Microarray-based copy number analysis of neurofibromatosis type-1 (NF1)-associated malignant peripheral nerve sheath tumors reveals a role for Rho–GTPase pathway genes in NF1 tumorigenesis†
Communicated by Paolo M. Fortina
Abstract
Neurofibromatosis type-1 (NF1) is associated with the growth of benign and malignant tumors. Approximately 15% of NF1 patients develop malignant peripheral nerve sheath tumors (MPNSTs), underlining the need to identify specific diagnostic/prognostic biomarkers associated with MPNST development. The Affymetrix Genome-Wide Human single-nucleotide polymorphism (SNP) Array 6.0 was used to perform SNP genotyping and copy number alteration (CNA), loss-of-heterozygosity (LOH), and copy number neutral–LOH (CNN–LOH) analyses of DNA isolated from 15 MPNSTs, five benign plexiform neurofibromas (PNFs), and patient-matched lymphocyte DNAs. MPNSTs exhibited high-level CNN-LOH, with recurrent changes occurring in MPNSTs but not PNFs. CNN–LOH was evident in MPNSTs but occurred less frequently than genomic deletions. CNAs involving the ITGB8, PDGFA, Ras-related C3 botulinum toxin substrate 1 (RAC1) (7p21-p22), PDGFRL (8p22-p21.3), and matrix metallopeptidase 12 (MMP12) (11q22.3) genes were specific to MPNSTs. Pathway analysis revealed the MPNST-specific amplification of seven Rho–GTPase pathway genes and several cytoskeletal remodeling/cell adhesion genes. In knockdown experiments employing short-hairpin RAC1, ROCK2, PTK2, and LIMK1 RNAs to transfect both control and MPNST-derived cell lines, cell adhesion was significantly increased in the MPNST cell lines, whereas wound healing, cell migration, and invasiveness were reduced, consistent with a role for these Rho–GTPase pathway genes in MPNST development and metastasis. These results suggest new targets for therapeutic intervention in relation to MPNSTs. Hum Mutat 33:763–776, 2012. © 2012 Wiley Periodicals, Inc.
Introduction
Neurofibromatosis type-1 (NF1; MIM# 162200) is a familial cancer syndrome that affects one in 3,500 individuals worldwide. It is caused by inactivation of the NF1 tumor suppressor gene (MIM# 162200) located at 17q11.2. Consistent with Knudson's two-hit hypothesis, NF1 patients harboring a heterozygous germline NF1 mutation develop neurofibromas upon somatic mutation of the second wild-type NF1 allele. The benign and malignant peripheral nerve sheath tumors (MPNSTs) that develop in NF1 patients are a hallmark of the disease, with cutaneous neurofibromas being the commonest benign tumors, and MPNSTs, a type of sarcoma, contributing to malignancy [Bennett et al., 2009; Upadhyaya and Cooper, 1998; reviewed by Upadhyaya, 2010]. NF1 individuals are predisposed to develop a variety of other cancers, such as optic gliomas, pheochromocytomas, gastrointestinal stromal tumors, and myeloid malignancies. NF1 is also characterized by a plethora of nontumor clinical features, including dermal pigmentary changes, hamartomatous lesions of the iris, bone deformities, and cognitive deficit.
Neurofibromatosis type-1 patients exhibit an 8–13% lifetime risk of developing an MPNST, usually from within a preexisting plexiform neurofibroma (PNF) or a focal subcutaneous neurofibroma [Brems et al., 2009a; Evans et al., 2002; Spurlock et al., 2010; Tucker et al., 2005 & Carrolle & Ratner, 2008]. Although most MPNSTs develop in adult NF1 patients, they can also occur in much younger affected individuals, emphasizing the need for early detection of MPNST development given that these aggressive tumors frequently recur and can metastasize to the lungs and other organs. Radical surgery, usually in combination with radiotherapy, constitutes the optimum treatment, especially if chemotherapy is also given in parallel to inhibit metastasis. Although early detection would facilitate both the identification and clinical management of those NF1 patients most likely to develop malignant tumors, a prerequisite for early detection is the availability of diagnostic and prognostic biomarkers associated with MPNST development [reviewed by Upadhyaya, 2011].
The difficulties inherent in the early clinical and pathological diagnosis of NF1–MPNSTs underline the need for a clearer definition of the molecular basis of NF1-associated malignancy. Somatic biallelic inactivation of both NF1 alleles was first demonstrated in NF1–MPNSTs more than 20 years ago [Skuse et al., 1989], an observation consistent with NF1 being a tumor suppressor gene. Homozygous NF1 inactivation was also subsequently detected in benign cutaneous and PNFs as well as in MPNSTs when loss-of-heterozygosity (LOH) analysis was performed [Glover et al., 1991; John et al., 2000; Legius et al., 1993; Rasmussen et al., 2000], indicating that additional genetic changes are required for malignant transformation to occur.
Although the functional loss of neurofibromin represents the primary event in tumorigenesis, the precise role of other genetic lesions in promoting the malignant transformation of benign neurofibromas is poorly understood. However, the recent introduction of microarray-based technologies allows us to simultaneously analyze multiple genes and genomic regions (at both the RNA and DNA level) in a single experiment. Such array-based analyses provide us with an unrivaled opportunity to profile concomitant copy number and gene expression changes associated with a given tumor state. Several studies have already analyzed tumor DNA and/or RNA in searches for potential molecular signatures capable of differentiating between MPNSTs and benign neurofibromas [Miller et al., 2006, 2010; Mantripragada et al., 2008, 2009; reviewed by Upadhyaya, 2011]. Array-based analyses have a major advantage in that they can be used to detect copy number variation/alterations, which can play a key role in tumorigenesis. Copy number variants (CNVs) constitute heritable genomic alterations that range in size from one kilobase up to several megabases in size. Although CNVs make a substantial contribution to interindividual variation, acquired somatic copy number alterations (CNAs) are increasingly being identified in many different cancers [Albertson, 2006; Kuiper et al., 2010; Rothenberg and Settleman, 2010; Shlien and Malkin, 2009, 2010]. As with other tumor-associated genetic changes, such CNAs (by virtue of the genes they contain) can confer a selective advantage with respect to cellular proliferation, survival, and clonal expansion. Various microarray-based methodologies have been developed to detect both CNVs and CNAs genome wide. Initially, comparative genomic hybridization arrays (array-CGH) constructed from overlapping human bacterial artificial chromosome (BAC) clones were recruited, but more recently, combined high-density single-nucleotide polymorphism (SNP) and CNV genotyping arrays have been employed. Such high-density microarrays, often containing several million individual markers, can now be used to simultaneously assess both heritable copy number variation and somatic CNA-based genomic changes associated with a specific tumor type [Lebron et al., 2011].
Recent studies of NF1-associated benign and malignant tumors, which have aimed to identify tumor-specific diagnostic and prognostic markers, have detected extensive somatic copy number changes in DNA derived from NF1–MPNSTs that were absent from the DNA of benign PNFs [Kresse et al., 2008; Mantripragada et al., 2008, 2009]. The present study has extended this analysis by providing high-resolution information from a combined genomic CNA analysis with data from an LOH screen of both PNF and MPNST tumor DNAs from 20 unrelated NF1 patients using the Affymetrix Genome-Wide Human SNP 6.0 Array.
Materials and Methods
Patient Samples and DNA Preparation
This project was approved by the local Ethics committee. Informed consent for sample collection was obtained according to protocols approved by this committee. Diagnosis of MPNSTs was made by the individual referring centers. All available information on the nature and grading of the tumors under study (15 MPNSTs) is provided in Supp. Table S1. Tumor biopsies and matched lymphocyte samples were obtained from the 20 unrelated NF1 patients. DNA from the tumors and matched lymphocyte samples was extracted as previously described [Upadhyaya et al., 2008a, 2008b]. DNA samples were checked to ensure compliance with the stringent spectrophotometric and electrophoretic quality control (QC) criteria required by the Affymetrix SNP 6.0 Array platform.
Affymetrix SNP 6.0 Array Analysis
The Affymetrix Genome-Wide Human SNP Array 6.0 was used as per the manufacturer's instructions. This Affymetrix array offers whole-genome coverage as well as superior resolution, and permits the simultaneous assessment of unbalanced chromosomal defects such as CNA, LOH, and acquired copy number neutral (CNN)–LOH. Details are given in the Supp. Methods.
Genomic Segmentation algorithms or Hidden Markov Model (HMM) (Affymetrix White Paper; http://www.affymetrix.com/support/technical/whitepapers.affx) were used to detect the copy number variation. The minimal number of genomic markers was set at 50 for both algorithms. Most copy number analysis was performed using the segmentation method, with the P value threshold set at 0.001 and a signal-to-noise ratio of ≥0.5, to attain the level required for positive CNA detection. In these copy number analyses, overlapping CNA regions were identified on the basis of the overlap of physical location intervals at each CNA boundary, either in NF1–MPNSTs or benign PNFs. All CNA and LOH analyses, including CNN–LOH, were performed in Partek Genomics Suite 6.4. Most CNA analysis entailed paired analysis involving the comparison of tumor DNAs against the matched constitutional patient lymphocyte DNAs, whereas unpaired CNA analysis was only used in median absolute pairwise difference (MAPD) calculation and QC to compare tumor DNA against reference data derived from the 270 normal individuals catalogued in the HapMap database (International HapMap Consortium, 2005). All LOH and CNN–LOH analyses were paired using matched lymphocyte DNAs as the baseline. Each positive tumor-specific CNA was assessed to determine the genomic region, overlap, or recurrence between samples, as well as the identity of the genes residing within the corresponding genomic regions. For CNN–LOH analysis, two separate workflows were used. One of these was the allele-specific copy number (ASCN) analysis in Partek that used both the CEL files containing probe intensity data and CHP files containing SNP call results together to assess copy number allele imbalance. The other workflow involved the comparison of paired CNV analysis using CEL files and paired LOH analysis (using both CEL files and CHP files) to identify regions with CNVs and LOH, respectively, and then to identify regions of CNN–LOH. The results from the two separate workflows were then merged, generating a consensus in CNN–LOH identification. Biological pathway analysis was performed in GeneGO's MetaCore (http://www.genego.com) using gene set enrichment analysis in conjunction with the gene lists from the corresponding CNA regions or LOH regions.
Quantitative Real-Time Polymerase Chain Reaction
From the derived Affymetrix array CGH data in the present study, the relative copy numbers of 14 genes found to be encompassed by tumor-specific CNAs were independently assessed by quantitative polymerase chain reaction (qPCR) of DNA and RNA isolated from the relevant tumor samples. The details are given in the Supporting Information.
Details of the short-hairpin RNA (shRNA) experiments, cell culture, Western blotting, immunohistochemistry, the wound healing assay, and the cell adhesion, invasion, and migration assays are given in the Supporting Information.
Results
Data Quality Assessment
In the present study, we analyzed the copy number and LOH state in DNA isolated from tumors and matched constitutional (lymphocyte) samples from 20 NF1 patients (including 15 MPNSTs and five PNFs) using the Affymetrix 6.0 SNP array. (NB. Throughout the text, the prefix M denotes MPNST, whereas the prefix P denotes PNF.) Because LOH and CNN–LOH analyses require satisfactory genotyping performance and consistent intensity measurement across genomic regions, we examined the SNP call rate to assess genotyping performance, MAPD to assess the suitability for using unpaired copy number analysis against the 794 normal HapMap samples (Supp. Fig. S1a–c), and principal component analysis (PCA) and clustering analysis for sample grouping and detection of sample outliers. Apart from one lymphocyte sample (patient M6) that failed to yield DNA of sufficient quality, all 39 samples successfully yielded data. The SNP call rates for 36 of the 39 arrays analyzed were 97–99%, and were therefore suitable for LOH analysis. The remaining three arrays yielded SNP call rates of 96.0–96.8% with acceptable genotyping performance and were therefore also included in the data analysis. All 39 DNA hybridizations yielded good MAPD scores, 0.23–0.37 (<0.4), thereby meeting the acceptance criterion for copy number analysis.
The clustering analysis (Fig. 1a and b) of copy numbers from the unpaired and paired analyses indicated that the MPNSTs represented a diverse group, with some samples (M10, M11, M12, M13, and M14) resembling PNF samples in terms of their copy number changes, whereas others (such as M2, M3, M7, M8, M9, and MX) were quite distinct from the PNF samples. After reviewing the PCA and clustering analysis results of copy numbers from the unpaired analysis, an apparent batch effect related to scan date (data production date) was noted. Three arrays, corresponding to M14 tumor and lymphocyte samples and M9 tumor samples produced on September 16, 2008 (Supp. Fig. S1a) deviated significantly from the rest of data due to the scan date batch effect. Although the paired analysis (Fig. 1a) placed M14 in the middle of the cluster, because both its tumor and matched lymphocyte samples were from the same batch, M9 would appear to be an outlier in the paired analysis because its tumor sample was profiled in the batch of September 16, 2008, but its matched reference lymphocyte sample was made in the other batch (August 28, 2008). Therefore, M9 was not used for any sample overlap and recurrent CNA analysis.
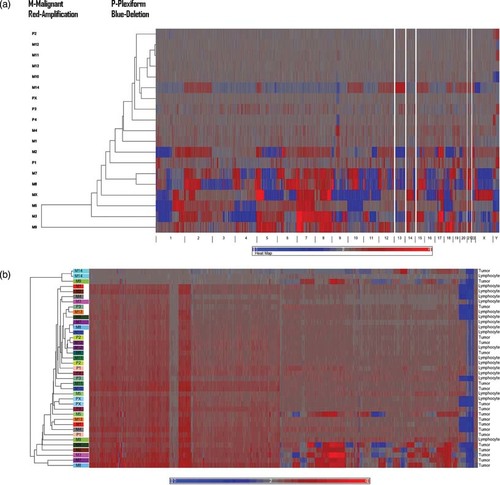
(A) Hierachical clustering of genome-wide copy number from the paired analysis. Global clustering view of copy number alterations across all NF1 tumor samples. M, malignant; P, plexiform neurofibromas amplification is indicated by red and deletion is denoted by blue. (B) Hierachical clustering of genome-wide copy number from the unpaired analysis compared with the HapMap reference. Amplification is indicated by red and deletion is denoted by blue.
The genome-wide CNA/CNV profiles associated with each tumor type and the matching lymphocyte DNA from the same patient were visualized using the heatmaps generated from hierarchical clustering analysis in Partek Genomics Suite 6.4. Each CNA profile was presented as a “heatmap” that depicted the copy number changes associated with each of the 23 chromosomes. As an example, the CNA heatmap for chromosome 5 is shown for the 13 MPNST tumor samples (top panel) and five PNF tumor samples pairwise in Figure 2; this serves to identify all copy number gains (in red) and all copy number losses (in blue) across the entire length of chromosome 5. Five PNF-harboring patients (bottom panel) displayed relatively few copy number changes, whereas the CNA profiles derived from five MPNSTs (top panel, M2, M3, M5, M6, M7, M8, and MX) were generally comparable (with genomic gains across the entire 5p arm and genomic losses across the 5q arm). However, the CNA profiles from several other MPNSTs (M4, M10, M11, M12, and M13) were quite varied, with several displaying CNA profiles with respect to chromosome 5 that were similar to those exhibited by benign PNFs. The genome-wide CNA profiles associated with the MPNSTs displayed a similar trend; however, these molecular profiles harbored two subgroups of malignant tumors, those that exhibited a large number of CNAs and those which exhibited relatively few CNA profiles and were comparable to the profiles characterizing benign PNFs (Figs. 1a and 2).
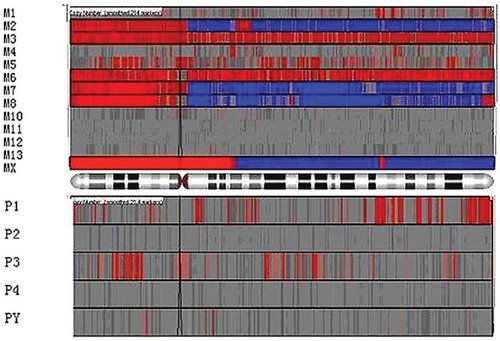
Copy number alteration (CNA) analysis of all NF1 tumor samples, both benign PNFs and malignant MPNSTs for chromosome 5. CNA heatmap for chromosome 5 is shown for the 13 MPNST tumor samples (top panel) and five PNF tumor samples pairwise; it identifies all copy number gains (in red) and all copy number losses (in blue) across the entire length of chromosome 5. Five PNF-harboring patients (bottom panel) displayed relatively few copy number changes, whereas the CNA profiles derived from five MPNSTs (top panel, M2, M3, M5, M6, M7, M8, and MX) were generally comparable (with genomic gains across the entire 5p arm and genomic losses across the 5q arm). However, the CNA profiles from several other MPNSTs (M4, M10, M11, M12, and M13) were quite varied, with several displaying CNA profiles with respect to chromosome 5 that were similar to those exhibited by benign PNFs.
CNV/CNA Identification by Paired and Unpaired Analysis
To determine whether any of the identified CNAs were specific to either the PNFs or the malignant MPNSTs, all tumor CNVs/CNAs were compared with the CNV profiles derived from the matched lymphocyte DNA samples from the same patients, as well as against the CNV profiles of the 794 normal HapMap samples. The Database of Genomic Variants (http://projects.tcag.ca/variation/) was queried for all CNV/CNA-harboring regions identified in the tumor and blood samples in order to exclude the possibility that they might constitute rare CNVs found in the normal population rather than true somatic events. To identify significant CNAs, we employed the criteria of 50 or more markers, a P value of 0.001 and a signal-to-noise ratio of no less than 0.5 for segmentation analysis. The CNA distributions in 14 MPNST and five PNF patients are summarized in Table 1 and Supp. Tables S2 and S3 for both the paired and unpaired analyses. The detailed results for individual patients are presented in Supp. Table S4. It would appear that CNAs in MPNSTs are much more frequent than in PNFs in the paired analysis (Table 1, Supp. Fig. S2a and b). The unpaired analysis yielded more CNAs in some samples than the paired analysis (Table 1), indicating that germline CNVs were evident in the unpaired analysis but not in the paired analysis. These results from the unpaired analysis did not include the X and Y chromosomes because the genders of individuals from the HapMap samples were different from those of the NF1 patients and hence inclusion of the sex chromosomes would have rendered the CNA analysis invalid. Therefore, the primary results and conclusion were based solely on the paired analysis. The unpaired analysis was mainly used to assess the difference between the somatic and germline changes. Among the results of the paired analysis, 41 CNAs had copy numbers greater than 4 or less than 0.5; 40 of these high-magnitude changes occurred in MPNST patients M2, M3, M7, M8, and MX including 13 amplifications on 18q21.2 in patient M3, two deletions on 17p11.2 in patient M2, and three deletions on 11p12 in MX. Only one amplification event was found in a PNF (P1 on 12p11.1).
| Patient ID | Tumor | Paired analysis number of CN gain | Paired analysis number of CN loss | Unpaired analysis number of CN gain | Unpaired analysis number of CN loss |
|---|---|---|---|---|---|
| M1 | MPNST | 1 | 8 | 18 | |
| M2 | MPNST | 37 | 61 | 35 | 65 |
| M3 | MPNST | 64 | 76 | 61 | 75 |
| M4 | MPNST | 9 | 23 | ||
| M5 | MPNST | 11 | 13 | 15 | 32 |
| M7 | MPNST | 68 | 102 | 50 | 63 |
| M8 | MPNST | 131 | 129 | 68 | 107 |
| M9 | MPNST | 61 | 23 | 39 | 34 |
| M10 | MPNST | 1 | 1 | 7 | 18 |
| M11 | MPNST | 7 | 25 | ||
| M12 | MPNST | 9 | 13 | ||
| M13 | MPNST | 9 | 9 | ||
| M14 | MPNST | 10 | 16 | 19 | 42 |
| MX | MPNST | 44 | 146 | 41 | 121 |
| P1 | PNF | 16 | 1 | 23 | 28 |
| P2 | PNF | 1 | 18 | 30 | |
| P3 | PNF | 6 | 20 | 24 | |
| P4 | PNF | 1 | 1 | 27 | 33 |
| PX | PNF | 26 | 38 |
Deletions and Amplifications
Of the total 1,022 CNVs/CNAs identified, there were more copy number losses (571; 55.87%) than copy number gains (451; 44.13%). Most CNAs (996/1022) were identified in MPNST tumor DNA, whereas only 26 CNAs were identified in benign PNF tumor DNA (Supp. Table S3). Recurrent or overlapping CNVs/CNAs were absent from PNFs (Supp. Table S3), except for one overlap on chromosome 4 in two patients, P1 and P3. Identification of the CNAs most likely to be involved in malignant transformation required the selection of recurrent CNAs (i.e., those CNAs found in at least three different MPNSTs from unrelated patients, and with copy number gains of ≥2.8 or losses of ≤1.5); employing these criteria, a total of 232 CNVs were selected for further analysis (all were among the MPNSTs because no such recurrent CNAs were observed among the benign PNFs). A subsequent assessment of the 232 recurrent MPNST-associated CNAs identified at least 2,972 genes that were contained within, or overlapped with, these variant genomic regions (Supp. Table S4, overlapping genes).
LOH Analysis and CNN–LOH analysis
The 19 pairs of samples were used to identify regions with CNN changes. Patient M6 was excluded owing to the lack of sufficient matched lymphocyte sample. In LOH analysis, a matched reference is required to eliminate potentially confounding genotypic differences from the reference sample. Two approaches were used to identify CNN changes (uniparental disomy) using Partek Genomics Suite 6.4. First, CNN–LOH was analyzed to identify regions where LOH was identified, yet total copy number equalled 2. Second, the ASCN was performed to identify CNN allele imbalance.
The 19 pairs of samples (corresponding to DNAs from 14 MPNSTs and five PNFs) were used in LOH analysis with the individually matched lymphocyte DNAs serving as reference samples in the paired analysis (Table 2).
| Categories of LOH or genes | LOH | LOH genes |
|---|---|---|
| Overall | 7,643 | 39,847 |
| Benign tumors | 78 | 117 |
| Overlap in benign tumors | 0 | 0 |
| Malignant tumors | 7,565 | 39,730 |
| Overlap in two MPNSTs | 8,159 | 15,122 |
| Overlap in five MPNSTs | 445 | 543 |
| Overlap in six MPNSTs | 40 | 529 |
Loss-of-heterozygosity results were obtained from paired analyses of matched tumor and lymphocyte DNAs from each patient using the SNP probe set in the Affymetrix SNP6.0 array. MPNST DNAs exhibited a higher level of LOH (with a considerable number of overlapping regions) as compared with benign tumors.
In total, 7,643 LOH regions were found that exhibited a heterozygosity rate of zero for at least two SNP markers. Supp. Table S3 summarizes the results obtained in LOH analysis and illustrates the very high levels of LOH exhibited by MPNST DNA as compared with the general lack of LOH, evident in benign PNF tumor DNAs. Among the MPNST DNA samples, a considerable number of overlapping LOH regions were observed, with at least 450 overlapping regions identified in five tumors and 40 overlapping regions identified in six tumors. In contrast, no overlap in LOH was found among the benign PNF tumors. In the paired copy number analysis, the 19 pairs of samples were used in HMM copy number analysis using their matched lymphocyte samples as a reference and a threshold of at least 50 SNP markers for a positive CNA to be ascertained. In the output of HMM copy number analysis, 1,310 regions with a copy number of 2 and a P value of ∼1.00 were then subsequently merged with the 7,643 LOH regions to identify overlaps. In total, 9,188 overlapping regions of potential CNN–LOH were identified. Following filtration of these 9,188 regions with a 0% heterozygosity call rate and the requirement for a minimum of 50 SNP markers in each region, the number of CNN–LOH regions was reduced to 512 because a majority of the LOH regions had fewer than 50 SNP markers. By further applying the criterion that each region must either occur in at least two unrelated patients, the CNN–LOH regions were reduced to 71 (CNN–LOH regions detected in two to four patients). The 71 identified regions were variously located on chromosomes 1, 4, 5, 9, 16, 17, 18, and X. The CNN allele imbalance approach was also applied in the analysis. Paired copy number analysis was performed using segmentation on the 19 pairs of samples. With the segmentation algorithms, at least 50 SNP markers, a P value of 0.001, and a signal-to-noise ratio of 0.5 set as threshold; 599 regions were found to have a P value of ∼1.00 or were unchanged in copy number in the segmentation. In the SCNA analysis, 3,768 regions are detected for SCNA imbalance. By overlapping the 599 CNVs from the total copy number analysis with the 3,768 regions from the SCNA analysis, we identified 578 regions of CNN imbalance with recurrence in two or more patients. Most of the 578 regions were identified in five or more patients.
Identification of Potential Genes Associated with MPNST Development
Biological pathway analysis
A variety of genes were contained within those CNAs that were found exclusively in MPNSTs. These included ITGB8 (7p15.3), PDGFA and Ras-related C3 botulinum toxin substrate 1 (RAC1) (7p22), PDGFRB (5q31-q32), and matrix metallopeptidase 12 (MMP12) (11q22.3). A biological pathway analysis was performed on the 1,521 genes that were associated with MPNST-specific CNAs, as identified by the MetaCore database (http://www.genego.com/metacore.php). MetaCore is an integrated software suite designed for the pathway analysis of array experimental data. The specific pathways deemed to be potentially involved are listed in Table 3 and Supp. Table S5, together with the map name, cell process, and hypergeometric P value found in the present study and in the entire pathway.
| Map | Cell process | P Value | Objects from LOH gene list | Total objects |
|---|---|---|---|---|
| Cell adhesion_ECM remodeling | Cell adhesion | 0.0064 | 6 | 51 |
| Apoptosis and survival_Lymphotoxin-beta receptor signaling | Immune response, apoptosis | 0.0109 | 5 | 41 |
| Immune response_Role of TLRs 3 and 4 in cell antiviral response: TICAM1-specific signaling pathways | Immune response | 0.0207 | 3 | 18 |
| Immune response _Oncostatin M signaling via JAK-Stat in mouse cells | Cytokine and chemokine mediated signaling pathway, immune response | 0.0207 | 3 | 18 |
| wtCFTR and delta508 traffic / Clathrin coated vesicles formation (norm and CF) | 0.0240 | 3 | 19 | |
| Immune response _Oncostatin M signaling via JAK-Stat in human cells | Cytokine and chemokine mediated signaling pathway, immune response | 0.0275 | 3 | 20 |
| Immune response _Oncostatin M signaling via MAPK in mouse cells | Cytokine and chemokine mediated signaling pathway, immune response | 0.0280 | 4 | 35 |
| Immune response_Oncostatin M signaling via MAPK in human cells | Cytokine and chemokine mediated signaling pathway, immune response | 0.0336 | 4 | 37 |
| Immune response _Histamine H1 receptor signaling in immune response | Immune response, G-protein coupled receptor protein signaling pathway | 0.0431 | 4 | 40 |
The details of these individual pathways are given in Supp. Figure S3. The main conclusion from this analysis was that the genomic aberrations present exclusively in MPNST DNA comprise both specific CNAs and widespread LOH, whereas no such genomic changes were evident in DNA from any benign PNF tumor.
A similar biological pathway assessment was undertaken on the 543 genes shown to be consistently affected by informative LOH changes in DNA from at least five MPNSTs from unrelated patients. The top 10 pathways identified are listed in Table 3. Many of these pathways are involved with cell growth, cell adhesion, signal transduction, and the immune response, and are therefore broadly representative of the systems frequently found to be involved in tumorigenesis and metastasis. This study has therefore identified a number of genes with potential links to the process of NF1 tumorigenesis, including genes directly involved in cytoskeletal remodeling and cell adhesion. For example, at least seven members of the Rho–GTPase gene family known to mediate cytoskeletal remodeling (ACTB, ACTG1, LIMK1, PRKCA, PTK2, RAC1, and ROCK2) (Supp. Fig. S3) were found to be associated with recurrent copy number gains in MPNST DNA. In addition, both ACTB and ACTG1 are cytoplasmic actin genes. Hence, their increased expression (as a likely consequence of gene duplication) may influence either the motility or the invasive properties of affected tumor cells (Table 3 and Supp. Table S5).
The relationship between copy number changes and gene expression was assessed by qPCR and a correlation was noted for the majority (∼90%) of the genes analyzed (Supp. Fig. S4a and b). All CNAs identified in this analysis were confirmed by real-time (RT) qPCR and at least seven genes in the Rho–GTPase pathway were found to be amplified (Supp. Fig. S4b). Four of these seven Rho–GTPase pathway genes have been shown to be involved in some of the features of NF1 tumorigenesis, namely, wound healing, cellular adhesion, migration, and invasion in this study. Alteration of the Rho–GTPase-encoding RAC1 gene has been identified in multiple solid cancer types including testicular, gastric, and breast cancer in addition to chronic myelogenous leukemia and primary human schwannoma cells [Harnois et al., 2003; Kaempchen et al., 2003].
Immunohistochemistry
For all four proteins, RAC1, ROCK2, LIMK1, and PTK2, greater intensity of cytoplasmic staining was observed in MPNSTs versus benign neurofibromas, both cutaneous and plexiform (data not shown). This finding is consistent with the results obtained by array and qPCR analysis. In neurofibromas, the vast majority of nuclei, blood vessels, and mast cells were stained, whereas in MPNST few nuclei were stained. The pattern of staining was similar for all four proteins in both the tumor types (data not shown) [Bartley & Ross, 2002].
Exploring the involvement of RAC1, ROCK2, PTK2, and LIMK1 in wound healing and cell adhesion, migration, and invasion
RAC1, ROCK2, PTK2, and LIMK1 were selected for further functional studies not only because they are key members of the Rho–GTPase pathway and have been implicated in many cancers [Malliri and Collard, 2003] but also because MPNST-specific copy number gains were observed for these genes. In addition, shRNAs for these four genes have been fully validated. All three MPNST cell lines ST8814, 507.1, and T529 used in this study exhibited copy number gains and overexpression of these genes (data not shown).
Wound healing is an intricate process in which the skin (or another organ–tissue) repairs itself after injury. In order to explore the potential role of the overexpression of RAC1, ROCK2, PTK2, and LIMK1 on wound healing, cells transfected with RAC1, ROCK2, PTK2, and LIMK1 knockdowns and scrambled control shRNA were seeded onto laminin and wounded with a pipette tip. Between 0 and 12 hr after wounding, the cells migrated across the wound. Wound healing was reduced in RAC1, ROCK2, PTK2, and LIMK1 knockdown cells by comparison with control scrambled shRNA (Fig. 3a; data for PTK2 and LIMK1 provided in Supp. Fig. S5a and b). Owing to the kinetics of wound healing, healing was deemed to be complete when no gap was observed in all shRNA cell lines by 24 hr, although healing occurred earlier (12–16 hr) in the control cell line than in the RAC1, ROCK2, PTK2, and LIMK1 knockdown cell lines (16–24 hr). The above experiment was repeated on a further three cell lines (ST8814, 507.1, and T529). All experiments were repeated a minimum of three times and the results generated from each of the three experiments were in concordance. MPNST cell doubling time is approximately 20.44 hr (ST8814) [Vahidnia et al. 2007]. Wound healing is unlikely to have been affected by cell proliferation as wound healing was observed by 12 hr in the scrambled control cell line.
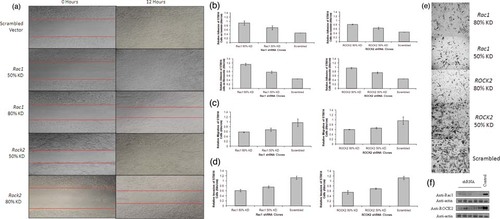
(A) Wound healing assay on MPNST cell line ST8814. Cells were seeded on laminin, syncronized in 1% FBS for 24 hr and wounded with a pipette tip. Wounds were followed up for 24 hr. Reduction in the rate of wound healing can be seen in the cell lines carrying RAC1 and ROCK2 knockdowns. Cells were imaged on a microscope at ×10 magnification (Leica). (B) Adhesion assay on MPNST cell line ST8814. (i) ST8814 cells transfected with RAC1 shRNA seeded on laminin, (ii) ROCK2 shRNA-transfected cells seeded on laminin, (iii) cells transfected with RAC1 shRNA on fibronectin, and (iv) ROCK2 shRNA-transfected ST8814 cells seeded on fibronectin. Adhesion was significantly increased in samples containing RAC1 and ROCK2 knockdowns in comparison with a cell line transfected with the scrambled control shRNA (P = 0.02) and (P = 0.05), respectively. The RAC1 knockdown results in more cell adhesion than the ROCK2 knockdown but this difference was not significant (P = 0.07). Adhesion on fibronectin was more efficient than that on laminin in cell lines carrying both RAC1 and ROCK2 knockdown, but again this observed difference was not significant (P = 0.2). All results were a readout of the absorbance of crystal violet (5 mg/ml) eluted with 1% SDS at 550 nm on a Genova MK3 Lifescience Analyser (Jenway). (C) Migration assay on ST8814 cell lines. (i) Migration of ST8814 cells transfected with RAC1 shRNA, (ii) migration of ST8814 cells transfected with ROCK2 shRNA. Migration of ST8814 cells was significantly reduced in cell lines carrying Rac1 and ROCK2 knockdowns in comparison with ST8814 cells transfected with a scrambled vector (P = 0.05 and P = 0.03, respectively). The reduction in migration was similar in cells transfected with both RAC1 and ROCK2. 1.34 × 10 fewer cells migrated through the membrane in the cell line with the RAC1 80% knockdown and 1.3 × 106 fewer cells migrated in the ROCK2 80% knockdown cell line as compared with the scrambled control. All results were a readout of the absorbance of crystal violet (5 mg/ml) eluted with 1% SDS at 550 nm on a Genova MK3 Lifescience Analyser (Jenway). (D) Invasion assay on ST8814 cell lines. (i) Invasion of ST8814 cells transfected with RAC1 shRNA. (ii) Invasion of ST8814 cells transfected with ROCK2 shRNA. The invasion activity of ST8814 cell lines was significantly reduced in those cell lines with both RAC1 and ROCK2 knockdown as compared with the scrambled control (P = 0.05 for both). RAC1 and ROCK2 knockdowns elicited similar reductions in invasive potential of the ST8814 cells. 1.85 × 106 fewer cells invaded the matrigel and membrane in the cell line with the RAC1 80% knockdown and 1.8 × 106 in the ROCK2 80% knockdown as compared with the control scrambled cell line. All results were a readout of the absorbance of crystal violet (5 mg/ml) eluted with 1% SDS at 550 nm on a Genova MK3 Lifescience Analyser (Jenway). (E) Invasion assay on ST8814 cells transfected with RAC1, ROCK2, and scrambled control shRNA. Cells were stained with crystal violet (5 mg/ml) and images were taken at ×10 magnification (Leica). RAC1 and ROCK2 knockdown elicited a reduction in the invasive potential of ST8814 cell lines as evidenced by the lack of RAC1 and ROCK2 shRNA-transfected cells on the underside of the membrane following 3 days incubation. All experiments were repeated a minimum of three times. Results were similar for all three MPNST cell lines studied (ST8814, 507.1, and T529). All statistical analysis was completed with SPSS v16 using independent samples t-test. (F) Western blot assay of cell lines transacted with 5 RAC1 and ROCK2 shRNA clones in addition to one cell line transfected with a scrambled control. β-actin control blots were completed to ensure expression was unaffected in genes not targeted by the shRNA.
Cell adhesion is a complex mechanism involved in a variety of processes including cell migration and invasion, embryogenesis, wound healing, and tissue remodeling. In order to explore the potential role of the overexpression of RAC1, ROCK2, PTK2, and LIMK1 on cell adhesion, cells were transfected with RAC1 and ROCK2 knockdown and control shRNAs on both laminin and fibronectin, and PTK2 and LIMK1 with control shRNA on laminin. Adhesion was significantly increased in cell lines containing RAC1, ROCK2, PTK2, and LIMK1 knockdowns (P = 0.02, P = 0.05, P = 0.001, and P = 0.002, respectively) by comparison with cell lines transfected with the scrambled control shRNA (Fig. 3b) (data for PTK2 and LIMK1 in Supp. Fig. S4a and b). RAC1 knockdown resulted in greater cell adhesion than ROCK2 knockdown, and LIMK1 knockdown resulted in greater cell adhesion than PTK2 knockdown. However, these differences were not statistically significant, P = 0.07 and P = 0.12, respectively. Adhesion on fibronectin appeared to be more efficient than that on laminin in cell lines harboring both RAC1 and ROCK2 knockdowns, but again this difference was not statistically significant (P = 0.20).
Cell migration is a highly integrated, multistep process that plays an important role in the progression of cancer. In order to explore the potential role of the overexpression of RAC1, ROCK2, PTK2, and LIMK1 on cell migration, the migration of cells transfected with RAC1, ROCK2, PTK2, and LIMK1 and control shRNA was assessed by seeding the cells onto polycarbonate membranes [de Pril et al., 2009]. Cell migration was significantly reduced in RAC1, ROCK2, PTK2, and LIMK1 knockdown cell lines in comparison with cells transfected with a scrambled vector (P = 0.05, P = 0.03, P = 0.025, and P = 0.027, respectively) (Fig. 3c). The reduction in migration was similar in cells that were transfected with RAC1, ROCK2, PTK2, and LIMK1 (Fig. 3C) (data for PTK2 and LIMK1 in Supp. Fig. S5). Approximately 40% reduction in cell migration was observed in 80% knockdowns of RAC1, ROCK2, PTK2, and LIMK1 and approximately 25% reduction in 50% knockdowns of RAC1, ROCK2, PTK2, and LIMK1. Results were representative of all cell lines and replicates (ST8814, 507.1, and T529).
Tumor cell invasion into the basement membrane is an important step in the multistep process of tumor metastasis. The potential role of the overexpression of RAC1 and ROCK2 on the invasive properties of cells transfected with RAC1 and ROCK2 knockdowns and control shRNAs was assessed by seeding cells onto matrigel basement membrane on polycarbonate membranes. A significant reduction in the invasive potential of cell lines transfected with both RAC1 and ROCK2 knockdown shRNA was observed by comparison with the scrambled control (P = 0.05 for both) (Fig. 3d and e) This analysis was not performed for PTK2 and LIMK1. RAC1 and ROCK2 knockdowns elicited a similar reduction in invasive potential; an approximately 45% reduction in cell invasion was observed with both RAC1 and ROCK2 80% knockdown clones as compared with the scrambled control-transfected cell line. Similarly, a 25% reduction in invasion was seen in both genes carrying 50% knockdown clones. All experiments were repeated and the results were comparable in all three cell lines (ST8814, 507.1, and T529) tested. In summary, both RAC1 and ROCK2 knockdown resulted in a reduction in wound healing, cell migration, and invasiveness and an increase in cell adhesion.
Validation of specific gene knockdown using shRNA clones was completed by western blot analysis followed by densitometry using Image J software (http://rsbweb.nih.gov/ij/) (Fig. 3f).
Discussion
Neurofibromin, the NF1 gene product, is a tumor suppressor protein whose main function is to downregulate Ras activity in the cell. The mutational inactivation of the NF1 gene, leading to loss of functional neurofibromin, therefore, results in the constitutive activation of cellular Ras, as well as the associated RAS/RAF/MAPK signaling pathway, leading to increased cell growth and proliferation and an increased likelihood of tumorigenesis [Guha et al., 2006; Upadhyaya, 2010]. The majority of the somatic NF1 mutations identifiable in both benign and malignant NF1-associated tumors serve to inactivate the normal NF1 allele [Knudson, 1971]. However, the presence of biallelic NF1 inactivation in both benign and malignant tumors indicates that constitutive Ras activation cannot fully explain the malignant transformation of a benign PNF to an MPNST and that additional genetic (or epigenetic) alterations are required to drive such transformation. In order to better understand the basis of tumorigenesis in NF1, the detailed characterization of all malignant tumor-associated genetic/genomic defects will be required. However, the somatic inactivation of cancer-associated genes may involve many different mutational mechanisms, from point mutations to large genomic rearrangements and even epigenetic changes [Bignell et al., 2010; Stratton et al., 2009; Kops et al., 2005; Rajagopalan et al., 2004. In NF1 tumors, somatic microlesions as well as LOH and CNAs are likely to play a key role in malignant transformation (reviewed by Upadhyaya, 2011)].
Both LOH analysis and/or direct somatic mutation screening at the NF1 locus have been reported for many NF1-associated tumors (both benign and malignant), including cutaneous and PNFs, MPNSTs, pheochromocytomas, myeloid leukemias, gastrointestinal tumors, glomus tumors, and bone tumors [Serra et al., 2000; Upadhyaya et al., 2004, Garcia-Linares et al., 2011; Bausch et al., 2007; Side et al., 1997; Stewart et al., 2007, 2008; Maertens et al., 2006; Brems et al., 2009b, Stevenson et al., 2006]. Such genetic analyses of MPNSTs have clearly shown that biallelic NF1 gene inactivation only occurs in the component Schwann cells or glial cells of these NF1-related tumors. What is still puzzling, however, is why only 10–15% of PNFs undergo malignant transformation to MPNSTs even though both cutaneous and PNFs have essentially the same cellular constitution. Alterations of other regions of the genome have also been detected in several NF1-related tumors including MPNSTs; these are known to involve the TP53, CDKN2A, and RB1 genes [Brems et al., 2009a; Cichowski and Jacks, 2001]. In contrast to the specific chromosomal translocations found in many synovial (and other types of) sarcoma, no consistently specific pathogenic chromosomal aberrations are known to be associated with MPNSTs, although a number of small recurring chromosomal alterations have been reported. Indeed, several cytogenetic studies have noted that MPNSTs possess complex karyotypes characterized by a wide spectrum of chromosomal aberrations including translocations and duplications as well as numerical gains and losses [Bridge et al., 2004; Fang et al., 2009]. Frequent deletions of the 9p21 region containing the CDKN2A (p16) gene have been reported in several NF1–MPNSTs [Cairns et al., 1995; Frahm et al., 2004; Nielsen et al., 1999; Perrone et al., 2003; Sabah et al., 2006]. At least two other regions on chromosome 17, in addition to that containing the NF1 gene at 17q11.2, appear to be involved in NF1 tumorigenesis, with gains at 17q21-q22 potentially accounting for the reported upregulation of the TOP2A gene from this chromosomal region in patients harboring MPNSTs and who have a poor disease outcome [Skotheim et al., 2003]. Loss of the 17p13 region, encompassing the TP53 gene, has been noted not only in NF1–MPNSTs [Legius et al., 1994; Lothe et al., 2001] but also in soft sarcomas and other cancers [Borrás et al., 2011; Latres et al., 1994]. In similar vein, aberrant expression of proteins of the Rb pathway has been associated with soft-tissue sarcomas including MPNSTs [Mawrin et al, 2002].
The identification of tumor-specific genomic copy number changes in a range of different cancers [Weir et al., 2004; Suzuki et al., 2008; Chen and Chen, 2008; Dear, 2009; Kuiper et al., 2010; Park et al., 2010; Barretina et al., 2010] has led to the increased use of SNP and CNV/CNA microarray platforms to screen tumor DNAs. Previous studies aimed at identifying the global genetic changes present in benign and malignant NF1 tumors have variously applied gene expression profiling and RT-PCR analysis to RNA from both benign and malignant tumors in order to identify specific gene expression signatures associated with malignancy. Significant alterations in the expression of many genes have been reported in MPNSTs, including the TP53, RB1, CDKN2A (p16INK4A and p14/ARF), CDKN1B (p27/KIP1), TWIST1, SOX9, SOX10, TNC, TRIO, NKD2, IRX2, tenascin C and tenascin XB, and EGFR loci [Lévy et al., 2004; Miller et al., 2006, 2009; Lévy et al., 2007 reviewed by Upadhyaya, 2011]. Although CNAs have been reported in many primary tumors, few such studies of NF1 tumors have so far been undertaken. Initially, we constructed an exon-based cancer gene microarray that specifically targeted 57 different genes whose expression had previously been reported to be significantly altered in MPNSTs [Mantripragada et al., 2008]. Subsequently, a 32K BAC array was used to analyze DNA from both benign neurofibromas and MPNSTs, thereby providing a genome-wide analysis of large [up to 150 kb] copy number changes potentially associated with progression to malignancy [Mantripragada et al., 2009] with an average resolution of 100 kb. However, the relatively low resolution of this array platform meant that many smaller (≤100 kb) genomic variants were unlikely to be identified, leading us here to repeat such a genome-wide analysis using the newly available Affymetrix Genome-wide SNP 6.0 platform, which permits the simultaneous high-resolution analysis of both CNA and LOH.
In a number of such microarray CNA studies, significant variability using different array platforms has been obtained when similar tumor types are analyzed [Dear, 2009]. Whether such variability arises from the different microarray platforms (BAC, PAC, SNPs, and oligonucleotides) used, or from the variable size, number, and genomic distribution of the probes in the microarray, or whether it relates to the inherent genetic heterogeneity of the tumors themselves, is often difficult to determine. We have found, however, that the Affymetrix SNP 6.0 platform provides both increased resolution (with an average interprobe distance of only 700 bp) and significantly greater genome-wide coverage (with 0.9 × 106 SNP probes and 0.95 × 106 CNV probes) by comparison with the older 32K BAC array [Kresse et al., 2008; Mantripragada et al., 2008, 2009].
A total of at least 17 different studies have now attempted to assess the chromosomal and genomic copy number changes associated with NF1-related tumors (see Table 4). Taken together, these studies indicate that genes involved in cell cycle regulation, cell growth, cell differentiation, and motility are specifically altered in terms of their copy number in MPNSTs. The present study has not only served to confirm these initial findings but also, courtesy of its higher resolution, identified additional genes that appear to be involved in MPNST development. For example, across chromosome 17, we find that several regions exhibit recurring copy number changes in MPNSTs; apart from the obvious 17q11.2 region containing the NF1 gene, two quite distinct regions at 17q22 and 17q24-q25 were also found to be frequently amplified in MPNSTs, whereas one region at 17p13 was consistently deleted in MPNSTs. The 17q24-25 region, which contains the BIRC5 gene, a gene previously reported to be amplified in MPNSTs [Storlazzi et al., 2006] and consistently upregulated in many other tumors [Kanwar et al., 2010], was found to be amplified in four MPNSTs. Survivin, the BIRC5 gene product, is a central inhibitory protein of apoptosis that also regulates mitosis. The 17q22 region, amplified in two MPNSTs in our study, contains the TOP2A gene known to play a role in cell proliferation [Skotheim et al., 2003]. The 17p13 region, which contains the TP53 gene, was found to be deleted in four MPNSTs but not in any of the benign tumors. Our CNA study also identified consistent amplification of the entire 5p region, reminiscent of findings from a previous CNA analysis of soft-tissue sarcomas [Adamowicz et al., 2006]. This latter study, which also analyzed 10 NF1-associated MPNSTs, found that most of the 5p13-p15.3 region was amplified in several different MPNSTs. Several of the 5p13-p15.3 region genes, including TRIO, IRX2, and NKD2, were also found to exhibit increased expression specifically in MPNSTs [Adamowicz et al., 2006]. TRIO is a member of the large family of guanine nucleotide-exchange factors that activate Rho–GTPases, specifically the RAC1 and RhoA Rho family GTPases, by accelerating GDP/GTP exchanges. TRIO has been implicated in the control of cytoskeleton organization, transcriptional regulation, cell cycle progression, apoptosis, vesicle trafficking, and cell–cell adhesion, whereas significant TRIO gene amplification has been previously associated with both rapid tumor cell proliferation and increased invasive tumor growth in bladder cancer [Zheng et al., 2004].
| Gene or chromosomal changes | ||||
|---|---|---|---|---|
| References | Tumors studied | Analytical methods used | Gains | Losses |
| Lothe et al. (1995, 1996) | Neurofibromas (15) MPNSTs (9) | 17q LOH study FISH analysis | – | 17q allelic imbalance 4/6 NF1-associated MPNSTs |
| Lothe et al. (1996) | MPNSTs (7) | Chromosome CGH | Increased 17q24-ter copy number in 5 of 7 tumors | Loss of 13q14-q21 in 6/10 MPNSTs |
| Kourea et al. (1999) | MPNSTs (11) Primary tumors (8) Recurrent tumors (3) | INK4A cDNA probe on Southern blot | – | INK4A deletions in ?? MPNSTs |
| Nielsen et al. (1999) | MPNSTs (6) | Multiplex PCR | – | Homozygous CDKN2A deletions in 3 MPNSTs |
| Schmidt et al. (2000) | MPNSTs (14) | Chromosome CGH | Gains on Chr 7; 8q; 15q; 17q | – |
| Skotheim et al. (2003) | MPNSTs (14) MPNSTs (16) MPNSTs (44) | Combined analyses 1) Chr:17-specific cDNA microarray 2) FISH analysis 3) Tissue microarray | TOP2A overexpressed in most MPNSTs | – |
| Levy et al. (2004) | MPNSTs (9) PNF (16) Dermal NF (10) | Real-time quantitative PCR on tumor RNA | MK167, BIRC5, MMP13, MMP9, TERT, TERC, TOP2A, SPP2, FOXM1, FOXA2, HNF3B, HMMR, CXCL5, OSF2, CCNE2, EPHA7 and TP73 only upregulated in MPNST | ITGB4, CMA1, LICA1, DHH, S100B, ERBB3, RASSF2, TPSB and SOX10 all downregulated only in MPNSTs |
| Adamowicz et al. (2006) | Soft-tissue sarcomas (34) included MPNSTs (10) | Chr:5p BAC microarray | TRIO, IRX2, and NKD2 all located at 5p15.3 are amplified | – |
| Miller et al. (2006) | Primary MPNSTs (44) MPNST cell lines (8) Normal Schwann cells (7) | Gene expression profiling (Affymetrix chip) | EDGRE, SOX9, TWIST1 | SOX10, CNP, PMP2 |
| Storlazzi et al. (2006) | MPNSTs (28) Primary tumors (25) Recurrent tumors (2) Metastatic tumor (1) | Interpase FISH analysis of 17q with BAC probes | ERBB2 and TOP2A in 17q12>17q25 amplicon that also contains BIRC5 | – |
| Kresse et al. (2008) | High-grade MPNSTs (7) | Array CGH | LOXL2, TOP2A, ETV4, and BIRC5 | – |
| Tabone-Eghanger et al. (2008) | MPNSTs (42) | FISH | EGFR | – |
| Mantripragada et al. (2008) | MPNSTs (35) PNFs (16) Derm NFs (8) | Targeted gene array | ITGB4, PDGFRA, MET, HGF and PP73 | NF1, HMMR, MMP13, INK4A and CDKN2B |
| Perrone et al. (2009) | Sporadic MPNST (11) NF1 MPNST (16) | COMBINED ANALYSES 1) FISH 2) Gene activation 3) Mutational analysis | PDGFRA and PDGFRB EGFR copy number | – |
| Mantripragada et al. (2009) | MPNSTs (24) | 32K BAC genomic array | NEDL1, AP3B1, and CUL1 | CFKN2A and CDKN2B |
| Miller et al. (2009) | MPNST cell lines (13) Neurofibromal Schwann cells (22) Primary MPNSTs (6) | Gene expression profiling (Affymetrix chip) Affymetrix | SOX9, TWIST1 | – |
| Yu et al. (2011) | Primary MPNSTs (125) | Affymetrix Genome-Wide 500K SNP | CDK4, FOXM1, SOX5, NOL1, MLF2, FKBP4, TSPAN31 | ERBB2, MYC, TP53 |
Our earlier study found that the hepatocyte growth factor (HGF) gene, the cellular HGF receptor (MET) gene, and the platelet-derived growth factor receptor A (PDGFRA) gene were concomitantly amplified (despite their being located on different chromosomes) in a number of MPNSTs but not in PNFs or cutaneous neurofibromas [Mantripragada et al., 2008]. The HGF–PDGFRA–p70S6K pathway is known to play an essential role in tumor angiogenesis, and rapamycin treatment could therefore be considered as a means to limit tumor growth by inhibiting p70S6K. In the present study, MET displayed a copy number of 2.8–3.0 in M3, M5, and MX, whereas PDGFRA displayed a copy number of 3–4 in MX (but only 1.2–1.5 in M3 and M7).
Our previous 32K BAC array-based analysis [Mantripragada et al., 2009] of MPNST DNA revealed that genomic gains (62%) were much more frequent than genomic losses (38%) in these malignant tumors. A number of genes were also consistently amplified in MPNSTs; indeed, the NEDL1, AP3B1, and CUL1 genes located at 7q, 5q14, and 7q36 were amplified in some 60% of tumors [Mantripragada et al., 2009]. The most frequently deleted region, at 9p21, which encompasses the CDKN2A, CDKN2B, and MTAP genes, was, however, only deleted in some 30% of tumors. Several of the abovementioned six genes have also been implicated in a number of other cancers [Goh et al., 2011; Mizuarai et al., 2011] and they may, therefore, prove to have potential diagnostic, prognostic, and even therapeutic relevance in the context of NF1 tumorigenesis.
The results of the Affymetrix SNP 6.0 analysis described here extended our previous findings [reviewed by Upadhyaya, 2011] based on different sets of benign and malignant tumors with recurrent MPNST-specific amplified CNAs being found at 1q24, 1q34, 17q23.2, and 8p23. These copy number changes have already been noted to affect a variety of different genes including ITGB4, BIRC5, MET, ETV4, TWIST1, TRIO, NKD2, IRX1, IRX2, IRX4, EGFR, SOX9, HOXB5-9, TOP2A, MMP13, TP53, CDKN2A, RB1, TNC, HMM, CDK4, and FOXM1 (Table 4). The use of the Affymetrix SNP 6.0 Array in this study led to the identification of several additional genes that were consistently amplified in MPNSTs, including the ITGB8, PDGFA, and RAC1 genes (all at 7p21.1-p22) and the PDGFRL gene (8p22-p21.3), and the MMP12 gene (11q22.3), which was deleted in MPNSTs.
The amplification in MPNSTs of two related integrin genes, ITGB4 at 17q25 and ITGB8 at 7p21.1-p22, is intriguing because integrin signaling regulates the metastatic cancer phenotype [Guo, 2004], mainly through synergistic interactions with several critical signaling pathways, including the Ras/Raf/MAPK pathway, the Rho-effector pathway, and the epidermal growth factor receptor pathway [Schubbert et al., 2004]. The RAC1 gene is a member of the Rac subfamily of the large family of Rho–GTPases directly involved in cell signaling and the regulation of cell growth, cell–cell adhesion, cytoskeletal reorganization, and the activation of protein kinases. RAC1 protein interacts with the T-cell lymphoma invasion and metastasis-inducing protein 1 and both are upregulated in nasopharyngeal carcinoma [Gao et al., 2001; Wu et al., 2007; Qi et al., 2009; Worthylake et al., 2000]. The MMP12 gene is one of several MMP genes located at 11q22.3. As with most other MMP proteins, activated MMP12 degrades soluble and insoluble elastin, and is directly involved in extracellular matrix protein degradation [Shapiro, 1998, 1999]. MMP12 has been reported to be involved in glioma cell invasion in the brain, apparently through its direct interactions with tenascin C [Erickson and Bourdon, 1989]. Changes in MMP12 gene expression might therefore be expected to accompany increased invasiveness of central nervous system (CNS) tumors. As the PDGF signaling system is upregulated in many cancers, the finding of coamplification of the PDGFA and PDGFRL genes in four MPNSTs in the present study may well be functionally significant. A previous investigation of the PDGF pathway in MPNSTs also noted increased expression of PDGFRA and PDGFRB, as well as the EGFR gene where increased gene expression and protein phosphorylation was found to correlate with gains in EGFR copy number [Perrone et al., 2007]. A more recent study also reported increased EGFR expression and PDGFRA amplification in MPNSTs and noted that both features were associated with a poor prognosis [Tabone-Eglinger et al., 2008]. Once activated, EGFR and PDGFR stimulate tyrosine kinase activity and intracellular pathways that control proliferation and differentiation.
The present study identified a much larger number of deleted genomic regions in MPNSTs than the two previous studies [Kresse et al., 2008; Mantripragada et al., 2009]. However, we failed to identify any relationship between the grade of MPNSTs (Supp. Table S1) and the CNA profile. Furthermore, the copy number profiles generated from the individual MPNSTs appear to identify two distinct subgroups of malignant tumors: those with a major CNA profile (tumors M2, M3, M6, M7, M8, and M15) and those with a minor CNA profile (tumors M4, M10, M11, M12, and M13) comparable with the copy number profiles obtained from benign PNFs. It is perhaps difficult to reconcile these data pertaining to inter-MPNST heterogeneity, especially as several of the MPNSTs in both subgroups were classified as high-grade tumors. Similarly, the finding of a high-CNA profile associated with a benign PNF (P116) derived from a sporadic NF1 patient is puzzling, although one explanation could be that this PNF was in transition toward MPNST development. This disparity in CNA profiles between apparently high-grade MPNSTs may also be a consequence of the considerable somatic genetic heterogeneity associated with these malignant tumors [reviewed by Upadhyaya, 2011] and further highlights the significant diagnostic and therapeutic challenges posed by these histologically complex and genetically highly heterogeneous tumors [Stasik & Tawfik, 2006; Spurlock et al., 2010].
Loss-of-heterozygosity and CNAs are only two of the many genetic mechanisms potentially involved in NF1 tumorigenesis. Hence, MPNSTs lacking a significant number of CNAs may harbor microlesions and/or aberrant methylation of gene promoter regions. However, the significant degree of LOH found in most MPNSTs, LOH that is essentially absent from benign neurofibromas, confirms our previous finding that many NF1-associated MPNSTs harbor large somatic NF1 deletions [Upadhyaya et al., 2008; Botillo et al., 2009]. Furthermore, CNN–LOH analysis indicated that CNN–LOH is less common than LOH (allelic imbalance) in the development of MPNSTs and this concurs with our earlier finding.
This study is the first to assess both LOH and CNA within the same panel of MPNST and PNF tumors. This approach has yielded new insights into the biological pathways involved in the malignant transformation of PNFs in NF1. This study has also identified a number of candidate genes that may be involved in MPNST development. Further studies of these genes should now be performed with a larger set of tumor samples, both to confirm the copy number changes and to seek additional types of tumor-associated mutation within these genes.
It is unclear what would be the net effect of the observed CNAs on cellular properties and how this would impact upon tumorigenesis. A better understanding of how such CNAs influence the expression of the genes they encompass is required in order to assess their possible role in tumorigenesis. Such studies are ongoing, one of which directly assessed the impact that SNPs and CNVs have on gene expression, finding that less than 20% of gene expression variability was directly attributable to CNVs of ≥40 kb in size [Henrichsen et al., 2009a, 2009b]. In addition, at least half of the genes whose expression was directly influenced by a CNV were actually located outside the relevant CNV [Henrichsen et al., 2009a, 2009b], an indication that many CNVs affect gene regulatory sequences located at some distance from the target gene. A genetic pathway represents a set of interactions occurring within and between a group of genes that are dependent upon each other's individual functions in order to make the aggregate function of the network available to the cell. Gene pathways are composed of multiple genes with coordinated functions. Thus, a genetic mutation disrupting the function of one gene in a pathway necessarily breaks the informational connection between genes acting before and after the mutant gene in that pathway. Several new insights into pathways potentially involved in NF1 tumorigenesis were obtained during the course of this study.
The principal requirements for cancer metastasis to occur include tumor cell adhesion, migration, cell invasion, and wound healing. The present study identified CNAs affecting several genes from the Ras-homologous (Rho)–GTPase signaling pathway (ACTB, ACTG1, LIMK1, PRKCA, PTK2, RAC1, ROCK2, and TRIO) specifically in MPNSTs. The potential involvement of this signaling pathway in tumor development is likely to be important because Rho–GTPases are key regulators of actin reorganization, cell motility, cell–cell and cell–extracellular matrix adhesion, cell cycle progression, gene expression, and apoptosis [Etienne-Manneville and Hall, 2002], processes that are involved in both malignant transformation and metastasis. Many of the copy number gains observed in our study may therefore be directly implicated in the malignant transformation from benign PNF to malignant MPNST through the promotion of cell proliferation, motility, and metastasis and by the specific alteration of the cellular pathways that serve to reorganize the cytoskeletal structures. Functional analysis of MPNST cell lines transfected with shRNAs for RAC1, ROCK2, PTK2, and LIMK1 indicated that targeted knockdown of at least four of the genes that map to the Rho–GTPase pathway reduces the malignant potential of these cells as evidenced by a significant reduction in wound healing, invasion, and migration and an increase in cell adhesion. On this basis, we propose that it would be worth exploring whether the use of specific inhibitors of individual Rho functions could be used to provide therapeutic benefit in patients with MPNSTs [Fritz & Kaina, 2006]. The use of such an inhibitor in a prostate carcinoma cell line has been shown to inhibit the proliferation, anchorage-dependent growth, and invasiveness of these tumor cells [Gao et al., 2004].
The use of DNA copy number data to explore the process of malignant transformation, as well as to subclassify different tumors, does however have several limitations, the most obvious being that many such tumor-specific CNAs involve multiple genes, many of which are unlikely to be involved in the process of tumorigenesis. Hence, the identification of diagnostically and prognostically applicable genetic markers in tumors is always going to be challenging. Indeed, this is clearly demonstrated in the present study, which revealed that no single gene, or even multiple gene set, was consistently altered in all MPNSTs as compared with PNFs. Other mutational mechanisms will therefore need to be considered in NF1-assocated tumorigenesis, for example, microlesions, increased microsatellite instability, and even epigenetic changes that may influence gene expression either specifically or globally. A major problem with many of the high-resolution analytical technologies recently applied to the study of human tumor genomes is the plethora of somatic mutation data they create. This mass of data then requires careful dissection to identify the very few “driver” mutations from among the many accompanying “passenger” mutations [Stratton et al., 2009]. Unfortunately, this is especially problematic in tumors such as MPNSTs, which exhibit both complex chromosomal abnormalities and significant cellular heterogeneity. A comprehensive study of a large panel of both NF-associated and sporadic MPNSTs, and that integrates copy number change measurements, gene expression analysis, and biologically relevant functional analyses, is clearly warranted. Such studies should follow the example of the comprehensive integrated genomic analyses that the Cancer Genome Atlas programme have applied to define a range of cancer genomes [Ding et al., 2008; Parsons et al., 2008]. It is intriguing that these and other similar studies consistently identify the presence of inactivating NF1 gene mutations in many of these tumors including ovarian serous carcinomas [Sangha et al., 2008; Kuo et al., 2009], acute myelogenous leukemia [Mullally and Ebert, 2010], neuroblastoma [Hölzel et al., 2010], lung cancer [Bignell et al., 2010, Ding et al., 2008; Kan et al., 2010], as well as in various other cancers [Kan et al., 2010]. These studies serve to highlight the central role that neurofibromin plays in the function of normal cells and supports the view that the loss of this function is a major cause of tumorigenesis. As somatic gene amplification, paralleled by increased gene expression, is a hallmark of many cancers, the generation of comprehensive maps of copy number variation [Bergamaschi et al., 2006; Lee et al., 2008] and their associated gene expression changes should contribute to a better understanding of the mechanisms through which CNAs promote tumorigenesis. Current challenges involve developing high-resolution methods for determining the association of CNAs with biological function in common cancer.
In summary, we have identified specific genomic changes in MPNSTs as reflected by CNV, LOH, and CNN–LOH analyses. Some 1,521 genes were found to be associated with the specific CNAs in MPNSTs, several of which represent excellent candidates for involvement in tumorigenesis, including novel changes involving ITGB8, PDGFA and RAC1 (all 7p21-p22), PDGFRL (8p22-p21.3), and MMP12 (11q22.3). We also report that allelic imbalance in NF1-associated MPNSTs is more prevalent than CNN–LOH, consistent with the view that imbalances of expression are critically important in tumorigenesis. Our other key finding was the involvement of several genes of the Rho–GTPase pathway, which could prove to have a pivotal role in the regulation of processes involved in NF1 tumorigenesis and metastasis. shRNA for RAC1, ROCK2, PTK2, and LIMK1 were used in adhesion, migration, and wound healing assays and also RAC1 and ROCK2 in cell invasion in control and MPNST-derived cell lines. The results of these assays clearly demonstrated that targeted knockdown of such genes is capable of reducing the malignant potential of these cells. Taken together, we have identified several new potential targets for therapeutic intervention during MPNST development.
Acknowledgements
We thank all the patients who have contributed to this study. We are grateful to Dr. Paul Northcote for his comments. We thank Cancer Research UK for their financial support. We are grateful to Dr. Jim Neal for his advice on IHC work and Chris Lovejoy for technical assistance.
Disclosure Statement: The authors declare no conflict of interest.