Characteristics of Patients With Chronic Hepatitis B Virus Infection With Genotype E Predominance in Burkina Faso
S: upported by the National Cancer Institute (grant K01CA234324 to B.M.N.), a Kathryn H. and Roger Penske Career Development Award (to B.M.N.), National Institutes of Health (NIH) (grants UL1TR000135 and T32 HL105355 to E.A.M; R01 CA186566, P50 CA210964, P30 CA15083, and UL1 TR002377 to L.R.R.), an IDeA Networks of Biomedical Research Excellence award (P20GM103424), NIH National Institute of General Medical Sciences (administrative grant 3P20GM103424-18S1 to J.C.C.), and Pietro Annigonni Biomolecular Research Center, Ouagadougou, Burkina Faso, West Africa.
Portions of this manuscript have been published in abstract form in Gastroenterology 2018 May;154(Suppl. 1):S1130-1.
Study design, contents, and conclusions are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Potential conflict of interest: Dr. Roberts consults for and received grants from Wako Diagnostics; he advises and received grants from Bayer, Gilead, and Exact Sciences; he consults for Medscape, Axis, and Onclive; he advises Grail, NACCME, QED, and Tavec; he received grants from Ariad, BTG International, and Redhill Biopharma. The other authors have nothing to report.
Abstract
Hepatitis B virus (HBV) genotype E (HBV-E) accounts for the majority of chronic hepatitis B (CHB) infections in West Africa. We aimed to determine factors associated with HBV-E-induced hepatocellular carcinoma (HCC) in West Africa. Data on patients from Burkina Faso who were hepatitis B surface antigen positive (HBsAg+) and had CHB were analyzed. HBV viral load and hepatitis B e antigen (HBeAg) status were measured in 3,885 individuals with CHB without HCC (CHB HCC−) and 59 individuals with CHB with HCC (CHB HCC+). HBV genotyping was performed for 364 subjects with CHB HCC− and 41 subjects with CHB HCC+. Overall, 2.5% of the CHB HCC− group was HBeAg+ compared with 0% of the CHB HCC+ group. Of the 364 patients who were CHB HCC− with available genotyping, the frequencies of HBV genotypes E and C/E were 70.3% and 12.9%, respectively. Age (odds ratio [OR] for older age, 1.08; 95% confidence interval [CI], 1.06-1.10 per 1-year increase in age), male sex (OR, 2.03; 95% CI, 1.11-3.69), and HBV viremia (OR, 1.48; 95% CI, 1.31-1.67 per 1 log10 IU/mL) were each associated with HCC diagnosis. Patients with genotype E had a lower HBeAg prevalence (6.3% vs. 14.9%), lower HBV viral load, and higher prevalence of cirrhosis (14.5% vs. 4.8%) than patients with genotype C/E. Conclusion: HBV-E is the most common circulating strain (70.3%) in West African patients. HCC was associated with older age, male sex, and high HBV viral load. It is expected that these results will further inform guidance on clinical management of HBV infection in West Africa.
Abbreviations
-
- −
-
- negative
-
- +
-
- positive
-
- ALT
-
- alanine aminotransferase
-
- AST
-
- aspartate aminotransferase
-
- CERBA
-
- Pietro Annigoni Biomedical Research Center
-
- CHB HCC−
-
- chronic hepatitis B without hepatocellular carcinoma
-
- CHB HCC+
-
- chronic hepatitis B with hepatocellular carcinoma
-
- CHB
-
- chronic hepatitis B
-
- CI
-
- confidence interval
-
- HBcAb
-
- hepatitis B core antibody
-
- HBeAb
-
- hepatitis B e antibody
-
- HBeAg
-
- hepatitis B e antigen
-
- HBsAg
-
- hepatitis B surface antigen
-
- HBV
-
- hepatitis B virus
-
- HBV-E
-
- hepatitis B virus genotype E
-
- HCC
-
- hepatocellular carcinoma
-
- IQR
-
- interquartile range
-
- OR
-
- odds ratio
-
- PCR
-
- polymerase chain reaction
Chronic hepatitis B (CHB) virus infection is among the major causes of hepatocellular carcinoma (HCC), the fourth leading cause of cancer-related mortality in Africa.(1-3) The risk of HCC development among persons with CHB is exacerbated by dietary exposure to aflatoxin B1 and heavy alcohol consumption.(1, 2) The prevalence of hepatitis B virus (HBV) in sub-Saharan Africa is approximately 6%, which is well above the global prevalence of 4%.(1, 4) We and others have reported a high prevalence of HBV in Burkina Faso, ranging from 9% to 15%.(5-7) Furthermore, it has been shown that HCC is the third leading cause of cancer-related mortality in Burkina Faso.(8) The high HBV prevalence in Burkina Faso, as in many other African countries, is due mainly to the relatively recent introduction of HBV vaccination and the failure to identify high-risk groups with active HBV infection, resulting in dramatically high rates of transmission in the perinatal period or early childhood.(5-7, 9-19)
HBV genotype E (HBV-E) is the most common strain of HBV in West Africa, whereas genotype A is prevalent in East Africa, and genotypes B, C, and D are frequently detected in patients in East Asia and South Asia.(20-23) Although it is now well established that HBV-C is associated with a higher incidence of HCC than genotype B in Asia,(24) little is known about the natural history and HCC risk of patients with CHB induced by HBV-E.(22, 25) HBV-E infection is almost exclusively restricted to West Africa, and HCC risk among patients with HBV-E versus other genotypes remains poorly studied.
Several studies on CHB infections have demonstrated that high HBV viral load is associated with a higher risk of developing HCC.(26-28) Current guidelines for HBV treatment have set the threshold for initiating therapy for patients who are hepatitis B e antigen (HBeAg) positive with CHB infections and elevated alanine aminotransferase (ALT) levels at an HBV DNA level of ≥20,000 IU/mL and the threshold for patients who are HBeAg negative at a level of ≥2,000 IU/mL.(29, 30) These treatment recommendations are based on data from the Risk Evaluation of Viral Load Elevation and Associated Liver Disease/Cancer-Hepatitis B Virus (REVEAL-HBV) study, which reported results and natural history for a large prospective cohort of patients with CHB infection in Taiwan.(31) However, the optimal cut-off values for treatment of CHB infections may differ by viral genotype and host factors, which may vary in different geographic regions.
Therefore, to address the scarcity of data on patients infected with HBV-E in West Africa, we conducted a cross-sectional study in a large cohort of patients from Burkina Faso who were hepatitis B surface antigen positive (HBsAg+) to determine their clinical and serologic characteristics, such as HBV DNA level and HBV genotype, and the associations of these characteristics with HCC.
Patients and Methods
Study Population and Data Abstraction
This study was approved by the Institutional Review Board of the Burkina Faso National Ethics Committee for Health Research (reference no. 2015-01-006; January 14, 2015). Patients were recruited and tested for HBV viral load from October 1, 2014, through August 30, 2017, at Pietro Annigoni Biomedical Research Center (CERBA) in Ouagadougou, Burkina Faso, West Africa. CERBA provides free HBV viral load testing as well as affordable medical care for low-income patient populations. The protocol for referring patients for HBV viral load testing required that all patients be diagnosed with CHB, confirmed by at least two HBsAg tests at 6-month intervals. A total of 4,163 patients who were HBsAg+ were identified from the CERBA database. Excluded patients included 17 without viral load data and 202 who received treatment before the viral load assay. In total, 3,944 patients with CHB infection (HBsAg+) were included, comprising 3,885 patients without HCC (CHB HCC−) and 59 with HCC (CHB HCC+) (Fig. 1).
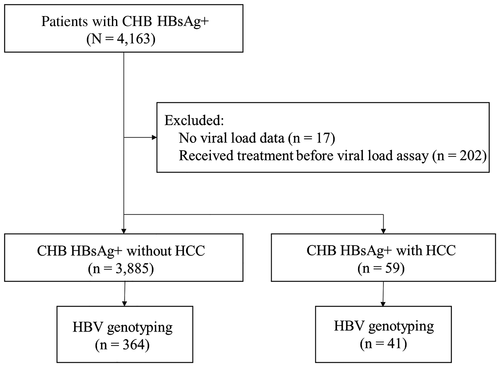
Characteristics of patients with CHB, such as age at diagnosis of CHB and HCC, sex, and cirrhosis status, were collected at the time of serologic testing.(26) Clinical data on HCC status and HBV treatment were obtained from electronic health records of the Department of Gastroenterology and Hepatology of the Yalgado Ouedraogo National Hospital in Ouagadougou. Cirrhosis and HCC were diagnosed based on radiologic findings. Patients did not routinely have imaging for surveillance because of its limited availability. They were seen by local providers once every 6 to 12 months after the diagnosis of CHB to monitor their clinical symptoms. Patients were referred for ultrasonography or cross-sectional imaging only if cirrhosis was suspected due to jaundice, abnormal liver size, ascites, dilated superficial abdominal veins, or elevated liver enzymes; HCC was suspected due to elevated liver enzymes or a palpable mass; or they had decompensated liver disease.
Specimen Collection and HBV DNA Extraction
Blood samples were collected aseptically into 10 mL ethylenediaminetetraacetic tubes and centrifuged for 15 minutes at 2,000g. HBV viral DNA was extracted from 200 μL of plasma, using a QIAamp DNA Blood Mini Kit (QIAGEN GmbH, Hilden, Germany).
HBV, Hepatitis C Virus, and Human Immunodeficiency Virus Serologic Markers
Laboratory assays, including HBsAg, HBeAg, hepatitis B e antibody (HBeAb), hepatitis B core antibody (HBcAb), aspartate aminotransferase (AST), ALT, human immunodeficiency virus (HIV) antibody, and antibody to hepatitis C virus, were performed. HBV serologic testing for HBsAg, HBeAg, HBeAb, and HBcAb was performed by immunoassay (bioMérieux, Boxtel, the Netherlands).
HBV Viral Load
HBV DNA was quantified in 20-µL reactions containing 5 μL of HBV-infected patient DNA sample using an Oasig 2X quantitative polymerase chain reaction (PCR) Mastermix (Primerdesign Ltd., United Kingdom). An ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA) was used with the following thermocycling conditions: 95°C for 10 minutes, followed by 50 cycles of 95°C for 10 seconds and 60°C for 1 minute. HBV-specific primers used in this reaction targeted a highly conserved region of the HBV surface gene, which provides accurate detection of genotypes A to H. For these reactions, HBV-plasmid DNA with 10-fold serial dilution was used to generate a standard curve. The data were analyzed using the 7500 Fast Software version 2.1 (Applied Biosystems), and HBV viral load was expressed in IU/mL. The detection limit was set to 20 IU/mL (sensitivity and specificity >95%). The quantitative HBV-specific PCR assays were routinely standardized using the World Health Organization standards for HBV DNA (National Institute for Biological Standards and Control code 97/750).
HBV Genotypes and Subgenotypes
HBV genotyping is not routinely performed for patients in Burkina Faso and was performed on a randomly selected subset in our study. HBV DNA was used in a multiplex PCR to identify HBV genotypes A to F based on the method described by Chen et al.(32) The GeneAmp PCR System 9700 (Applied Biosystems) was used to amplify HBsAg gene sequences with the following PCR conditions: activation (95°C, 15 minutes), 35 cycles of denaturation (94°C, 1 minute), primer annealing (58°C, 1 minute), and extension (72°C, 1 minute), followed by a final extension (72°C, 10 minutes). PCR products were separated by electrophoresis in 2% agarose gels, and DNA was visualized under ultraviolet light. We sequentially first used primers specific for genotypes A, B, and C, followed by a second PCR with primers specific for genotypes D, E, and F. A result of two or more genotypes, for example, C/E, indicates that genotype C was found in the first PCR and E in the second PCR and is reported as having recombinant genotypes E and C. Our group has successfully used this approach and analysis of HBV pre-S1/S2 regions using Sanger sequencing to confirm the predominance of HBV-E in Burkina Faso.(22, 23)
Statistical Analysis
Baseline characteristics were compared between patients with and without HCC and reported as percentages for categorical variables and median (interquartile range [IQR]) for continuous variables. Continuous data, including age, AST, ALT, HBV DNA viral load, and log10 viral load, were compared using the Mann-Whitney U test. Categorical data, including sex, HBV serology, cirrhosis status, and coinfection status, were compared using the χ2 or Fisher’s exact test. The associations of baseline characteristics with cirrhosis, HCC, and HBV viral load were analyzed by univariate and multivariable logistic regression. All analyses were performed using JMP version 10 software (SAS Institute Inc., Cary, NC). P < 0.05 was considered statistically significant.
Results
Characteristics of Burkina Faso Patients Who Were CHB HCC+ or CHB HCC−
There were 59 patients who were CHB HCC+ and 3,885 patients who were CHB HCC−. Patients who were CHB HCC+ were significantly older (median, 43; IQR, 34-50; range, 25-73 years) than patients who were CHB HCC− (median, 32; IQR, 26-39; range, 1-85 years; P < 0.001) (Table 1). The percentage of male patients was higher in the CHB HCC+ cohort (n = 39; 66.1%) than in the CHB HCC− cohort (n = 1,717; 44.2%; P < 0.001). In the CHB HCC− cohort, 10 patients had multiple infections, including 5 with HBV/HCV (0.13%) and 5 with HBV/HIV (0.13%). No coinfections were found in the CHB HCC+ cohort. Fifty-four of the 3,885 (1.4%) CHB HCC- patients had cirrhosis, while 17 of 33 (51.5%) CHB HCC+ patients with available cirrhosis status had cirrhosis (P < 0.001).
| Clinical Characteristics* | HBsAg+ | P Value | Patients With Available Data | |
|---|---|---|---|---|
| No HCC (n = 3,885) | HCC (n = 59) | (CHB HCC‒/CHB HCC+) | ||
| Age (years) at HBV diagnosis, median (IQR) | 32 (26-39) | 43 (34-50) | <0.001 | 3,843/59 |
| Age, years | NA | 3,843/59 | ||
| <20 | 227 (5.9) | 0 | ||
| 20-29 | 1,326 (34.5) | 5 (8.5) | ||
| 30-39 | 1,410 (36.7) | 16 (27.1) | ||
| 40-49 | 574 (15.0) | 17 (28.8) | ||
| 50-59 | 217 (5.6) | 14 (23.7) | ||
| ≥60 | 89 (2.3) | 7 (11.9) | ||
| Male sex | 1,717 (44.2) | 39 (66.1) | <0.001 | 3,885/59 |
| Cirrhosis | 54 (1.4) | 17 (51.5) | <0.001 | 3,885/33 |
| Liver enzyme levels, median (IQR) | ||||
| AST, IU/L | 25 (19-34) | 90.5 (39.2-197.5) | <0.001 | 2,168/58 |
| AST ≥30 IU/L | 770 (35.5) | 54 (93.1) | <0.001 | |
| ALT, IU/L | 21 (14-31) | 59.5 (36.0-102.8) | <0.001 | 2,190/58 |
| ALT ≥30 IU/L | 615 (28.1) | 48 (82.8) | <0.001 | |
| AST/ALT ratio, median (IQR) | 1.2 (0.9-1.6) | 1.4 (1.1-2.7) | 0.002 | 2,161/58 |
| AST/ALT <1.0 | 603 (27.9) | 11 (19.0) | <0.001 (for all classes) | |
| AST/ALT 1.0-<1.5 | 927 (42.9) | 22 (37.9) | ||
| AST/ALT 1.5-<2.0 | 365 (16.9) | 5 (8.6) | ||
| AST/ALT ≥2.0 | 266 (12.3) | 20 (34.5) | ||
| Serologic testing | ||||
| HBcAb+ | 3,280 (99.6) | 59 (100) | 1.00 | 3,294/59 |
| HBeAg+ | 96 (2.5) | 0 (0) | 0.40 | 3,807/58 |
| HBeAb+ | 3,593 (95.8) | 58 (100) | 0.52 | 3,752/58 |
| Genotypes | 364/41 | |||
| A | 1 (0.3) | 0 | NA | |
| B | 0 | 0 | NA | |
| C | 21 (5.8) | 1 (2.4) | 0.36 | |
| A/B/C | 2 (0.5) | 0 | NA | |
| B/C | 8 (2.2) | 0 | NA | |
| E | 256 (70.3) | 32 (78.0) | 0.83 | |
| A/E | 4 (1.1) | 2 (4.9) | 0.60 | |
| B/E | 1 (0.3) | 0 | NA | |
| C/E | 47 (12.9) | 5 (12.2) | 0.76 | |
| D/E | 1 (0.3) | 0 | NA | |
| A/C/E | 4 (1.1) | 1 (2.4) | 0.32 | |
| B/C/E | 14 (3.8) | 0 | NA | |
| C/D/E | 2 (0.5) | 0 | NA | |
| C/D/E/F | 3 (0.8) | 0 | NA | |
| HBV viral load, IU/mL, median (IQR) | 502 (0-6,475) | 83,593 (1,405-830,902) | <0.001 | 3,885/59 |
- * Values are numbers (%) unless otherwise indicated.
- Abbreviation: NA, not applicable.
The median serum aminotransferase levels (AST and ALT) reflective of active hepatitis were significantly higher in the CHB HCC+ group in than the CHB HCC− group (median AST, 90.5; IQR, 39.2-197.5 vs. median, 25.0; IQR, 19.0-34.0; P < 0.001, respectively, and median ALT, 59.5; IQR, 36.0-102.8 vs. median, 21.0; IQR, 14.0-31.0; P < 0.001, respectively). The median AST/ALT ratio was 1.4 (IQR, 1.1-2.7) in the CHB HCC+ group and 1.2 (IQR, 0.9-1.6) in the CHB HCC− group (P = 0.002).
Of the patients who were CHB HCC−, 2.5% (96/3,807) were HBeAg+ and 95.8% (3,593/3,752) were HBeAb+. The distribution of HBeAg+ by age was 4% (9/225), <20 years; 4.1% (54/1,317), 20-29 years; 1.9% (26/1,398), 30-39 years; 1.2% (7/569), 40-49 years; and 0% (0/298), ≥50 years. All 58 patients who were CHB HCC+ with available data were HBeAg− and HBeAb+. There was no significant difference in HBeAg or HBeAb serologic status between patients who were CHB HCC− and CHB HCC+ (P = 0.40 and 0.53, respectively).
Factors Associated With Cirrhosis in Patients Who Were CHB HCC−
Of the CHB HCC- patients, the 54 patients with cirrhosis were significantly older and had significantly higher HBV viral loads than the 3,831 patients without cirrhosis, with an odds ratio (OR) of 1.08 (95% confidence interval [CI], 1.06-1.11; P < 0.001) per 1-year increase in age and 1.26 (95% CI, 1.12-1.43; P < 0.001) per 1 log10 IU/mL increase in viral load (Table 2). Male sex was not significantly associated with cirrhosis status (OR, 1.26; 95% CI, 0.71-2.28; P = 0.44).
| Clinical Characteristics | OR (95% CI) for Cirrhosis* | P Value | OR (95% CI) for HCC | P Value |
|---|---|---|---|---|
| Age increase, per 1 year | 1.08 (1.06-1.11) | <0.001 | 1.08 (1.06-1.10) | <0.001 |
| Male sex | 1.26 (0.71-2.28) | 0.44 | 2.03 (1.11-3.69) | 0.02 |
| HBV viral load, per increase of log10 (log IU/mL) | 1.26 (1.12-1.43) | <0.001 | 1.48 (1.31-1.67) | <0.001 |
- * Among patients without HCC.
Factors Associated With HCC in Patients With CHB
Comparing CHB HCC+ and CHB HCC- patients, older age, male sex, and higher HBV viral load were significantly associated with HCC (OR, 1.08; 95% CI, 1.06-1.10; P < 0.001 per 1-year increase in age; OR, 2.03; 95% CI, 1.11-3.69; P = 0.02 for male sex; and OR, 1.48; 95% CI, 1.31-1.67; P < 0.001 per 1 log10 IU/mL increase in viral load) (Table 2).
HBV Viral Load of Patients Who Were CHB HCC+ or CHB HCC−
The median HBV viral load of patients who were CHB HCC+ (median, 83,593; IQR, 1,405-830,902 IU/mL) was significantly higher than that of patients who were CHB HCC− (median, 502; IQR, 0-6,475 IU/mL; P < 0.001) (Fig. 2).
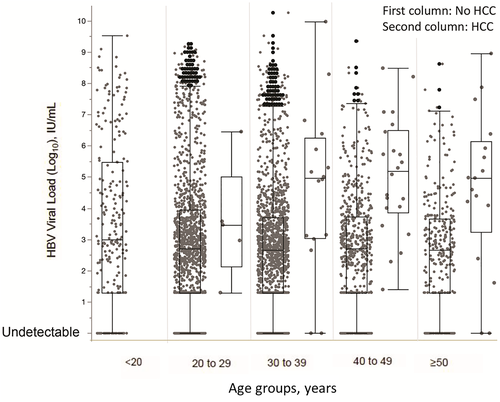
Comparing patients who were CHB HCC− and those who were CHB HCC+, we found a significantly higher percentage of patients with HBV viral load ≥2,000 IU/mL (73.3% vs. 34.4%) and ≥20,000 IU/mL (60.0% vs. 18.7%) in the CHB HCC+ group than the CHB HCC− group (P < 0.001 and P < 0.001, respectively).
Among the CHB HCC− cohort, there were 1,337 patients (34.4%) with an HBV viral load ≥2,000 IU/mL, 728 patients (18.7%) with an HBV viral load ≥20,000 IU/mL, and 946 (24.4%) patients with undetectable viral DNA.
Among the CHB HCC+ cohort, there were 44 patients (73.3%) with an HBV viral load ≥2,000 IU/mL, 36 patients (60%) with an HBV viral load ≥20,000 IU/mL, and 4 patients (6.8%) with undetectable viral DNA.
Among the age groups, HBV viral loads of patients with HCC were significantly higher than those in patients without HCC, except in patients from 20 to 29 years of age. When patients in different age groups were classified by viral load, we found that individuals <20 years of age had the highest percentage of viral load ≥20,000 IU/mL (n = 80, 35.2%). Patients from 20 to 29 years of age had the second highest percentage of viral load ≥20,000 IU/mL (n = 272, 20.5%) compared with <15% of patients in the other age groups. Most patients had viral loads of 60 IU/mL or less (30.4%-35.3% across all age groups). There were significant differences in viral load among age groups (P = 0.004). We also observed similar trends of differential viral loads among different age groups between male and female patients (Fig. 3). However, with regression analysis, HBV viral load was not correlated with age (r = −0.028; 95% CI, −0.059-0.003).
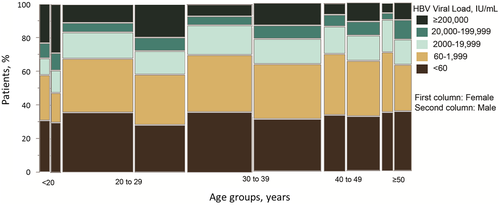
Characteristics of CHB in Patients With Different HBV Genotypes
We performed HBV genotyping in 364 patients who were CHB HCC− and 41 patients who were CHB HCC+. Characteristics of patients with genotype E, other genotypes excluding genotype E, and nongenotyped patients are shown in Table 3. Of the 364 patients who were CHB HCC−, 256 (70.3%) had genotype E, 47 (12.9%) had genotype C/E, 21 (5.8%) had genotype C, and 14 (3.8%) had genotype B/C/E. Among 41 patients with HCC with genotyping, genotype E was found in 32 patients (78.0%). The other less common genotypes are shown in Table 1. There was no difference in numbers of patients with genotype E, C, or C/E among patients who were CHB HCC‒ and CHB HCC+ (P = 0.83, 0.36, and 0.76, respectively).
| Clinical Characteristics* | Patients Who Were HBsAg+, CHB HCC‒ | P Value | P Value | ||
|---|---|---|---|---|---|
| Genotype E (n = 256) | Genotype C/E (n = 47) | Genotype C (n = 21) | For E vs. C/E | For E vs. C | |
| Age (years) at HBV diagnosis, median (IQR) | 33 (27-41) (missing 3) | 28 (25-38) (missing 0) | 34 (24-29) (missing 0) | 0.021 | 0.634 |
| Age, years | 0.143 | 0.923 | |||
| <20 | 5 (2.0) | 4 (8.5) | 0 | ||
| 20-29 | 89 (35.2) | 21 (44.7) | 8 (38.1) | ||
| 30-39 | 92 (36.4) | 13 (27.7) | 8 (38.1) | ||
| ≥40 | 67 (26.5) (missing 3) | 9 (19.1) (missing 0) | 5 (23.8) (missing 0 ) | ||
| Male sex | 126 (49.2) (missing 0) | 25 (53.2) (missing 0) | 11 (52.4) (missing 0) | 0.617 | 0.781 |
| Cirrhosis | 37 (14.5) (missing 0) | 5 (10.6) (missing 0) | 1 (4.8) (missing 0) | 0.474 | 0.163 |
| HBeAg+ | 16 (6.3) (missing 1) | 7 (14.9) (missing 0) | 2 (9.5) (missing 0) | 0.061 | 0.584 |
| HBeAb+ | 232 (92.1) (missing 4) | 39 (83.0) (missing 0) | 18 (85.7) (missing 0) | 0.069 | 0.352 |
| Liver enzyme levels, median (IQR) | |||||
| AST, IU/L | 28 (21-39) | 30 (20-47) | 28 (21-39) | 0.569 | 0.196 |
| AST ≥30 IU/L | 121 (47.5) (missing 1) | 24 (51.1) (missing 0) | 11 (52.4) (missing 0) | 0.649 | 0.664 |
| ALT, IU/L | 24 (16-38) | 34 (16-52) | 31 (19-61) | 0.141 | 0.050 |
| ALT ≥30 IU/L | 99 (38.8) (missing 1) | 25 (53.2) (missing 0) | 13 (61.9) (missing 0) | 0.066 | 0.041 |
| AST/ALT ratio, median (IQR) | 1.17 (0.92-1.49) (missing 1) | 1.09 (0.85-1.30) (missing 0) | 1.29 (0.87-1.54) (missing 0) | 0.236 | 0.629 |
| AST/ALT <1.0 | 77 (30.2) | 18 (38.3) | 77 (30.2) | 0.551 (for all classes) | 0.792 (for all classes) |
| AST/ALT 1.0-<1.5 | 115 (45.1) | 21 (44.7) | 8 (38.1) | ||
| AST/ALT 1.5-<2.0 | 42 (16.5) | 6 (12.8) | 3 (14.3) | ||
| AST/ALT ≥2.0 | 21 (8.2) | 2 (4.3) | 3 (14.3) | ||
| HBV viral load, IU/mL, median (IQR) | 13,424 (1,463-310,013) | 19,496,410 (1,248,321-80,494,145) | 939,129 (9,800-2,795,475) | <0.001 | 0.003 |
- * Values are numbers (%) unless otherwise indicated.
Among the patients who were CHB HCC−, we compared the 256 patients with genotype E to the 47 patients with genotype C/E and the 21 patients with genotype C (Table 3). There were fewer patients who were HbeAg+ in the genotype E group (6.3%) than the genotype C/E (14.9%) or genotype C (9.5%) groups (P = 0.06 and P = 0.58, respectively). More patients with genotype E (14.5%) than patients with genotypes C/E (10.6%) or C (4.8%) had cirrhosis (P = 0.47 and P = 0.16, respectively). The HBV viral load was lower in the genotype E group (median, 13,424; IQR, 1,463-310,013 IU/mL) than the C/E group (median, 19,496,410; IQR, 1,248,321-80,494,145 IU/mL;P < 0.001) or the C group (median, 939,129; IQR, 9,800-2,795,475 IU/mL; P = 0.003).
Characteristics of CHB With Genotype E With and Without HCC
Of the 256 patients who were CHB HCC− with exclusively genotype E, there were 16 (6.3%, 1 missing) patients who were HBeAg+ and 232 (92.1%, 4 missing) who were HBeAb+. None of the 32 patients with HCC were HBeAg+ (Supporting Table S1). Patients with HCC had significantly elevated levels of AST and ALT and a higher AST/ALT ratio compared with the patients without HCC (P < 0.001, P < 0.001, and P = 0.004, respectively; Supporting Table S1). HBV viral load was not significantly different between patients with and without HCC (P = 0.107).
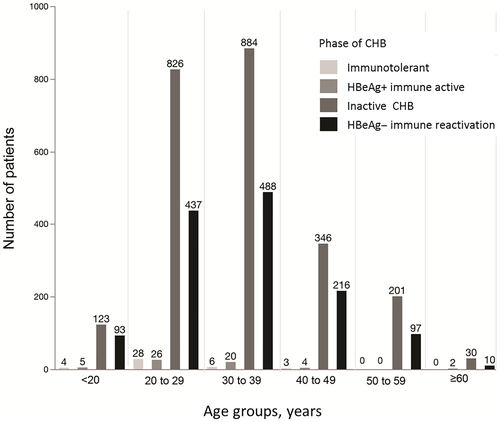
Patient Classification by Phase of CHB Infection
Patients were classified into four phases of chronic HBV infection according to the 2016 American Association for the Study of Liver Diseases (AASLD) guidelines: (1) immunotolerance, (2) HBeAg+ immune active, (3) inactive CHB viral infection, and (4) HBeAg− immune reactivation.(31) When strictly classified according to AASLD criteria, only 1,317 (33.9%) patients could be categorized. Patients who did not meet the inclusion requirements of any specific group were categorized on the basis of presence of HBeAg, elevated ALT level, and viral load, respectively. The number of patients in each phase by age group is shown in Fig. 4, with most patients categorized in the inactive CHB and HBeAg− immune reactivation phases.
Discussion
This study reports one of the largest cohorts of patients with CHB infection in Africa and identifies HBV-E as the most prevalent circulating HBV genotype. The next most commonly prevalent strains were HBV-C and recombinant genotypes C/E. Additionally, these genotypes were associated with low frequencies of HBeAg+ and relatively low HBV viral load, particularly HBV-E. Of patients who were CHB HCC−, 2.5% were HBeAg+, whereas none of the patients who were CHB HCC+ were HBeAg+. Previous studies of CHB genotype E infection in sub-Saharan Africa showed HBeAg+ prevalence ranging between 8% and 19%.(33, 34) This large variability between countries could be potentially explained by differences in modes of transmission and/or the natural history of different genotypes. The clearance of HBeAg strongly correlates to mode of transmission as the risk of mother-to-child HBV transmission is much lower in mothers who are HBeAg− than those who are HBeAg+ in sub-Saharan Africa, with a risk of 38% in HBeAg+ versus 5% in HBeAg− for mothers who did not use HBV prophylaxis.(33, 35) Patients with CHB genotype E who have a high HBV viral load also have a higher rate of vertical transmission.(36) It is now generally accepted that horizontal transmission within the household during childhood is the primary mode of HBV transmission in sub-Saharan Africa.(37) The predominance of this mode of HBV transmission could be due to the natural history of CHB genotype E as we also found that genotype E has a lower prevalence of HBeAg+ than other genotypes, including genotypes C and C/E.
We showed that HCC was associated with older age, male sex, and high HBV viral load in patients with CHB infection, findings that have been reported in the literature.(16-19, 26-28) In contrast to studies that showed HBeAg+ to be a risk factor for patients with CHB to develop HCC, none of the patients who were CHB HCC+ in our study were HBeAg+.(26, 27, 38) However, in this study, HBeAg positivity was determined mainly at diagnosis of HCC and only infrequently when patients were diagnosed with CHB. In addition, HBeAg seroconversion beyond the fourth decade of life has been associated with increased HBV viral load, liver cirrhosis, and HCC.(39) We were unable to confirm whether HCC was diagnosed after a late-onset seroconversion or if the patients had been HBeAg− since childhood.
Genotypes E, C/E, and C, respectively, were predominant in our study. Patients with genotype E had significantly more cirrhosis. However, only a few patients with HBV-E were HBeAg+, and patients with HBV-E had lower HBV viral loads than patients with genotypes C/E or C. This indicates the existence of different characteristics of HBV infection among patients with different genotypes. Nonetheless, the clinical significance of either the lower viral load or the correlation with cirrhosis or HCC development will require further longitudinal study. Because of the geographic distribution of CHB genotype E, available comparisons of genotype E to other genotypes are limited. A small study in Sudan compared CHB genotypes C and D with genotype E and found that patients with genotype E had higher HBeAg+ levels and higher HBV viral loads than those with genotype D.(40)
Current guidelines use HBeAg status, HBV viral load, and elevated ALT level as indications for treatment of patients with HBV without cirrhosis.(29, 31) Interestingly, our patients presented with a lower prevalence of HBeAg+ and lower HBV viral loads when compared with patients enrolled in the REVEAL-HBV study.(31) This suggests that the current classification of CHB infection based on viral load, HBeAg status, and liver enzyme levels may have limited applicability to patients from West Africa, particularly Burkina Faso.
This is the first large study of patients with CHB infection from West Africa, an area uniquely known to have HBV-E. Although HBV-E was predominant in our cohort, we do not have extensive genotyping results, and genotyping was performed on samples with high HBV viral loads; thus, further studies are crucial for identifying HBV genotypes and recombinant strains circulating in the country. Moreover, cirrhosis and HCC in this cohort were likely underreported because of the limited availability of surveillance imaging modalities for screening. The actual cirrhosis and HCC status of the patients with CHB is likely higher than the numbers we reported. Complete clinical data for patients with CHB and HCC were limited, including data for other risk factors, such as cirrhosis, dietary aflatoxin B1 exposure, alcohol consumption, and smoking status. Longitudinal follow-up of patients from the time of HBV diagnosis to the development of HCC will be needed to provide sufficient clinical and genomic information to conclusively determine the factors associated with the risk of developing HCC. Importantly, this study provides for the first time useful data on the characteristics of patients with CHB with and without HCC at the time of diagnosis in an HBV-endemic area of West Africa.
In Burkina Faso, CHB genotype E was the most common circulating strain (70.3%); HBeAg prevalence was low (2.5%). HCC was associated with older age, male sex, elevated liver enzyme levels, and high viral load. Patients with genotype E had relatively lower HBeAg positivity, higher cirrhosis prevalence, and lower HBV viral load compared with genotypes C/E and C. This study once again highlighted the need for more studies to characterize the strains of HBV circulating in West Africa and their association to chronic HBV, cirrhosis, and the early onset of HCC in sub-Saharan Africa.
Acknowledgment
We thank Dr. Murray H. Brilliant for critical review of this manuscript.




