Haploinsufficiency of SOX5 at 12p12.1 is associated with developmental delays with prominent language delay, behavior problems, and mild dysmorphic features†
Communicated by Ravi Savarirayan
Abstract
SOX5 encodes a transcription factor involved in the regulation of chondrogenesis and the development of the nervous system. Despite its important developmental roles, SOX5 disruption has yet to be associated with human disease. We report one individual with a reciprocal translocation breakpoint within SOX5, eight individuals with intragenic SOX5 deletions (four are apparently de novo and one inherited from an affected parent), and seven individuals with larger 12p12 deletions encompassing SOX5. Common features in these subjects include prominent speech delay, intellectual disability, behavior abnormalities, and dysmorphic features. The phenotypic impact of the deletions may depend on the location of the deletion and, consequently, which of the three major SOX5 protein isoforms are affected. One intragenic deletion, involving only untranslated exons, was present in a more mildly affected subject, was inherited from a healthy parent and grandparent, and is similar to a deletion found in a control cohort. Therefore, some intragenic SOX5 deletions may have minimal phenotypic effect. Based on the location of the deletions in the subjects compared to the controls, the de novo nature of most of these deletions, and the phenotypic similarities among cases, SOX5 appears to be a dosage-sensitive, developmentally important gene. Hum Mutat 33:728–740, 2012. © 2012 Wiley Periodicals, Inc.
Introduction
Molecular cytogenetic techniques, such as array comparative genomic hybridization (aCGH), have precipitated a change in diagnostic emphasis from phenotype to genotype. Traditionally, identification of genetic causes of a syndrome first required ascertainment of multiple patients with similar phenotypes followed by a search for the underlying genetic cause. In contrast, techniques such as aCGH allow for identification of patients with similar genotypes followed by characterization of the associated phenotype. This genotype-first approach [Shaffer et al., 2007] is the only way to appreciate how similar genetic changes can lead to a phenotypic spectrum that may include nonspecific features, be variably expressed, and have overlapping features that may be found in other syndromes.
To increase the likelihood of identifying previously uncharacterized copy-number imbalances that may be causing a phenotypic spectrum of nonspecific neurodevelopmental features, we constructed whole-genome microarrays with enhanced coverage of over 500 functionally significant genes including transcription factors and other developmentally important genes. This has facilitated identification of intragenic, disease-causing deletions [Rosenfeld et al., 2009a, b; Talkowski et al., 2011b]. In our laboratory, another of the targeted genes, located on 12p12.1, SRY-box 5 (SOX5; MIM# 604975), has had multiple, small, apparently de novo deletions identified in patients referred for clinical aCGH testing.
SOX5, along with SOX6 and SOX13, encode members of the SOXD family of transcription factors. SOXD proteins play a role in multiple developmental pathways, including cartilage formation [Aza-Carmona et al., 2011; Lefebvre et al., 1998] and nervous system development [Kwan et al., 2008; Lai et al., 2008; Lefebvre, 2010]. There are three major SOX5 transcription products, two long forms (NM_006940.4 and NM_152989.2) that code for proteins similar in size to those coded for by SOX6 and SOX13 (NP_008871.3 and NP_694534.1, respectively) and a unique short form (NM_178010.1, encoding NP_821078.1; Fig. 1) [Kiselak et al., 2010]. In humans, the long forms are highly expressed in chondrocytes and striated muscles [Ikeda et al., 2002] and have been seen in the fetal brain [Wunderle et al., 1996], while the short form is expressed mainly in the testes [Wunderle et al., 1996]. Mouse studies have shown the long and short forms to be expressed in the brain [Kiselak et al., 2010]. Mouse models support a role for long Sox5 and Sox6 in chondrogenesis [Smits et al., 2001] and in the development of neocortical projection neurons [Kwan et al., 2008; Lai et al., 2008]. Homozygous loss of long Sox5 (through deletion of a coding exon specific to the long transcripts) leads to respiratory distress causing death at birth, apparently due to cleft palate and a small thoracic cage. Complete knockout of Sox6 is frequently lethal at birth; a short sternum is the only apparent skeletal defect observed, although severe dwarfism develops postnatally. Inactivation of both genes is lethal three days before birth, with restricted skeletal growth and ossification [Dy et al., 2008; Smits et al., 2001]. The short Sox5 protein also functions as a transcription factor that drives testis-specific gene expression [Blaise et al., 1999; Budde et al., 2002; Kiselak et al., 2010; Xu et al., 2009] and likely plays a major role in the formation and function of motile cilia in brain, lung, testis, and sperm [Kiselak et al., 2010].
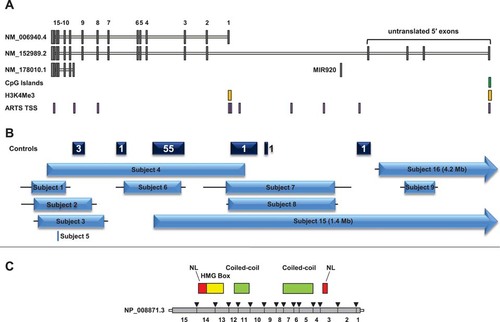
SOX5 transcripts, genomic environment, protein structure, and partial SOX5 deletions in this cohort. (A) SOX5 has two long transcripts (NM_006940.4 and NM_152989.2) and one short transcript (NM_178010.1). Gray boxes represent exons, and coding exons are numbered 1–15. MicroRNA gene MIR920 within SOX5 is also shown. The green box represents the CpG island at the SOX5 promoter; yellow boxes represent consensus trimethylated histone H3K4 sites, a mark of transcriptional regulation (Birney et al., 2007); and purple boxes represent TSSs as identified by ARTS (accurate recognition of transcription starts; Sonnenburg et al., 2006) (B) Dark blue boxes represent locations of deletions found in a control cohort (Cooper et al., 2011). Numbers represent number of deletions within the indicated interval; deletions were not necessarily identical or encompassing the entire block. Four deletions included exons: three overlapped the first exon of the short isoform, one of which also deleted exon 9 of the long form, and the other removed untranslated exon 4. Light blue boxes represent the minimum size of the partial SOX5 deletions reported in this paper; horizontal lines extend through gaps between probes on the arrays to show the maximum possible deletion size. Subject 5 (DGAP189) had a reciprocal translocation with a breakpoint at the location indicated. (C) Protein domains in NP_008871.3 relative to its spliced transcript (NM_006940.4). Red regions are nuclear localization (NL) domains.
Each of the long transcripts is associated with a separate promoter region and transcription start site (TSS), supported by the presence of H3K4Me3 histone modifications, a mark commonly associated with the promoter regions of actively transcribed genes [Bernstein, 2002; Ng et al., 2003; Pokholok et al., 2005; Santos-Rosa et al., 2002; Schneider et al., 2004; Schubeler et al., 2004] (Fig. 1). The two long isoforms have slightly different translation start sites, resulting in the inclusion of 13 additional amino acids at the N-terminus of NP_008871.3 (the protein product encoded by NM_006940.4; Fig. 1C). The short transcript includes only 7 exons from the 3′ end of the gene, encoding a smaller protein containing the high mobility group (HMG) domain, which is involved in DNA binding and some interaction with other proteins [Aza-Carmona et al., 2011], and only one of the two coiled-coil domains found in the larger protein, which allow for homo- and heterodimerization necessary for the dimer to bind to some paired DNA binding sites (Fig. 1). The TSS for the short transcript does not have a consensus H3KMe3 mark [Birney et al., 2007], which may be due to its more restricted expression [Kiselak et al., 2010].
The role of SoxD genes in many developmental pathways is well established in mouse and suggests that alterations of SOXD genes in humans could impact human disease [Lefebvre, 2010]; however, no genetic studies to date have established such a link. Therefore, to understand how SOX5 alterations may contribute to disease, we analyzed the chromosomal abnormalities and phenotypes in a series of 16 subjects with structural variations disrupting SOX5, including an individual with autism that we previously reported with a small, apparently de novo, intragenic SOX5 deletion [Rosenfeld et al., 2010].
Materials and Methods
Subject Ascertainment
Subjects were identified after referral for clinical molecular cytogenetic testing, either to Signature Genomic Laboratories, Seattle Children's Hospital, Pittsburgh Cytogenetic Laboratories, Nantes University Hospital, or Hôpital Jean Verdier, or through enrollment in the Developmental Genome Anatomy Project (DGAP). Informed consent was obtained to publish the subject photographs shown here, according to protocols approved by IRB-Spokane.
Molecular Cytogenetics
Oligonucleotide-based aCGH was performed on subjects 2, 3, 6, 11, 14, and subject 6's mother using a 105K-feature whole-genome microarray (SignatureChip Oligo Solution [OS] version 1, custom-designed by Signature Genomics; manufactured by Agilent Technologies, Santa Clara, CA) as previously described [Ballif et al., 2008]. Oligonucleotide-based aCGH was performed on subjects 1, 8, 9, 10, 12, 13, and subject 9's father using a 135K-feature whole-genome microarray (SignatureChipOS version 2, custom-designed by Signature Genomics; manufactured by Roche NimbleGen, Madison, WI) as previously described [Duker et al., 2010]. DNA from subject 4 was analyzed using Illumina HumanHap 300 single nucleotide polymorphism (SNP) microarray (Illumina, San Diego, CA); DNA from subject 7 was analyzed using an Agilent oligonucleotide-based 105K whole-genome microarray (SignatureSelect OS version 1.0); DNA from subject 15 was analyzed using an Agilent oligonucleotide-based 180K-feature whole-genome microarray; DNA from subject 16 was analyzed using a RocheNimbleGen oligonucleotide-based 135K-feature whole-genome CGX microarray, all according to manufacturers' instructions.
Fluorescence In Situ Hybridization
Metaphase fluorescence in situ hybridization (FISH) analysis was performed using a bacterial artificial chromosome (BAC) clone from the abnormal region as determined by aCGH to visualize the abnormalities as previously described [Traylor et al., 2009]. When available, parental samples were also assayed for the abnormal region detected by aCGH in the proband, using FISH.
Characterization of Translocation Breakpoint through Next Generation Sequencing of Customized Large-Insert Libraries
Subject 5 (DGAP189) was sequenced using a custom large-insert jumping library for Illumina sequencing as previously described [Talkowski et al., 2011a]. In brief, 20 μg of genomic DNA from subject 5 was sheared to ∼3.5-kb fragments that were size selected, end repaired, and ligated to cap adaptors containing an EcoP15I restriction site and a GT overhang. Fragments were circularized with an oligonucleotide containing an AC overhang, a subject-specific bar code, and a single biotinylated thymine at the circularization junction. Circularized fragments were restriction digested, and fragments containing the biotinylated base were captured onto streptaviden beads, purified, and Illumina paired-end adaptors were ligated. The sample was run on a single lane of a HiSeq 2000 (Illumina), using paired-end 25-bp sequencing. Reads were aligned with Burrows–Wheeler Alignment Tool [Li and Durbin, 2009] then processed with SAMtools [Li et al., 2009] and BamStat, a customized program designed to isolate anomalous read pairs indicating a chromosomal rearrangement [Talkowski et al., 2011a].
Analysis of Recurrent 12p12.3p11.23 Deletions
The proximal and distal breakpoint interval sequences were compared using Basic Local Alignment Search Tool (BLAST) sequence similarity [Altschul et al., 1990], and all sequence alignments were manually visualized for stretches of high sequence identity. Analysis for repeats within these breakpoint intervals were then performed using RepeatMasker (http://www.repeatmasker.org).
Results
Molecular Information
We identified eight subjects with heterozygous deletions that only involved SOX5 and ranged in size from 72 kb to 466 kb. Most deletions involved at least some of the coding exons and/or a region likely to be involved in transcription initiation, while subject 9's deletion only involved two of the 5′ untranslated exons. In addition, in subject 5 (DGAP189), with an apparently balanced de novo translocation [46,XX,t(11;12)(p13;p12.1)dn], we identified a translocation breakpoint within SOX5; sequencing revealed a 9-bp deletion at the breakpoint in intron 11 (chr12:23,602,450–23,602,458) and a 16-bp deletion at the breakpoint in 11p13 (chr11:35,033,903–35,033,918). No genes were present within 50 kb on either side of the 11p13 breakpoint. Depending on the abnormality size and location, the translocation or deletions are predicted to impact different protein isoforms to varying degrees (Fig. 1, Tables 1 and 2). However, it is not always known which protein isoforms will be altered by the deletions. The deletions in subjects 7 and 8 remove the transcription initiation site of NM_006940.4, so while this likely prevents expression of that isoform, it is not known whether the other long transcript is affected. The deletion of two untranslated exons in subject 9 would alter the 5′ untranslated region of NM_152989.2, though it is uncertain if this ultimately affects gene expression or protein translation.
| Predicted normal expression of isoform | |||||||
|---|---|---|---|---|---|---|---|
| Subject | Deletion size (kb) | Coding exons deleted | 5′ UTR | Long forma promoter region | Longestb | Longa | Shortc |
| 1 | 80 | 11–15 or 10–15 | + | + | − | − | − |
| 2 | 137 | 9–15 | + | + | − | − | − |
| 3 | 156 | 8-15 | + | + | − | − | − |
| 4 | 466 | 1–15 | + | − | − | − | − |
| 5 | Trans | None | + | + | − | − | − |
| 6 | 133 | 4–6 or 3–6 | + | + | − | − | + |
| 7 | 255 | 1 | + | − | ? | − | + |
| 8 | 255 | None or 1 | + | − | ? | ? | + |
| 9 | 72 | None | − | + | − | + | + |
| 15 | 1406 | 1–3 | − | − | − | − | ? |
| 16 | 4196 | None | − | + | − | ? | ? |
- aNM_006940.4
- bNM_152989.2
- cNM_178010.1
- Abbreviations: +, present; −, absent; ?, uncertain; kb, kilobase pairs; trans, translocation; UTR, untranslated region.
| Subject 1 | Subject 2 | Subject 3 | Subject 4 | Subject 5 | Subject 6 | Subject 7 | Subject 8 | Subject 9 | |
|---|---|---|---|---|---|---|---|---|---|
| Gender | Male | Female | Male | Female | Female | Male | Female | Male | Female |
| Age | 2.5y | 3.5y | 15.5y | 11y | 32y | 4y | 4.3y | 7.7y | 3.5y |
| Deletion coordinatesa | 23,528,946–23,608,782 | 23,532,993–23,670,480 | 23,543,231–23,699,047 | 23,565,524–24,031,096 | 23,602,450–23,602,458 | 23,745,849–23,879,303 | 23,986,652–24,241,246 | 24,001,784–24,256,835 | 24,406,625–24,478,141 |
| Deletion size | 80 kb | 137 kb | 156 kb | 466 kb | 9 bpb | 133 kb | 255 kb | 255 kb | 72 kb |
| Number of genes involved | 1 (SOX5) | 1 (SOX5) | 1 (SOX5) | 1 (SOX5) | 1 (SOX5) | 1 (SOX5) | 1 (SOX5) | 1 (SOX5) | 1 (SOX5) |
| Inheritance | De novo | De novo | De novo | De novo | De novo | Maternal | Unknown | Unknown | Paternal |
| Growth | Hemihypertrophy | FTT, resolved | Tracking <3rd | ||||||
| Weight percentile | 75th–90th | 3rd | 10th | 3rd–10th | 50th–75th | <3rd (22m) | 72nd | >97th | <3rd |
| Height percentile | 25th–50th | 10th–25th | 10th | 25th | 50th–75th | NS | 71st | 90th | <3rd |
| OFC percentile | 25th | <2nd (45.4 cm) | 10th | 3rd | 50th–75th | 25th (22m) | NS | 50th | 50th |
| Neurological | |||||||||
| DD/ID | + | Global developmental disorder; functioning at 12-24m level at 4y | IQ 45–70 | Moderate-severe ID | Mild global delays; IQ testing borderline to mild ID; low-average nonverbal intelligence | Moderate global DD | Motor delays; Bayley III scores (30m): composite 16th percentile; motor 12th | Slight motor delays; IQ 60 | Early motor delays; intellectual development average to advanced |
| Speech delay | Moderate-severe speech delay | Severe mixed receptive/expressive language disorder | First word at 2y; two-word phrases at 4y | No language; had 2 words at 5–6y but lost them | + | Severe speech delay; no words at 4y | Present; Bayley III language score 6th percentile | First words at 4.5y | − |
| Behavior problems | PDD; self injury | Aggressive; stereotypies (rocking, hand flapping, spinning, clapping, self injury); severe hyperkinesis | PDD/atypical autism; aggressive; mood instability | Stereotypies (rocking, hand motions); avoids eye contact; occasional aggression | − | − | − | ADHD; behavior problems in school | − |
| Hypotonia | − | − | In face and lower limbs | − | NS | Truncal | − | Slight | + |
| Seizures | − | + | − | − | + | − | − | − | − |
| Brain malformations | NS | Mild periventricular leukomalacia | − | − | − | Prominent subarachnoid space | NS | NS | NS |
| Other neurologic features | Clumsiness; left facial drooping | Poor articulation | Dyspraxia; awkward gait | Articulation difficulties | Poor balance; decreased strength | ||||
| Ophthalmologic features | − | Intermittent exotropia | − | Strabismus | Myopia | Blue sclerae; strabismus | Left intermittent esotropia; astigmatism | Strabismus | Amblyopia |
| Dysmorphic features | + | − | + | + | + | − | − | + | + |
| Head | Flat occiput; frontal bossing | − | − | Narrowed forehead | − | Mild frontal bossing | − | Brachycephaly; frontal bossing; displaced occipital hair whorl | Frontal bossing |
| Auricular region | Crumpled ear lobule | − | Small, simple ears | Thick, hemmed ears | − | − | − | − | − |
| Periocular region | − | − | − | − | − | − | Epicanthal folds | − | Deep-set eyes |
| Midface | Broad and low nasal bridge; upturned nose with bulbous tip | − | Low nasal bridge | − | − | − | − | − | Hypoplastic |
| Perioral region | Accentuated, prominent philtral ridges; prominent and full lips | − | − | − | Crowded teeth; high and narrow palate; micrognathia | − | − | Short philtrum; crowded teeth; rounded lower facies | − |
| Musculoskeletal anomalies | |||||||||
| Hands and feet | Flat feet | − | High arched feet | Overlapping toes | − | − | − | Flat feet | − |
| Back and spine | Scapular winging | − | Butterfly vertebrae of thoracic spine; mild scoliosis | − | − | − | − | − | Lumbar scoliosis |
| Other | Genu valgum; outturned ankles | Reduced muscle bulk | |||||||
| Additional features | |||||||||
| Heart defects | Heart murmur | − | − | − | − | − | − | − | AV canal; secundum ASD; coarctation of aorta; PDA |
| Genital abnormalities | Shawl scrotum; cryptorchidism | − | − | − | − | − | Hypopigmentationof labia | − | − |
| Other | Hepatomegaly | Chronic constipation | Thyroglossal duct cyst | G-tube for feeding | |||||
| Family history | NS | Noncontributory | NS | Noncontributory | Noncontributory | Mother has short stature, borderline microcephaly, moderate ID, and did not speak until 8y; sister (2.5y) carries deletion and has moderate global DD (walking at 22m), strabismus, and is nonverbal | Maternal history of substance abuse | Mother with LD | Father and paternal grandmother carry deletion and are healthy |
- aChromosomal coordinates based on UCSC Genome Browser 2006 hg18 build.
- bSubject 5 had a reciprocal translocation with a breakpoint in SOX5 and a small deletion at the breakpoint at coordinates indicated.
- Abbreviations: +, feature present; −, feature absent; ADHD, attention deficit-hyperactivity disorder; ASD, atrial septal defect; AV, atrioventricular; bp, base pair(s); DD, developmental delay; FTT, failure to thrive; G-tube, gastric feeding tube; ID, intellectual disability; IQ, intelligence quotient; kb, kilobase pairs; LD, learning disability; m, month(s); NS, not specified; OFC, occipitofrontal circumference; PDA, patent ductus arteriosus; PDD, pervasive developmental delay; y, year(s).
We identified seven additional subjects with 12p deletions encompassing multiple genes including SOX5, ranging from 1.4 Mb to 12.1 Mb and including 8 to 63 genes (Fig. 2, Table 3). Two of these deletions have breakpoints within SOX5, one (in subject 15) between coding exons 3 and 4 of NM_006940.4 and extending 5′ and the other (in subject 16) between untranslated exons 3 and 4 of NM_152989.2 and extending 5′ (Fig. 1). Therefore, while subject 15's deletion is predicted to impact both long isoforms of the gene, subject 16's deletion may only impact NM_152989.2. However, it should be noted for both of these deletions that it is not known what effect, if any, deletion of the promoter region 5′ of the untranslated exons has on expression of the shorter transcripts (Table 1).
| Subject 10 | Subject 11 | Subject 12 | Subject 13 | Subject 14 | Subject 15a | Subject 16 | |
|---|---|---|---|---|---|---|---|
| Gender | Female | Female | Female | Male | Female | Male | Female |
| Age | 6y | 4.25y | 4.5m | 7y | 9y | 3y | 10m |
| Deletion coordinatesb | 14,427,395–26,520,296 | 14,696,149–25,145,536 | 17,785,732–26,583,409 | 17,785,732–26,583,409 | 23,087,676–28,745,337 | 23,815,999–25,222,173 | 24,346,835–28,542,656 |
| Deletion size | 12.09 Mb | 10.45 Mb | 8.80 Mb | 8.80 Mb | 5.66 Mb | 1.41 Mb | 4.20 Mb |
| Number of genes involved | 63 | 52 | 41 | 41 | 31 | 8 | 30 |
| Inheritance | Unknown | Unknown | Not maternal | Unknown | Unknown | De novo | Paternal |
| Growth | |||||||
| Weight percentile | 10th | 34th (22m) | 3rd–5th | 49th | 25th | 25th-50th | <3rd (6.72 kg) |
| Height percentile | 10th | 4th (22m) | 10th–25th | 53rd | 10th–25th | 75th | <3rd (65 cm) |
| OFC percentile | 3rd | 29th (22m) | <3rd (38.1 cm) | −1 SD | 75th | 10th | 3rd–5th |
| Neurological | |||||||
| DD/ID | Moderate ID | Global and severe: walked at 4y | Moderate-severe DD | Mild ID; PPVT receptive score 65 (−2.3 SD) | + | Moderate DD | + |
| Speech delay | Greatest delays in expressive speech; no words and 5 signs at 6y | No words yet | NA | Nonverbal; expressive speech disorder | + | First words at 3y | NA |
| Behavior problems | Hyperactivity; anxiety | Hand twirling but social | NA | ADHD | Compulsive; ritualistic; distractible | Autistic; hyperactivity | NA |
| Hypotonia | − | + | − | + | Severe | + | − |
| Seizures | − | − | + | − | − | − | − |
| Brain malformations | Mild ventriculomegaly with prominent cortical sulci, suggesting volume loss | Hypoplastic CC; mild cerebral volume loss; mild prominence of lateral ventricles | NS | Chiari I malformation | − | Short and thick CC | NS |
| Other neurologic features | Brisk DTRs | Intermittent adducted thumbs; constant tongue thrust | Speech dyspraxia; moderate-severe bilateral SNHL | ||||
| Ophthalmologic features | − | − | Blue sclerae | − | Strabismus; optic nerve hypoplasia | Myopia; strabismus | - |
| Dysmorphic features | + | + | + | + | Mild | − | + |
| Head | Metopic ridge; bitemporal grooves | Mild frontal bossing; positional plagiocephaly | Sparse hair | Mild hair upsweep | Low facial tone | − | − |
| Auricular region | − | − | Low-set ears; familial Darwinian tubercles; small ear lobules; soft cartilage | Protruding and large ears | − | − | − |
| Periocular region | Short, upslanted palpebral fissures | Epicanthal folds; small glabellar hemangioma | Minimal synophrys; upslanting palpebral fissures | − | − | − | − |
| Midface | Prominent, boxy nasal tip; alar hypoplasia | Midface hypoplasia | High and wide nasal bridge; square/tubular | Small nares and alae | Prominent nasal bridge; small alae; | − | Midface hypoplasia |
| nose; long columella; broad nasal tip | broad nasal tip; midline nasal dimple | ||||||
| Perioral region | − | Short philtrum | Short philtrum; broad and short uvula | Downturned upper lip; straight lower lip; malpositioned teeth | Narrow palate | − | − |
| Musculoskeletal anomalies | |||||||
| Hands and feet | Mild ulnar drift of hands; bilateral thenar hypoplasia; adducted thumbs; 2 parallel thenar creases; progressive toe contractures; progressive valgus great toe deformity | 1–2 syndactyly on right hand; medially deviated and broad right index finger; narrow left palm; hypoplastic right thenar eminence; limited motion of fingers; right clubfoot | Arachnodactyly; hyperconvex nails; deep plantar creases; minimal clinodactyly of second and third toes | Single right palmar crease; prominent fingertip pads; short second toes | Short fingers and metacarpals; short toes; deviated second fingers; short thumbs; broad great toes; cone-shaped epiphyses on phalanges | − | − |
| Back and spine | − | Congenital fusion C5–C7 causing torticollis | − | Scoliosis/kyphosis | − | − | − |
| Other | Early metopic fusion | Hypermobile; lack of muscle control of right face at birth | Hip laxity | Prominent sternum | Congenital torticolis | Rhizomelia | |
| Additional features | |||||||
| Heart defects | − | VSD | − | − | Slight arrhythmia | − | − |
| Genital abnormalities | − | − | Anteriorly placed anus | − | − | − | − |
| Other | Alternating constipation/ diarrhea; eczema | Deep sacral cleft with sacral dimple; hypoplastic and inverted nipples | Chronic diarrhea; low-set nipples | Feeding difficulties | GERD; laryngomalacia; eczema | ||
| Family history | Father is borderline microcephalic; otherwise noncontributory | Mother had hiatal hernia and was congenitally “pigeon-toed”; paternal family history of clubfoot and VSD | Maternal half sib with DD; maternal family history of LD/ID and psychiatric disease; father has ADHD and aggression; paternal cousin autistic | NS | NS | NS | Father has low weight (111 lb, 5′6′′), reportedly has short digits, poor dentition, Asperger-like features, recurrent fevers |
- aID 256754 in the DECIPHER Database (http//decipher.sanger.ac.uk/).
- bChromosomal coordinates based on UCSC Genome Browser 2006 hg18 build.
- Abbreviations: +, feature present; −, feature absent; ADHD, attention deficit-hyperactivity disorder; CC, corpus callosum; DD, developmental delay; DTRs, deep tendon reflexes; GERD, gastroesophogeal reflux disease; ID, intellectual disability; LD, learning disability; m, month(s); Mb, megabase pairs; NA, not applicable; NS, not specified; OFC, occipitofrontal circumference; PPVT: Peabody Picture Vocabulary Test; SD, standard deviation; SNHL, sensorineural hearing loss; VSD, ventricular septal defect; y, year(s).
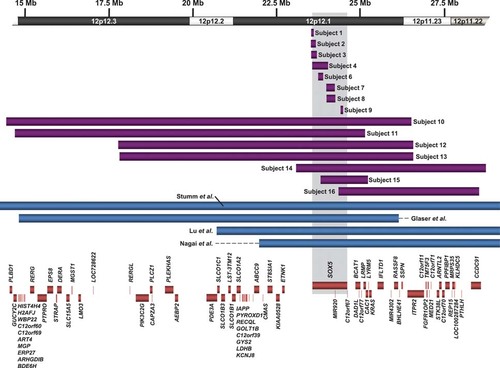
Schematic of deletions in this cohort and molecularly defined deletions in the literature. Deletions in this cohort are shown in purple, and deletions from the literature are shown in blue. The boxes represent the minimum size of the abnormalities, and the horizontal dashed lines extend through gaps in coverage to show the maximum possible sizes. Genes within the region are represented by orange boxes.
No other clinically significant gains or losses of DNA were identified in any of the 16 subjects.
Two subjects (12 and 13) had apparently identical 12p12.3p11.23 deletions. Query of Signature's database of abnormalities revealed two additional cases carrying this apparently identical deletion, one referred for developmental delay (DD) and microcephaly and the other referred for pituitary dwarfism, lack of coordination, pervasive developmental delay (PDD), attention deficit-hyperactivity disorder (ADHD), and optic nerve abnormality. No additional follow-up clinical information was available. The similarity in the breakpoints of these alterations suggests that underlying genomic architecture may play a role in mediating these recurrent deletions. The aCGH results refined the intervals containing the distal and proximal breakpoints to approximately 49 kb (chr12:17,755,660–17,785,732) and 30 kb (chr12:26,583,349–26,632,432), respectively. A search for repeats within these breakpoint intervals using RepeatMasker in the reference sequence (Build 36, hg18) showed enrichment for long and short interspersed repeats (Supp. Fig. S1). Specifically, L1MA4 repetitive elements with high sequence identity are present within the breakpoint intervals (Supp. Fig. S2), which may be mediating recurrent deletions via nonallelic homologous recombination (NAHR), as has been proposed for long stretches of highly homologous sequences such as long interspersed elements (LINEs) and Alus [Deininger and Batzer, 1999; Han et al., 2008; Shaw and Lupski, 2004].
FISH, using BAC probes to the deleted region, confirmed the deletion in all subjects, including diminished signals for the smallest deletions in which the BAC probes used in FISH are larger than the deletion intervals. Parental FISH testing indicated the deletions in subjects 1–4 and 15 were apparently de novo in origin; additionally, subject 12's mother did not have the deletion, while her father was unavailable for testing. Three deletions were inherited. Two of these segregated with a developmental phenotype in the family, one from a more severely affected mother and was also present in an affected sister (subject 6) and one from an affected father (subject 16). The third was inherited from an apparently normal father (subject 9) (Tables 2 and 3). For the parents of subjects 6 and 9, aCGH confirmed that the deletions were identical in parents and children. In addition, FISH revealed that the healthy paternal grandmother of subject 9 also carried the deletion. All other parents were unavailable for testing.
Clinical Information
Clinical information is presented for subjects 1–9 in Table 2 and for subjects 10–16 in Table 3. Major features for the nine subjects with abnormalities limited to SOX5 include developmental delay/intellectual disability (DD/ID) (9/9), speech delay (8/9), behavior problems (5/9), strabismus (6/9), mild dysmorphic appearance (6/9), brain anomalies (2/5), seizures (2/9), and genital anomalies (2/9) (Table 4). Behavioral aspects include aggressive behavior in subjects 2–4, self-injurious behavior in subject 1, and ADHD in subject 7. Subjects 2 and 4 demonstrated stereotypies but were not formally assessed for autism, while subject 1 received a diagnosis of PDD from his therapists and primary care physician, and subject 3 had a diagnosis of PDD and atypical autism through the Treatment and Education of Autistic and Communication Handicapped Children (TEACCH) program [Mesibov and Shea, 2010], which uses assessment batteries including the Childhood Autism Rating Scale (CARS-2) and Psychoeducational Profile—Third Edition (PEP-3). Some minor dysmorphic features were noted in all but subject 2, with the only common feature of frontal bossing seen in 4/9 subjects (Fig. 3). Skeletal system involvement was noted as butterfly vertebrae in one and scoliosis in two subjects.
| SOX5-only abnormalitiesa | Large deletions in this reportb | Moleculary characterized deletions in the literaturec | Cytogenetically defined 12p12.1 deletionsd | |
|---|---|---|---|---|
| Short stature | 1/8 | 1/7 | 3/3 | 5/8 |
| Failure to thrive/low weight | 3/9 | 1/7 | 1/3 | 9/9 |
| Microcephaly | 2/8 | 2/7 | 1/1 | 6/8 |
| Developmental delay/intellectual disability | 9/9 | 7/7 | 3/3 | 9/9 |
| Speech delay | 8/9 | 5/5 | 3/3 | 5/6 |
| Behavior problems | 5/9 | 5/5 | 0/2 | 1/7 |
| Brain abnormalities | 2/5 | 4/5 | 1/2 | 1/2 |
| Hypotonia | 4/8 | 4/7 | 0/3 | 3/9 |
| Seizures | 2/9 | 1/7 | 0/4 | 1/9 |
| Optic nerve atrophy | 0/9 | 1/7 | 0/3 | 2/9 |
| Strabismus | 6/9 | 2/7 | 0/3 | 3/9 |
| Abnormal hearing/auditory canals | 0/9 | 1/7 | 1/3 | 2/9 |
| Dysmorphic features | 6/9 | 6/7 | 3/4 | 9/9 |
| Frontal bossing | 4/9 | 1/7 | 0/4 | 1/9 |
| Blue sclerae | 1/9 | 1/7 | 0/4 | 1/9 |
| Abnormal nasal bridge | 2/9 | 2/7 | 3/4 | 4/9 |
| Low-set ears | 0/9 | 1/7 | 3/4 | 6/9 |
| Micro/retrognathia | 1/9 | 0/7 | 1/4 | 9/9 |
| Cleft lip and/or palate | 0/9 | 0/7 | 2/4 | 0/9 |
| Short/broad neck | 0/9 | 0/7 | 1/4 | 3/9 |
| Sparse or abnormal hair | 0/9 | 1/7 | 1/4 | 2/9 |
| Irregular teeth/oligodontia | 0/9 | 1/7 | 1/3 | 4/9 |
| Brachydactyly | 0/9 | 2/7 | 2/4 | 5/9 |
| Clinodactyly/deviated fingers or toes | 1/9 | 4/7 | 2/4 | 5/9 |
| Craniosynostosis | 0/9 | 1/7 | 0/4 | 2/9 |
| Spinal abnormalities | 1/9 | 1/7 | 0/4 | 0/9 |
| Scoliosis | 2/9 | 1/7 | 0/4 | 2/9 |
| Other skeletal anomalies | 2/9 | 4/7 | 3/4 | 4/9 |
| Congenital heart defects | 1/9 | 1/7 | 2/4 | 3/9 |
| Genital abnormalities | 2/9 | 0/7 | 0/4 | 3/9 |
| Renal abnormalities | 0/9 | 0/7 | 2/4 | 1/9 |
- aSubjects 1–9 in this study.
- bSubjects 10–16 in this study.
- cProbands with SOX5-containing deletions reported in (Bahring et al., 1997; Glaser et al., 2003; Lu et al., 2009; Nagai et al., 1995; Stumm et al., 2007).
- dProbands with 12p12.1-containing deletions reported in (Boilly-Dartigalongue et al., 1985; Fryns et al., 1990; Magenis et al., 1981; Magnelli and Therman, 1975; Malpuech et al., 1975; Mayeda et al., 1974; Orye and Craen, 1975; Tenconi et al., 1975).

Physical features of subjects with SOX5 deletions. (A) Subject 1 at 2.5 years of age. Note broad and low nasal bridge, upturned and bulbous nose, prominent and full lips, and accentuated, prominent philtral ridges. (B–C) Subject 2 at 3 years of age. Note microcephaly and nondysmorphic appearance. (D–E) Subject 10 at 4.5 years of age. Note upslanting palpebral fissures and boxy nasal tip. Feet are shown post-surgical correction of valgus deformity of the great toes and progressive toe contractures.
Major features for the seven subjects with larger, SOX5-encompassing deletions include: DD/ID (7/7), speech delay (5/5), behavior problems (5/5), dysmorphic features (6/7; Fig. 3), clinodactyly/deviated fingers or toes (4/7), skeletal anomalies (4/7), and brain malformations (4/5). No aggressive behavior was noted in this group (Tables 3 and 4).
Case-Control Comparison
Among 24,081 probands tested with oligonucleotide-based microarrays at Signature Genomics between February 2008 and April 2011, seven deletions within SOX5 were identified; excluding the deletions in subject 8, which may or may not involve a coding exon, due to gaps in probe coverage, and subject 9, which only includes untranslated exons, five of these are known to include coding exons. In addition, one deletion immediately 5′ of exon 1 was identified in a parental sample (chr12:24,001,784–24,041,797); this healthy parent's affected child did not carry this deletion. Ten additional larger deletions, involving all or part of SOX5 and additional genes, were identified during this time period. In comparison, in one series of 8,329 control subjects studied on high-resolution Illumina genome-wide SNP arrays (mostly with >550,000 probes) with denser coverage of SOX5 than our arrays [Cooper et al., 2011], 62 deletions were identified in SOX5. Most were intronic, one involved an untranslated exon, and three involved coding exons (Fig. 1). Unlike the coding exon deletions in cases, these control deletions may still allow the production of functional long SOX5 isoforms. No whole-gene deletions were detected. Unfortunately, comparison of deletion frequency in cases to controls is complicated by incomplete knowledge of how the deletions affect expression of the various SOX5 isoforms.
Discussion
SOXD genes—SOX5, SOX6, and SOX13—encode transcription factors that play important roles in the development of many systems and processes such as cell proliferation, differentiation, terminal maturation, and survival. Although no known association of these genes with human disease has been previously noted, it has been hypothesized that such associations will be observed due to the critical role of these genes in a large number of pathways [Lefebvre, 2010]. Furthermore, predictive modeling shows SOX5 and SOX6 as being likely haploinsufficient [Huang et al., 2010]. A survey of aCGH results among patients referred for clinical testing in our laboratories shows multiple cases with deletions affecting SOX5, including apparently de novo intragenic deletions. Interestingly, almost no cases of deletions or small duplications involving the coding regions of SOX6 or SOX13 have been observed in our patient populations, with only one exception of an intragenic, apparently de novo duplication in SOX6 in a male referred for DD, autism spectrum disorder, and morbid obesity. This difference in the number of copy number variants affecting these genes may be due to critical developmental functions performed by SOX6 and/or SOX13 that cannot be compensated for by SOX5, or it may reflect a greater susceptibility of the SOX5 locus to rearrangement. Our analysis of SOX5 abnormalities suggests that haploinsufficiency of this gene results in speech delays, behavioral problems, and minor dysmorphic features.
Abnormalities Involving Only SOX5
SOX5 encodes three major transcription products, and the phenotypic consequences of intragenic deletions may depend upon the protein isoforms affected. Subjects 1–4 have de novo deletions that are predicted to result in loss of the primary DNA-binding domain and lead to haploinsufficiency of all three protein isoforms. The de novo translocation in subject 5 would lead to expression of truncated versions of all three protein isoforms that lack the primary DNA-binding domain. The deletions in subject 6 and his affected mother and sister may prevent expression of functional long forms of SOX5 through two mechanisms: either by inducing a translational frameshift by removing exons 4–6 or by losing a putative coiled-coil domain partially encoded by these same exons and presumably critical to homo- and heterodimerization potential. This frameshift would not be predicted if exon 3 is also included in the deletion; however, loss of the coiled-coil domain would still be expected. Deletions in subjects 7 and 8 may eliminate proximal regulatory elements of NM_006940.4, thereby preventing effective transcription initiation. Potential effects, if any, of this deletion on the expression of the other isoforms cannot be predicted without further characterization of the regulation of SOX5 transcription (Fig. 1, Table 1). Subjects 1–8 demonstrate DD, with greatest delay in speech. In addition, subjects 1–4 and 8 also demonstrate behavior problems, including a diagnosis of PDD in subjects 1 and 3; behavior problems were not noted in subjects 5 or 6. This may be due to variable expressivity, as subjects 5 and 6 are predicted to have altered expression of different isoforms (Table 1).
To help interpret the clinical significance of copy number variations in our patient population, comparisons need to be made to rearrangements in the gene observed in a control population [Cooper et al., 2011; Girirajan and Eichler, 2010; Sharp, 2009]. A deletion similar to those observed in subjects 7 and 8 was detected in one control sample (Fig. 1) and in a healthy parent in our clinical aCGH testing population. The deletion in the control sample retained approximately 3 kb of sequence upstream of exon 1, whereas the deletion in subject 7 removed this 3-kb region as well as the first exon, and due to gaps between probes, it is unknown if this region is deleted in subject 8 and the parent. The presence of the sequence upstream of exon 1 may allow for normal gene expression to continue, although SOX5 expression levels were not assayed in these subjects. Interestingly, a deletion affecting both exon 9 of NM_006940.4 and the TSS of the short form was observed in one control individual, and two additional control individuals had deletion of the TSS of the short form. While deletion of exon 9 would not be predicted to cause a frameshift in the larger protein products, deletion of the TSS of the short form would be expected to cause reduced expression of this transcript. This may suggest that haploinsufficiency for the short form alone is not generally detrimental to normal phenotypic development. Because the short and long proteins each have distinct tissue-specific expression patterns and contain different functional domains [Kiselak et al., 2010], it is unlikely that short and long forms can completely compensate for each other's functions, and it remains possible that adding haploinsufficiency of the short form to haploinsufficiency of the long forms can further impact phenotypic expression.
Unlike the deletions in subjects 1–4 and 6, subject 9's deletion was inherited from a phenotypically normal father and grandmother. The deletion removes two untranslated exons, and a very similar deletion affecting the fourth untranslated exon was observed in one control individual. Deletions within the 5′ untranslated region may not affect expression of the gene or may only affect one of the long forms, leaving the other long form intact. Subject 9's phenotype is milder than seen in our other subjects; the child does not have delays in language. Therefore, it is possible that her phenotype may not be caused by the deletion in SOX5, or that it may be attributed to reduced penetrance or variable expression.
Larger Deletions Containing SOX5
We attempted to determine if the phenotypic observations in subjects 1–9 were also seen in subjects with larger deletions containing SOX5. Subjects 10–14 had whole-gene deletions (Fig. 2). Subjects 15–16 had deletions of the 5′ end of the gene that remove a TSS and the control region and, therefore, should result in haploinsufficiency of at least one of the long forms (Fig. 1, Table 1). It is not known if this would affect expression of all products. All of the subjects older than one year with large deletions including SOX5 have speech delay, consistent with the effects of SOX5 haploinsufficiency observed in subjects 1–8. In addition, all subjects older than one year demonstrate some type of abnormal behaviors, including subject 15 and subject 16's father, who was described to be Asperger-like. This is similar to what was observed in subjects 1–5, where behavioral problems were seen in most subjects with haploinsufficiency for the long and short isoforms of SOX5 (4/5).
Additional genes within these large deletions may be contributing to these subjects' phenotypes. Consistent with this hypothesis, we observed that the subjects with larger deletions that include 30–63 genes tend to show more dysmorphic features and have more musculoskeletal anomalies (Tables 3 and 4). In the literature, common features reported among individuals with 12p12 deletions include DD/ID, short stature, microcephaly, brachydactyly, clinodactyly, and dysmorphic features including low-set ears, broad nasal bridge, and microretrognathia [Bahring et al., 1997; Boilly-Dartigalongue et al., 1985; Fryns et al., 1990; Glaser et al., 2003; Lu et al., 2009; Magenis et al., 1981; Magnelli and Therman, 1975; Malpuech et al., 1975; Mayeda et al., 1974; Nagai et al., 1995; Orye and Craen, 1975; Stumm et al., 2007; Tenconi et al., 1975] (Table 4). Behavior problems have only been described in one 13-month-old male with poor psychosocial contact [Orye and Craen, 1975], although a majority of these cases were identified through traditional cytogenetic techniques, and the inclusion of SOX5 in the deleted intervals is uncertain. The brachydactyly observed in these individuals is type E, with shortening of the metacarpals and metatarsals, and, along with the short stature and oligodontia seen in some of these individuals, may be due to the deletion of PTHLH within 12p11.22 [Klopocki et al., 2010]. In our series, subjects 14 and 16 are deleted for this gene; subject 14 demonstrates brachydactyly, and subject 16 has short stature. There is also an autosomal-dominant hypertension with brachydactyly syndrome (MIM# 112410) due to an inversion of a minimum ∼450-kb segment immediately distal to SOX5 and containing no known protein-coding genes but containing putative microRNA-coding gene(s) that show altered splicing in inversion carriers [Bahring et al., 2008]. Expression of SOX5 is not altered in these individuals [Bahring et al., 2004]. This suggests a gain-of-function mechanism for this disease, and consistent with that, the individuals in our cohort with deletions of SOX5 and not PTHLH do not demonstrate brachydactyly. However, in our cohort we do not have information on hypertension, which has been described in an individual with a deletion of 12p11.22 [Bahring et al., 1997].
In summary, deletions within SOX5 result in prominent speech delay and frequently in behavior problems. Larger deletions that include all of SOX5 or that remove the 5′ regulatory region, which may or may not alter expression of all protein isoforms, also show language delay, behavioral problems, and more dysmorphic features. These findings support the role of SOX5 in human neurodevelopment. Complete haploinsufficiency of SOX5, with roles in chondrogenesis, may only occasionally result in skeletal abnormalities, such as the butterfly vertebrae and scoliosis in some of our subjects with deletions. Haploinsufficiency of SOX5 may be compensated for by SOX6 so that the resulting phenotype is milder than may have been hypothesized for the developmentally important SOXD family of genes [Lefebvre, 2010]. Further research into how various SOX5 deletions impact the function of the SOX5 protein isoforms and identification of additional individuals with SOX5 abnormalities will be helpful in understanding further how loss of this developmentally important gene contributes to neurodevelopmental disease.
Acknowledgements
The authors thank the patients and families who contributed clinical information for this study; Erin Dodge (Signature Genomic Laboratories) for her critical editing and preparation of the manuscript; and Beth Torchia, Carrie Hanscom, and Shahrin Ahsan for their technical assistance. E.E.E. is an investigator of the Howard Hughes Medical Institute. J.A.R., N.J.N., R.A.S., B.C.B., and L.G.S. disclose the following possible conflict of interest: they are employees of Signature Genomic Laboratories, PerkinElmer, Inc E.E.E. is a scientific advisory board member of Pacific Biosciences, Inc. and SynapDx Corp. All other authors have no conflict of interest to report.