Classification of mismatch repair gene missense variants with PON-MMR†
Communicated by A. Jamie Cuticchia
Abstract
Numerous mismatch repair (MMR) gene variants have been identified in Lynch syndrome and other cancer patients, but knowledge about their pathogenicity is frequently missing. The diagnosis and treatment of patients would benefit from knowing which variants are disease related. Bioinformatic approaches are well suited to the problem and can handle large numbers of cases. Functional effects were revealed based on literature for 168 MMR missense variants. Performance of numerous prediction methods was tested with this dataset. Among the tested tools, only the results of tolerance prediction methods correlated to functional information, however, with poor performance. Therefore, a novel consensus-based predictor was developed. The novel prediction method, pathogenic-or-not mismatch repair (PON-MMR), achieved accuracy of 0.87 and Matthews correlation coefficient of 0.77 on the experimentally verified variants. When applied to 616 MMR cases with unknown effects, 81 missense variants were predicted to be pathogenic and 167 neutral. With PON-MMR, the number of MMR missense variants with unknown effect was reduced by classifying a large number of cases as likely pathogenic or benign. The results can be used, for example, to prioritize cases for experimental studies and assist in the classification of cases. Hum Mutat 33:642–650, 2012. © 2012 Wiley Periodicals, Inc.
Introduction
Lynch syndrome or hereditary nonpolyposis colorectal cancer (HNPCC) accounts for approximately 2–5% of colorectal cancers [Hampel et al., 2008; Lynch et al., 2009]. The patients are exposed in addition to colorectal cancer to some extracolonic cancers (endometrium, stomach, ovary, kidney, urinary tract, biliary tract, small intestine, brain, and skin tumors). The syndrome is caused by germline mutations in mismatch repair (MMR) genes. These genes are MLH1 (MIM# 120436), MLH3 (MIM# 604395), MSH2 (MIM# 609309), MSH6 (MIM# 600678), PMS1 (MIM# 600258), PMS2 (MIM# 600259), or TFGBR2 (MIM# 190182). The role of PMS1, TGFBR2, and MLH3 in Lynch syndrome is still elusive. MMR is an evolutionary conserved DNA repair system that recognizes and repairs base–base mispairs and insertion–deletion loops arising during DNA replication and recombination. MMR malfunction affects DNA stability, which can result in microsatellite instability.
Thousands of MMR variants have been identified and stored to databases including InSiGHT (http://www.insight-group.org) and MMR Gene Unclassified Variants (http://mmruv.info/), but the relevance to cancer has been verified just in a small number of cases. Even for experimentally studied cases, the situation may be confusing, for example, R217C variant in MLH1 has been classified as pathogenic [Fan et al., 2007], neutral [Takahashi et al., 2007; Trojan et al., 2002], and as having unknown effect [Ellison et al., 2001]. In addition to experimental methods, the pathogenicity of a variant can be predicted with bioinformatic methods [Thusberg and Vihinen, 2009]. Bioinformatic predictors provide valuable information faster, easier, and cheaper than laboratory methods.
Experts in the field have organized to International Agency for Research on Cancer (IARC) unclassified genetic variant working group to establish standards for the classification of variants, including the terminology, evaluation, and validation of data [Tavtigian et al., 2008]. IARC has suggested a five-tier classification system [Plon et al., 2008] based on the probability of being pathogenic derived from clinical, genetic, in vitro, in vivo, and in silico information. Only a small number of MMR variants have been classified so far. The most extensive effort for MMR genes and proteins is taken by the InSiGHT Interpretation Committee; however, results have not yet been published.
We developed a dedicated prediction tool for MMR missense variants and applied it to analyze 616 unclassified variants (UVs). We reduced the number of UVs substantially by classifying 81 MMR missense variants as disease related and 167 as neutral. The results can be utilized to prioritize variants for further experimental validation and diagnosis of Lynch syndrome and other cancers together with clinical and other information.
Materials and Methods
MMR Missense Variants
Altogether 784 MMR missense variants for Lynch syndrome patients were downloaded (January 27, 2011) from the International Society for Gastrointestinal Hereditary Tumours (InSiGHT) database at http://www.InSiGHT-group.org. The unique MMR variants were distributed to five MMR proteins as follows: MLH1 (287), MLH3 (18), MSH2 (226), MSH6 (156), and PMS2 (97).
Functional effects were used as the signs of the pathogenicity of the variants. Information about functional assays was searched from literature. The experimentally verified functional effects of MMR missense variants were collected from articles. The most widely applied methods in these studies included in vitro MMR activity [Christensen et al., 2009; Drost et al., 2010; Jäger et al., 2001; Kansikas et al., 2011; Kariola et al., 2004; Korhonen et al., 2008; Nyström-Lahti et al., 2002; Ollila et al., 2006a, b; Raevaara et al., 2004, 2005; Takahashi et al., 2007; Trojan et al., 2002]. Additional methods were in vivo DNA MMR assays in yeast [Ellison et al., 2001], yeast two-hybrid system [Fan et al., 2007; Ou et al., 2009], and RNA expression [Pagenstecher et al., 2006].
Some variants had been studied several times and if the reports disagreed on the effect, the conclusions of the latest, most extensive, consistent, and systematic studies of Kansikas et al. [Kansikas et al., 2011] and Takahashi et al. [Takahashi et al., 2007] were utilized. With the cases investigated by Kansikas et al., special attention was given to MMR activity, microsatellite instability, expression, and localization. Cases for which at least two methods agreed were classified as disease causing or tolerated. Variants analyzed by Takahashi et al. were grouped based on in vitro MMR activity by using 60% (as recommended by the authors) as a threshold. In their study, gene expression values varied too much to be informative and correlation with dominant mutation effect was so poor that the enzyme activity was only reliable information type similar to the other studies, where experimental results were used as the basis for the variant classification.
Studies of Kansikas et al. and Takahashi et al. unanimously agreed on the definition of all the 11 overlapping variants. The results of all predictive tools were excluded as unreliable and because their use would have been circuitous in a case of a novel prediction tool. Altogether, data was available for 168 functionally tested MMR missense variants, out of which 80 were pathogenic. This dataset had 123 variants in MLH1, 11 in MLH3, 27 in MSH2, and 7 in MSH6 protein. The remaining 616 unclassified MMR missense variants were distributed to proteins as follows: 164 for MLH1, 7 for MLH3, 199 for MSH2, 149 for MSH6, and 97 for PMS2. There were no missense variants in PMS1 and TFGBR2.
Prediction of Pathogenicity
Pathogenic-or-not-pipeline (PON-P) [Olatubosun et al., 2012] at http://bioinf.uta.fi/PON-P was utilized for the submission, prediction, and analysis of protein sequences and MMR missense variants with various bioinformatic prediction methods. Variant tolerance prediction methods included Mutation Taster [Schwarz et al., 2010], MutPred [Li et al., 2009], nsSNPAnalyzer [Bao et al., 2005], PhD-SNP [Capriotti et al., 2006], PMut [Ferrer-Costa et al., 2005], PolyPhen2 [Adzhubei et al., 2010], SIFT [Ng and Henikoff, 2003], SNAP [Bromberg and Rost, 2007], and SNPs&GO [Calabrese et al., 2009]. Sequence-based stability effect predictions were performed with SCPRED [Dosztányi et al., 1997], MUPRO [Cheng et al., 2006], and I-Mutant 3.0 [Capriotti et al., 2005], and structure-based predictions with SCide (stabilization centers) [Dosztanyi et al., 2003] and SRide (stabilizing residues) [Magyar et al., 2005] for MSH2 and MSH6 variants.
Structural disorder was predicted with MetaPrDOS [Ishida and Kinoshita, 2008], PrDOS [Ishida and Kinoshita, 2007], DISORPED2 [Ward et al., 2004], DisEMBL [Linding et al., 2003], DISPROT (VSL2P) [Peng et al., 2006], DISpro [Cheng et al., 2005], IUpred [Dosztanyi et al., 2005], and POODLE-S [Shimizu et al., 2007].
All the variants were entered to protein aggregation predictors Aggrescan [Conchillo-Sole et al., 2007] and Waltz [Oliveberg, 2010]. The interatomic contacts of variants in MSH2 and MSH6 protein structure were checked with CMA (Contact Map Analysis) [Sobolev et al., 2005], CSU (Contacts of Structural Units) [Sobolev et al., 1999], and RankViaContact [Shen and Vihinen, 2003].
The default parameters were utilized in all the prediction methods, and only the protein sequence and MMR missense variant were provided as input. Blastp [Altschul et al., 1997] was used to search for homologous sequences in NCBI nonredundant sequence database for all the MMR proteins. Multiple sequence alignments containing only full-length sequences were obtained with ClustalW [Chenna et al., 2003]. We selected sequences only with known functions and removed putative or hypothetical sequences. Conservation for each variant position in sequence alignment was determined with PAM250 and Blosum 62 amino acid substitution matrices.
Quality Parameters for Tolerance Prediction Methods
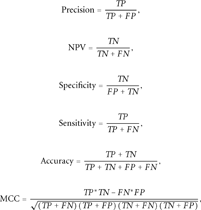
In order to be able to compare various methods with the different numbers of predicted cases, the numbers of negative cases were normalized to be equal with those for positive cases.
Novel Classifier
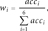

The pathogenicity score for PolyPhen2 was set to 0 for benign predictions and 1 for pathogenic predictions.
The pathogenicity scores were formulated such that the higher the reliability and probability of a prediction, the closer the pathogenicity score approaches 1 for pathogenic predictions, or 0 for neutral predictions. Lower reliability or probability induces the pathogenicity score to approach 0.5 in both cases.
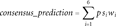
The upper and lower cutoff values were established such that variants on the evaluation set having pathogenicity score greater than the upper cutoff value 0.7615 are classified as pathogenic, those having scores lower than the lower cutoff value 0.351 are classified as neutral, and those in-between left unclassified.
Structural Effects of MSH2 and MSH6 Missense Variants
The effects of MSH2 and MSH6 missense variants were studied based on the structure of the heterodimer in PDB entry 2O8B [Warren et al., 2007]. Recognition of secondary structural elements in proteins was done with STRIDE [Heinig and Frishman, 2004] and visualization with program Pymol [Schrödinger, 2010].
Results
Our aim was to group previously unclassified MMR missense variants as pathogenic or neutral. To do this, we first investigated the suitability of a wide spectrum of prediction methods, in total 30 programs, to classify experimentally verified MMR variants. After finding deficiencies in prediction performance, we developed a novel classifier.
Testing Prediction Method Performance with Known MMR Missense Variants
We retrieved 168 experimentally verified MMR missense variants with known functional effect from the literature (Table 1) of which 80 were pathogenic and 88 neutral. The variants have highly biased distribution in the MMR proteins. MLH1 contains the majority (123 cases, 73%) of the variants.
The dataset of cases with functional information was utilized to test the suitability of a large number of bioinformatic prediction methods. The distinct prediction method categories included tolerance, stability, disorder, aggregation, interatomic contacts, and sequence conservation. Of these, only the tolerance prediction methods demonstrated correlation to experimental results and thus were employed in subsequent studies.
The performance of the tolerance prediction methods, as analyzed with six quality measures, is displayed in Table 2. The best individual method measured by accuracy (0.8) and MCC (0.61) is nsSNPAnalyzer followed by SNPs&GO, which has the highest precision (0.83) and specificity (0.86). Mutation Taster has relatively low accuracy (0.63) and MCC (0.37), but the best sensitivity (0.98) and NPV (0.93) values. None of the individual methods can provide highly accurate results alone.
| Mutation Taster | MutPred | nsSNPAnalyzer | PhD-SNP | PolyPhen | SIFT | SNAP | SNPs&GO | |
|---|---|---|---|---|---|---|---|---|
| TP | 80 | 77 | 67 | 71 | 75 | 70 | 73 | 59 |
| FP | 59 | 50 | 20 | 25 | 43 | 45 | 55 | 12 |
| TN | 25 | 36 | 50 | 61 | 43 | 41 | 31 | 74 |
| FN | 2 | 4 | 9 | 11 | 7 | 12 | 3 | 23 |
| Cases P/Na | 82/84 | 81/86 | 76/70 | 82/86 | 82/86 | 82/86 | 76/86 | 82/86 |
| Total numberb | 166 | 167 | 146 | 168 | 168 | 168 | 162 | 168 |
| Accuracyc | 0.63 | 0.68 | 0.80 | 0.79 | 0.70 | 0.66 | 0.64 | 0.79 |
| Precisionc | 0.58 | 0.61 | 0.77 | 0.74 | 0.64 | 0.61 | 0.57 | 0.83 |
| Specificityc | 0.30 | 0.42 | 0.71 | 0.71 | 0.50 | 0.48 | 0.36 | 0.86 |
| Sensitivityc | 0.98 | 0.95 | 0.88 | 0.87 | 0.91 | 0.85 | 0.96 | 0.72 |
| NPVc | 0.93 | 0.90 | 0.85 | 0.85 | 0.86 | 0.77 | 0.91 | 0.76 |
| MCCc | 0.37 | 0.43 | 0.61 | 0.58 | 0.45 | 0.36 | 0.39 | 0.59 |
- aNumber of experimentally verified pathogenic (P) and neutral (N) cases predicted by the program.
- bTotal number of cases predicted by the program.
- cCalculated from normalized numbers.
MMR Missense Variant Classification by Consensus Predictor
As only tolerance prediction methods correlated with the experimental MMR missense variant effects, we utilized them to develop our own method. For that purpose, we combined the predictions of five tolerance predictors: PhD-SNP, PolyPhen2, SIFT, SNAP, and SNPs&GO. We introduced pathogenicity score that is calculated from the classifications of individual classifiers and the reliability of these predictions. The cutoff values of the consensus predictor were optimized to be 0.351 and 0.7615. The optimized consensus predictor has improved accuracy (0.87), precision (0.81), specificity (0.77), sensitivity (0.97), NPV (0.65), and MCC (0.77) in comparison with the individual methods when testing with 95 variants for which it gave prediction pathogenic or neutral out of total 162 training variants as all the utilized programs could not predict the outcome of all the 168 cases.
The new predictor was used to classify the dataset of 616 variants with unknown effect. Predictions with high score were obtained for 248 variants (40.3%) of which 81 were predicted to be pathogenic and 167 neutral (Table 3). The MMR consensus classifier called PON-MMR (http://bioinf.uta.fi/PON-MMR) is freely available as part of the PON-P service.
| Pathogenic | Neutral | |||||||
|---|---|---|---|---|---|---|---|---|
| MLH1 | MSH2 | MSH6 | PMS2 | MLH1 | MLH3 | MSH2 | MSH6 | PMS2 |
| A21E | Y43C | L435P | E705K | I32V | V420I | T8M | A20V | A182T |
| R27P | D49V | G566R | S815L | E53A | V741F | A72L | N21S | S445T |
| N38K | L93P | C765W | C843Y | S95A | P844L | V102I | A25S | P446S |
| G67E | N127I | L792P | R127K | V971I | R106K | P42S | S455A | |
| G98R | L173R | C1158R | L135V | M141V | G54A | I462L | ||
| G98S | L175P | L166F | A189S | S65L | I462M | |||
| G101D | L310P | V213A | G203R | A81T | I462T | |||
| G101S | L310R | V213L | A207S | L147H | V467G | |||
| S106R | G338R | E320D | I216V | K185E | L468F | |||
| V113D | R359S | A353V | I237V | K187T | L468V | |||
| Y126N | L387P | T364A | K248E | E221D | R469I | |||
| G147R | Y408C | H381Y | N331D | N223S | P470S | |||
| I216S | L421P | L400V | P336S | I251V | E473V | |||
| L260R | L440P | K416E | V342I | T269S | S477F | |||
| L272S | V470E | D418E | N361S | G289D | H479Q | |||
| V303E | R524L | P435L | L390F | S315F | T485K | |||
| V384D | R524P | G454R | Q419K | A326V | D502E | |||
| A539D | R534C | M458K | T441P | F340S | I508L | |||
| Q542P | D603G | S459L | D487E | R361H | D510E | |||
| L559R | D603Y | K461N | G508S | R378K | T511A | |||
| F568I | H639Y | N468D | N547S | L396V | Y519C | |||
| L622P | C641G | D485H | S554T | I425V | A520V | |||
| G634R | G669R | R487Q | E561K | S532A | S523T | |||
| L636P | G669D | P496R | T564A | K610N | D526E | |||
| P640L | G669S | E515K | M592V | P623A | P540T | |||
| P640T | G669V | R522Q | N596S | R644S | N554H | |||
| F656S | P670L | E578G | Q629R | I669T | L571I | |||
| R659L | N671Y | A623S | A636V | E675D | A572T | |||
| W666R | G674R | N635S | T682A | Q698E | T573S | |||
| C680R | G674S | N645S | I770V | I725M | K581E | |||
| R725H | G683R | V647M | T803A | R761K | E583K | |||
| L749P | L687P | E668K | T807S | A787V | L585I | |||
| L749Q | M688R | L724M | N835H | V800A | S587D | |||
| Q690E | S860L | V800L | S587T | |||||
| G692R | A870G | D803G | I590L | |||||
| G692V | T905I | P831A | L594F | |||||
| C697R | T905R | V878A | L594V | |||||
| D748Y | I930M | I886V | T597S | |||||
| G751R | F985L | M600I | ||||||
| G827R | I1054F | M600L | ||||||
| P1073R | I629L | |||||||
| P1073S | E635N | |||||||
| P1082L | ||||||||
| Y1128C | ||||||||
| E1163V | ||||||||
| M1202T | ||||||||
| E1254D | ||||||||
| R1304K | ||||||||
| E1310D | ||||||||
| S1329L | ||||||||
Features of Pathogenic and Neutral Missense Variants
The distributions of the mutated (original) and mutant amino acids in the functionally verified set of 168 cases are biased both for pathogenic and neutral MMR missense variants. Among the pathogenic variants (Supp. Table S1), glycine and leucine occur more frequently in the original amino acid residues, whereas arginine and proline are overrepresented among the mutant residues. Alanine and isoleucine appear in excess among neutral variants in the original amino acids, while threonine and valine overrepresent in the mutant residues.
Pathogenic MMR variants have more substitutions from leucine to proline (12 cases) and glycine to arginine (11 cases) while neutral MMR variants have more substitutions from isoleucine to valine (7 cases) and asparagine to serine (7 cases). The numbers are too small for statistical analysis; however, they are in line with general variation distribution [Thusberg et al., 2011].
Structural Effects of MSH2 and MSH6 Missense Variants
We were able to inspect the structural effects of MMR missense variants only on MSH2 and MSH6, because protein three-dimensional (3D) structures are known just for these two proteins. We investigated the effects of the predicted pathogenic and neutral variants based on the protein dimer structure and paid attention to the location of the original residue on the protein surface or core, localization in secondary structures, possible sterical clashes of the substituted amino acid side chains, and effects on electrostatistics.
Altogether, we studied 109 variants of which 63 neutral ones were considered not to substantially affect the structure, for example, due to conservative substitutions, appearing on the protein surface. One of the MSH2 variants, N547S was predicted to be neutral although it participates in DNA binding and an alteration in it would be pathogenic. We concluded that at least 42 of 45 pathogenic variants (93%) may have serious effect, due to the introduction of structural strain, decreasing stability, missing interchain interactions or changing the DNA binding cleft (Fig. 1).
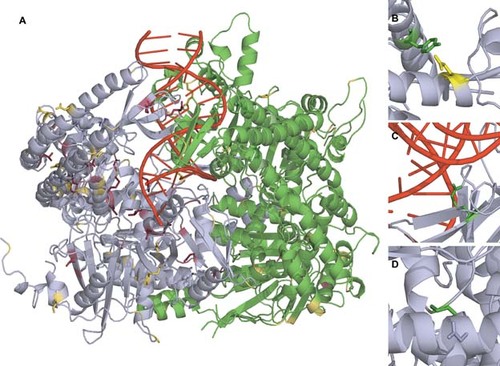
(A) MSH2-MSH6 protein dimer in PDB entry 2O8B with the positions of variants colored. MSH2 is in cyan and MSH6 in green. Variants predicted to be pathogenic are in red and neutral variants in yellow. Structure includes in addition a stretch of double stranded DNA in red. Examples of variation effects: (B) Variation of Y408 (green) to cysteine is likely harmful because the ionic interaction with E455 (yellow) in another α-helix is removed. (C) Substitution R524P (green) is considered as pathogenic because of the structure alteration and prevention of DNA recognition. (D) G692R (green) substitution appears in a tight turn. There is not sufficient space to fit the extended arginine side chain.
The locations of the predicted neutral and pathogenic variants, and some examples of effects are illustrated in the MSH2–MSH6 complex structure (Fig. 1). The structure is for a truncated version of MSH6, and thus only variants after sequence position 362 are visualized. As both chains contained some gaps, nine additional variants could not be studied at structural level.
Discussion
We classified MMR missense variants into pathogenic and neutral cases by utilizing a novel consensus predictor. First, we tested the performance of altogether 30 predictors in several categories including tolerance, stability, disorder, aggregation, interatomic contacts, and sequence conservation with 168 experimentally verified MMR variants. Only tolerance methods correlated with variant severity (i.e., pathogenicity). The methods had significant performance differences, for example, MCC varied from 0.36 to 0.61. The best individual method proved to be nsSNPAnalyzer; however, its performance was not considered sufficient. The novel method builds a consensus from the output of five tolerance methods and their reliability estimates. This method utilizes results from PhD-SNP, PolyPhen2, SIFT, SNAP, and SNPs&GO and classifies the variants as pathogenic, neutral, or UV. We did not include nsSNPAnalyzer in the new predictor as it cannot predict many of the variants due to missing 3D structure data for some of the MMR proteins in the ASTRAL database it uses. Previous studies indicated that the performance of tolerance [Thusberg et al., 2011] and protein stability [Khan and Vihinen, 2010] predictions vary significantly. With the new method, we were able to classify 81 variants as pathogenic and 167 as neutral, 368 remaining UVs. To the best of our knowledge, this is the largest bioinformatic effort to classify MMR missense variants.
The residue distribution among pathogenic and neutral MMR variants is biased. Residue alterations in the pathogenic variants include many substitutions to proline, which are generally pathogenic, because proline is a known protein secondary structure breaker. The probable reason for the high number in arginines among the mutated pathogenic residues is that four out of six codons for this amino acid contain the highly mutable CpG dinucleotide, a known mutational hotspot [Ollila et al., 1996]. Arginine substitutions remove the functionally important basic side chain. Another enriched amino acid among the pathogenic variants was glycine, which as the smallest amino acid appears frequently in tight turns where it cannot be replaced by any other residues. The observed amino acid substitution trends are consistent with those in protein secondary structures [Khan and Vihinen, 2007] and among known disease and benign variations [Thusberg et al., 2011].
PON-MMR classifies variants with the pathogenicity score higher than the upper cutoff value 0.7615 to be pathogenic and lower than the cutoff value 0.351 to be neutral, and those in between remain unclassified. This consensus prediction is calculated from the reliability and the probability of the prediction. Thus, we could not use the strict classification system that IARC recommends [Plon et al., 2008] for these variants.
As an independent study of the quality of the predictions we investigated the effect on the protein structure of two proteins, MSH2 and MSH6, for which 3D structures have been determined. This study of MSH2–MSH6 complex supported the predictions for 105 out of 109 variants. In the case of remaining four variants, we could not draw conclusive decision for three of them and one appears in DNA-binding site based on the structure, information that is not available for the predictors.
Numerous MMR missense variants have been identified from Lynch syndrome patients and investigated with experimental methods. In addition to the functional studies of missense, insertion, and deletion variants [Pagenstecher et al., 2006; Kansikas et al., 2011], the consequences of splicing in MMR genes have been studied [Betz et al., 2010]. PON-MMR was developed only for missense variants and, therefore, does not take into account other kinds of variants such as nonsense substitutions or mRNA splicing effects.
Some MMR missense variants have been classified previously with bioinformatic methods. Doss and Sethumadhavan [Doss and Sethumadhavan, 2009] predicted 125 MMR missense variants with SIFT, PolyPhen, and PupaSuite. Out of these, SIFT classified 22 and PolyPhen 40 variants as pathogenic. In addition, PupaSuite predicted the protein activity effects. They investigated MSH2 and MSH6 variants further based on protein structure. Chan et al. [Chan et al., 2007] classified 28 MLH1 and 14 MSH2 variants with SIFT, PolyPhen, and A-GVGD. They did not note major differences in the performance of the methods. In silico methods can be applied for the priorization or evaluation of variants, for example, in whole-genome scans.
The effects of MLH1 variants that disturb the MLH1–PMS2 dimerization have been analyzed by examining protein expression, dimerization, MMR activity, and bioinformatic predictions [Kosinski et al., 2010]. Of 19 MLH1 variants, they classified 15 as pathogenic and 4 as UVs. Due to controversial results in literature, three variants, which they predicted to be pathogenic, were neutral in our evaluation data set. We based the classifications on the extensive functional data (for details, see section “Materials and Methods”). Six variants, which they predicted to be pathogenic, agree with our evaluation set. They predicted L749P and R755W to be pathogenic, while we classified them as UVs. Three variants, UV in their classification, were part of our neutral evaluation set. We both classified the variant D601G as UV. One of their variants was not a missense variant.
Chao et al. [Chao et al., 2008] have developed a classification system for MMR variants called MAPP-MMR. We used our evaluation set to estimate MAPP-MMR, which has been trained only with 24 pathogenic and 26 neutral variants. We used 138 cases, not used for training with which the PON-MMR cutoffs were optimized as the test set. MAPP-MMR had accuracy of 0.83, precision of 0.92, specificity of 0.88, sensitivity of 0.80, NPV of 0.71, and MCC of 0.65 being in performance between PON-MMR and the tolerance predictors.
We compared the performance of MAPP-MMR and PON-P with cases for which both methods provided prediction, either pathogenic or neutral. MAPP-MMR cannot predict all the instances in the dataset. Finally, there were 96 pathogenic or neutral variants in the test set. The methods agreed on the pathogenicity of 84 variants (45 were neutral and 39 pathogenic) of which 76 were correct predictions. All the cases predicted as pathogenic were correct, but 8 cases predicted as neutral although the functional classification indicated them to be disease associated. PON-P was somewhat better than MAPP-MMR with cases on which the methods disagreed, further it can predict all the test cases unlike MAPP-MMR. The user interface between both PON-P and PON-MMR allows the submission of more than one case at a time and does not require a manual picking of normal and variant amino acids as MAPP-MMR provided on a commercial site. Further, in comparison to MAPP-MMR, PON-P provides instructions and explanation for predictions, features that are missing from MAPP-MMR.
We sampled the performance of generic PON-P, which is not optimized for MMR variants, with the evaluation set. Unlike other methods, PON-P provides a reliability measure, which can be utilized for evaluating the output. When, the reliability parameter was increased from 0.90 to 0.99 the MCC increased from 0.63 to 0.79 indicating the good performance of the method. Still, the dedicated PON-MMR is better as expected for a tool optimized for these proteins.
In silico methods have already been used [Kansikas et al., 2011; Plon et al., 2008] in combination with other methods for classifying MMR variants. PON-MMR could be used in these and similar UV classification schemes as one of the criteria for pathogenicity. The growing number of variants poses a need for more reliable prediction methods.
The PON-MMR consensus predictor was applied to classify over 600 MMR variants. This prioritization allows experimental scientists to concentrate on the most likely cases to verify the results. Results from PON-MMR or any other predictor or experimental method should not be used as the only evidence for pathogenicity. According to recent recommendations at least two independent indications are needed to make diagnosis [Kohonen-Corish et al., 2010]. PON-MMR can be applied in to Lynch syndrome and other cancers where MMR variants are involved.