no major clinical features of spinocerebellar ataxia type 2 (SCA2). In the light of this report, and because SCA2 is one of the most frequent SCAs in Spain [Infante et al., 2005; Pujana et al., 1999], we screened the ATXN2 expansion, by fragment analysis, as well as triplet-repeat primed PCR in our series of 161 Spanish HSP patients. We did not observe any instance of expanded ATXN2 alleles in these cases.
Machado–Joseph disease/spinocerebellar ataxia type 3
A fifth Machado–Joseph disease (MJD) clinical type was proposed by Sakai and Kawakami [Sakai and Kawakami, 1996], after observing two MJD patients presenting spastic paraplegia without cerebellar signs. Additional reports of families with different origins, namely, Japanese [Kaneko et al., 1997], German [Landau et al., 2000], Brazilian [Teive et al., 2001], and Chinese [Gan et al., 2009], clinically diagnosed with HSP, but presenting an expanded allele at the MJD locus (66–86 CAG repeats), were subsequently described. Moreover, the screening of the MJD mutation in a larger Chinese series of AD-HSP patients [Wang et al., 2009], revealed that expanded ATXN3 alleles (64–81 CAG repeats) were responsible for the disease in 13% of the studied AD-HSP families. We screened the ATXN3 expansion, by fragment analysis and triplet-repeat primed PCR (detailed methods available upon request), in 161 Spanish HSP patients. Only one patient tested positive, a boy with complicated HSP (spastic tetraplegia, borderline intellectual performance, and cerebellar ataxia) with onset in the first months of life. His brain magnetic resonance image showed bilateral frontoparietal demyelination, together with atrophy of the corpus callosum and slight atrophy of the upper cerebellar vermis. This patient had suffered a mild perinatal hypoxia, which seemed insufficient to justify the severe symptoms. An expanded ATXN3 allele, with 71 CAG repeats, was identified in this patient. A cerebellar syndrome, with onset in the third decade, was present in his father, whereas his grandfather had much milder and later-onset cerebellar manifestations. We hypothesize that the expanded MJD/SCA3 (spinocerebellar ataxia type 3) allele is probably contributing to the severe spastic ataxia of this patient. It can also be speculated that the expansion mutation may have caused a special vulnerability of the corticospinal tracts to a mild hypoxia. However, given the severe spasticity and thin corpus callosum, we ruled out SPG3A and SPG4 mutations, and the screening of additional HSP genes is currently underway. This case emphasizes the difficulties in dissecting the neurological phenotypes and the complexity of etiological evaluation in many cases.
Spinocerebellar ataxia type 7
A family clinically diagnosed with spastic paraplegia, in the absence of any definitive cerebellar or visual impairment, was reported by Linhares and colleagues [Linhares Sda et al., 2008] as having an expanded ATXN7 allele (38–43 CAG repeats), thus pointing to the CAG expansion at the spinocerebellar ataxia type 7 (SCA7) locus as an additional possible cause of HSP-like manifestations. In addition to this case report of HSP-like phenotype, and similarly to SCA1 and SCA3, spasticity is a clinical feature frequently observed in SCA7 patients [David et al., 1998; Durr, 2010; Giunti et al., 1999; Gu et al., 2000; Inaba et al., 2009; Johansson et al., 1998].
Friedreich ataxia
Gates and colleagues [Gates et al., 1998] described the first case of a male patient who, after being initially diagnosed with a spastic paraplegia, was later found to have two expanded Friedreich ataxia (FRDA) alleles (∼690/1040 GAA repeats), and subsequently diagnosed with an atypical FRDA phenotype. Later, additional patients were reported with the diagnosis of a spastic paraplegia-like syndrome, and for whom molecular analysis revealed the presence of two expanded FRDA alleles (>150 GAA repeats each) [Badhwar et al., 2004; Castelnovo et al., 2000; Lhatoo et al., 2001; Silvers and Felice, 2000; Webb et al., 1999]. Although point mutations are rare in the FRDA locus, there have been reports of compound heterozygous patients (GAA expansion + point mutation) presenting a phenotype that resembles spastic paraplegia [Bidichandani et al., 1997; Cossee et al., 1999; Diehl et al., 2010; McCabe et al., 2002]. Indeed, atypical FRDA presentations are reported for a considerable percentage of FRDA patients [Berciano et al., 2002]. Specifically, adult onset spastic ataxias represent a distinctive FRDA subtype [Berciano et al., 2002; Ragno et al., 1997]. Several phenotypes associated with spasticity may, therefore, be explained by mutations in the frataxin gene.
Potential Evolution of Diagnostic Algorythms in HSP
The broad genetic heterogeneity, together with the difficulties to clinically distinguish between HSP forms, and the advent of high-throughput sequencing technologies are rapidly changing the way in which the diagnosis of these diseases is approached [Ku et al., 2011; Tsuji, 2010]. For the molecular evaluation of ataxias and spastic paraplegias, the EFNS has produced diagnostic guidelines [Gasser et al., 2010]. Emphasizing the complexity of these groups of disorders, these guidelines provide a good orientation for genetic testing, according to the main symptoms. As a reflection exercise, we present herein two possible genetic diagnostic algorithms for HSP. These algorithms are not intended to be taken strictly (guidelines of these type are necessarily evolving in the light of advancements of knowledge and technologies) but as temptative orientation. They serve to illustrate what could be the future evolution of guidelines for the genetic study of heterogeneous groups of neurogenetic diseases from a “classical,” gene-by-gene, strategy, to a more “innovative” approach based on high-throughput techniques and the availability of databases of genetic variation. A classical “one-gene-at-a-time” algorithm is proposed in Figure 1A. In the light of the data reviewed in the preceding sections, we included the FRDA and SCA3/MJD testing as relevant steps in this potential algorithm. Only the main HSP genes and some triplet-repeat mutations have been included in the flow chart. Additional gene testing not mentioned in the figure can be considered in specific cases because including every potential gene associated with pyramidal signs would produce an unpractical, almost unmanageable algorithm. In this approach to the molecular testing of a genetically heterogeneous disease, the inheritance mode is often accounted firstly, and then screening steps are generally decided according to the main phenotypic features and mutation frequency. Technical questions are frequently also considered, such as the feasibility of mutation detection by conventional DNA analysis (e.g., Sanger sequencing, and multiplex ligation-dependent probe amplification). In most laboratories, this conventional diagnostic routine usually leaves most of the less frequent and/or less known genes unscreened. Whenever a new variant is found in a patient, further studies are often needed to address its pathogenic role, including the screening of a control population, segregation analysis in the family, effect predictions with bioinformatics tools, or even functional analysis. Because such an assessment of pathogenicity of each variant is beyond the reach of most diagnostic laboratories, curated databases are very much needed.
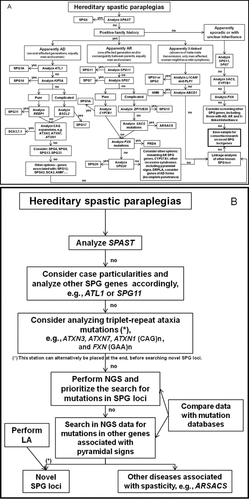
Diagnostic algorithms for HSPs of unknown cause. A: “Classical” algorithm; B: “Innovative” algorithm. Note: these algorithms are not intended to be taken as strict guidelines, they provide an idea of what could be the evolution from a standard protocol to a new approach on how to deal with this heterogeneous group of diseases; in specific cases, the priority genes may not necessarily follow these algorithms. AD, autosomal dominant; AR, autosomal recessive; SPG, spastic Gait; DRPLA, dentatorubral-pallidoluysian atrophy; SCA1, spinocerebellar ataxia type 1; SCA2, spinocerebellar ataxia type 2; SCA3/MJD, spinocerebellar ataxia type 3/Machado–Joseph disease; SCA7, spinocerebellar ataxia type 7; FRDA, Friedreich ataxia; ARSACS, autosomal recessive spastic ataxia of Charlevoix–Saguenay; AMN, adrenomyeloneuropathy; *especially if women are less affected than men; NGS, next generation sequencing; LA, linkage analysis.
A very different diagnostic scenario can be envisioned in the near future. Next-generation sequencing (NGS) prices are going down, at whole exome or even whole genome level, and it will not be cost-efficient to follow a classical “gene-by-gene” path. In Figure 1B, we suggest what the future diagnostic algorithm for HSP might look like. Conventional molecular screening techniques will still have a role, at least for some time, since they will be applied to test the most common genes in the first place—SPG4, SPG3A, and SPG11, for instance, although additional or alternative genes could be chosen at this station, depending on mode of inheritance, geographical frequency of mutations, and availability of the test. In the particular case of HSPs, we suggest that the screening of some triplet-repeat expansions may be considered afterward, especially when supported by clinical and electrophysiological clues, as this type of mutations is rarely detected by NGS. In addition, analysis of triplet-repeat expansions is generally available in most laboratories, and there is ample experience to guide the interpretation of these tests with the current knowledge. Furthermore, as HSP-look alike cases of SCAs have been reported occasionally, as reviewed above, it will only be through the screening of additional spastic paraplegia patients that we will know whether this clinical presentation could be more frequent than previously thought. After these first steps in which well-known and most common genes are tested, NGS will make its way to routine diagnostic protocols in neurogenetics. Both targeted-sequencing (gene panels) and whole exome or genome sequencing protocols may be established for this purpose. In the latter case, known disease-associated genes should be prioritized to search for mutations. Whole exome analyses would not only permit the search of mutations in known SPG loci, but would also enable the simultaneous search for mutations in other genes associated with spastic paraplegia, such as SACS [Depienne et al., 2007], CYP27A1 [Mignarri et al., 2011], and also the FMR1 premutation [Cellini et al., 2006; Jacquemont et al., 2005]. Thus, such a broader testing approach will likely be generalized in HSP and other heterogeneous groups of disorders because it will allow to increase the number of patients for whom the causative mutation can be identified, as well as to achieve an earlier diagnosis, before the full clinical picture has been manifested. In addition, this algorithm may lead to the discovery of new SPG loci. NGS is now transitioning from a research tool to a routine in genetic diagnostics. The major challenge, however, is still how to interpret the data, how to distinguish among the huge amount of sequence variants found in each individual the ones that are truly pathogenic. As clearly pointed by the work of Tucker and colleagues [Tucker et al., 2012], rigorous experimental follow-up to confirm mutation pathogenicity is needed. Although just the plain collection of sequence variants will be useful, well-curated locus-specific and disease-specific databases will constitute a crucial tool for NGS data interpretation in neurogenetics diagnosis.
Conclusions
The broad clinical spectrum of both HSPs and HAs, as well as the sometimes subtle phenotypic differences between these groups of disorders has been better known over time. Cases of triplet-repeat disorders presenting as HSP emphasize the extensive clinical and genetic overlap that can exist in neurogenetic diseases, specifically between HAs and HSPs. Thus, in addition to the SPG genes and other genes known to present with spasticity and ataxia, screening of triplet-repeat expansions should be considered in the workup of some patients with spastic paraplegia, especially the MJD/SCA3 locus in cases with AD complicated HSP, and the FRDA locus in cases with AR or sporadic spastic paraplegias. The number of genes and mutations known to cause these syndromes keeps expanding, posing a great challenge to any comprehensive attempt to build mutation databases and genotype-phenotype data collections, however making these databases even more necessary. The identification of the underlying causative mutation in a given patient is crucial for adequate diagnosis and genetic counseling, and for advancing our understanding of the underlying biological mechanisms. In the near future, diagnostic algorithms in neurogenetics will make use of high-throughput analysis techniques, combined with a systematic search in available databases. Therefore, the building of high-quality and comprehensive repositories of genotype-phenotype information will be of increasing relevance to improve diagnostic strategies.