Long-term reduction of jaundice in Gunn rats by nonviral liver-targeted delivery of Sleeping Beauty transposon†
Potential conflict of interest: Nothing to report.
Abstract
Asialoglycoprotein receptor (ASGPR)-mediated endocytosis has been used to target genes to hepatocytes in vivo. However, the level and duration of transgene expression have been low because of lysosomal translocation and degradation of the DNA and lack of its integration into the host genome. In this study we packaged the DNA of interest in proteoliposomes containing the fusogenic galactose-terminated F-glycoprotein of the Sendai virus (FPL) for targeted delivery to hepatocytes. After the FPL binds to ASGPR on the hepatocyte surface, fusogenic activity of the F-protein delivers the DNA into the cytosol, bypassing the endosomal pathway. For transgene integration we designed plasmids containing one transcription unit expressing the Sleeping Beauty transposase (SB) and another expressing human uridinediphosphoglucuronate glucuronosyltransferase-1A1 (pSB-hUGT1A1). The latter was flanked by inverted/direct repeats that are substrates of SB. In cell culture, FPL-mediated delivery of the E. coli β-galactosidase gene (LacZ) resulted in transduction of ASGPR-positive cells (rat hepatocytes or Hepa1 cell line), but not of ASGPR-negative 293 cells. Intravenous injection of the FPL-entrapped pSB-hUGT1A1 (4-8 μg/day, 1-4 doses) into UGT1A1-deficient hyperbilirubinemic Gunn rats (model of Crigler-Najjar syndrome type 1) resulted in hUGT1A1 expression in 5%-10% of hepatocytes, but not in other cell types. Serum bilirubin levels declined by 30% ± 4% in 2 weeks and remained at that level throughout the 7-month study duration. With histidine containing FPL, serum bilirubin was reduced by 40% ± 5%, and bilirubin glucuronides were excreted into bile. No antibodies were detectable in the recipient rats against the F-protein or human UGT1A1. Conclusion: FPL is an efficient hepatocyte-targeted gene delivery platform in vivo that warrants further exploration toward clinical application. (HEPATOLOGY 2009.)
Although recombinant viruses have been used extensively in gene therapy, nonviral vectors for gene transfer are of great interest because of a lower potential for toxic or immunological reactions. One benefit of nonviral vectors is the possibility of delivering genes to specific cell types. Targeted delivery to hepatocytes could provide some important benefits for the treatment of liver-based disorders. Rapid and efficient clearance of the therapeutic agent would reduce the required dose and spare other tissues of potential toxicity. Reduced exposure of potential antigens to antigen-presenting cells may abrogate the immune response to the gene/drug transfer vehicle and the transgene product.
Antibodies against cell surface proteins or ligands of cell surface receptors are being explored for endocytosis-based delivery of macromolecules to normal or tumor cells. For example, nucleic acids complexed with asialoglycoprotein-polylysine conjugates1, 2 or incorporated into galactocerebroside-containing liposomes3 have been used to deliver DNA to hepatocytes in vivo by way of the hepatocyte-specific asialoglycoprotein receptor (ASGPR). However, transgenes delivered by way of endocytosis are typically translocated to lysosomes, where they largely degraded by lysosomal enzymes. Interruption of this trafficking to lysosomes by disrupting microtubules prolongs transgene expression,4 but because the DNA is compartmentalized into cytoplasmic vesicles, transgene expression remains low.5 To take advantage of the hepatocyte-specificity of ASGPR, while circumventing lysosomal translocation, investigators have utilized the naturally galactose-terminated fusogenic F-glycoprotein of the Sendai virus envelope.6, 7 In the presence of hemagglutinin/neuraminidase (HN), another major protein of the Sendai virus envelope, F-protein binds to cell surface membranes ubiquitously, causing fusion.8 Vectors containing both HN and F proteins can deliver macromolecules to all cell types and, when injected intravenously, localize primarily to the spleen.8 Thus, these vectors are not desirable for liver-targeted gene transfer in vivo. In contrast, when HN is removed, F-protein binding requires interaction of its galactose-terminated carbohydrate moieties with ASGPR.6 Therefore, liposomes containing the F-protein, but not HN, bind specifically to hepatocytes. After the proteoliposome binds to hepatocyte surface ASGPR, fusogenic activity of the F-protein leads to deposition of its contents into the hepatocyte cytosol, bypassing the endocytotic pathway, and thereby enhancing DNA translocation to the nucleus.6
Here, we evaluated the utility of F-protein-containing proteoliposomes (FPL) in gene therapy for a hepatocyte-based inherited liver disease. The Gunn rat, a model of the potentially lethal human disease Crigler-Najjar syndrome type 1,9 exhibits life-long unconjugated hyperbilirubinemia because of the lack of uridinediphosphoglucuronate glucuronosyltransferase 1A1 (UGT1A1) activity in hepatocytes. To obtain a long-term effect, we used the Sleeping Beauty (SB) transposon system, which promotes transposition of the transgene into the host genome.10 We also tested the hypothesis that rapid clearance of FPL by hepatocytes would prevent the exposure of the F-protein to the host immune system, thereby obviating the immune response against the vector.
Abbreviations
ASGPR, asialoglycoprotein receptor; BDG, bilirubin diglucuronide; BMG, bilirubin monoglucuronide; FPL, F-protein-containing proteoliposomes; HN, hemagglutinin/neuraminidase; hUGT1A1, human uridinediphosphoglucuronate glucuronosyltransferase-1A1; IR/DR, inverted repeat/direct repeats; IRES, internal ribosomal entry site; RHA, Roman high avoidance; SB, Sleeping Beauty; UCB, unconjugated bilirubin.
Materials and Methods
Animals.
(Details of the Materials and Methods are in the Supporting Material.) Gunn rats of both genders (80-100 g initial body weight) were used.
Plasmid Constructs.
Three plasmid constructs were used: (1) pT2-pCAAGS-hUGT1A1//eIF-SB10 (Fig. 1) contains two transcription units, one expressing a hyperactive Sleeping Beauty transposase (a gift from Dr. Steve Yant)11 driven by the eIF4A1 promoter. A second transcription unit, flanked by the Sleeping Beauty inverted repeat/direct repeats (IR/DR) expressed human UGT1A1 (hUGT1A1) from a CAGGS promoter. As marker genes, we generated plasmid (2) pT2-pCAAGS-LacZ//eIF-SB10, expressing E. coli β-galactosidase and (3) pT2-pCAAGS-Luc//eIF-SB10, expressing firefly luciferase by replacing the human UGT1A1 sequence with the LacZ or firefly luciferase sequences, respectively.
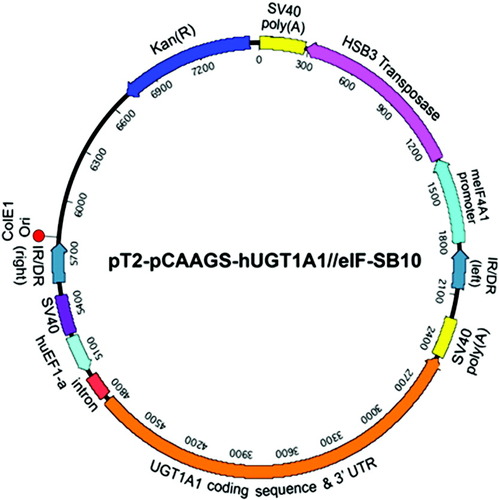
cis pT2-pCAAGS-hUGT1A1//eIF-SB10 plasmid construct expressing both human UGT1A1 and a hyperactive SB transposase (HSB3): human UGT1A1 is expressed from the hybrid SV40 enhancer-human elongation factor 1α (SV40EF1α) promoter and HSB3 is expressed from the mammalian intrinsic factor A1 (mIFA1) promoter. The UGT1A1 transcription unit, consisting of the promoter-enhancer, coding region, untranslated 3′LTR, and the SV40 polyadenylation signal (SV40 poly(A)) is flanked by the IR/DR that are substrates of HSB3. ColE1 Ori, E. coli site of origin; Kan(R), kanamycin resistance gene.
Generation of DNA-Loaded FPL.12
Sendai virus, grown in chicken embryo, was harvested by centrifugation of the allantoic fluid. The disulfide bonds were reduced by treatment with dithiothreitol (DTT), which was then removed by dialysis, thereby renaturalizing the F-protein, but not other viral proteins. The F-protein was solubilized in Triton X-100 and the DNA of interest was added. The DNA was entrapped into FPL by removing Triton X-100, using SM2 Biobeads (BioRad). In some experiments, to enhance the fusogenic activity of the FPL, histidine lipids (L-histidine(N,N-di-n-hexadecylamine)ethylamide) were added before removing Triton X-100.7
FPL-Mediated Transfection of Cultured Cells.
Gunn rat hepatocytes (isolated by collagenase perfusion of the liver), Hepa-1, an ASGPR-positive mouse hepatoma cell line, and ASGPR-negative 293 cells were incubated with FPL loaded with pT2-pCAAGS-LacZ//eIF-SB10 or pT2-pCAAGS-hUGT1A1//eIF-SB10 (2-4 μg / 2 mL of serum-free medium) for 2 hours. E. coli β-galactosidase activity was determined by cytochemical staining and hUGT1A1 expression was evaluated by immunocytochemical staining using a monoclonal antibody, WP1.13
Gene Transfer In Vivo.
For short-term tissue distribution studies we injected FPL, loaded with pT2-pCAAGS-Luc//eIF-SB10 (4 μg DNA in 0.25 mL by tail vein injection over 2 minutes) into four Gunn rats. The rats were killed 72 hours later and liver, kidney, spleen, brain, and lungs were harvested for determination of luciferase activity.14 For long-term metabolic studies, Gunn rats underwent tail vein injection with pT2-pCAAGS-hUGT1A1//eIF-SB10 (4 μg or 8 μg/day for 4 days) entrapped in FPL with or without histidine lipid. For comparison, the rats were injected saline or the naked DNA (8 μg). Serum bilirubin levels were measured at various timepoints. After 28 weeks, bile samples were collected by bile duct cannulation for bile pigment analysis by high pressure liquid chromatography.15 The rats were then sacrificed and tissues were harvested to determine the biodistribution of the transgene polymerase chain reaction (PCR) amplification of total DNA. Hepatic hUGT1A1 expression was determined by Western blot of liver homogenates and immunohistochemical staining of paraffin sections using WP1. UGT1A1 activity with bilirubin was determined as described.16
Transposition Site Analysis.
Transposition sites were recovered by two-step inverted PCR amplification of ligated chromosomal DNA.17 Chromosomal sequences flanking the insertion sites were matched with the rat genomic sequence data base to determine the insertion sites.
Evaluation of HSB3 Coding Sequence Persistence.
Total liver DNA from saline-treated control or treated Gunn rats was subjected to PCR amplification to determine if the coding sequence the HSB3 transposase persisted in the livers of the treated animals.
Evaluation of Humoral Immune Response to F-Protein or hUGT1A1.
Using whole Sendai virus proteins or hUGT1A1 expressed in Gunn rat fibroblasts as antigen, we evaluated the presence of antibodies in the sera of treated Gunn rats by Western blotting. As positive controls, we used an anti-Sendai virus polyclonal antibody or WP1, respectively.
Histological Examination of the Liver.
Paraffin-embedded liver sections were examined by hematoxylin-eosin staining to evaluate morphological injury or cellular infiltration.
Serum Alanine Aminotransferase (ALT) and Albumin Analysis.
These measurements were performed to evaluate liver injury and synthetic function after treatment.
Statistical Analysis.
Serum bilirubin levels at various timepoints after treatment were compared with those of control groups by Student's t test. P-values < 0.01 were considered significant.
Results
FPL-Mediated Gene Transfer into Cultured Cells Depended on the Presence of ASGPR.
To determine whether FPL-mediated gene transfer required cell-surface ASGPR, we transfected Hepa1 cells (an ASGPR+ mouse hepatoma cell line) or 293 cells (an ASGPR− human kidney epithelial cell line) with a plasmid expressing E. coli β-galactosidase (pT2-pCAAGS-LacZ//eIF-SB10). Ten to 20% of transfected Hepa1 cells exhibited the expression of bacterial β-galactosidase, whereas there was no expression of the marker gene in 293 cells (Fig. 2A -C). Next, we evaluated the ability of FPL to transfer the plasmid expressing human UGT1A1 (pT2-pCAAGS-hUGT1A1//eIF-SB10) into primary Gunn rat hepatocytes. Immunocytochemical staining showed that 15%-22% of the hepatocytes expressed human UGT1A1 after transfection (Fig. 2D,E).
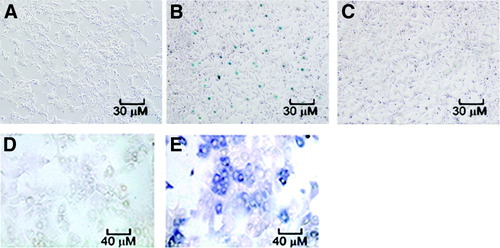
Transfection of cultured cells with FPL loaded with plasmid DNAs. (A-C) transfected cell stained for E. coli β-galactosidase expression. (A) Hepa1 cells, untransfected; (B) Hepa1 cells transfection with FPL loaded with pT2-pCAAGS-LacZ//eIF-SB10; (C) 293 cells transfected with FPL loaded with pCMV-LacZ. (D,E) Immunocytochemical staining for human UGT1A1. (D) Primary Gunn rat hepatocytes, untransfected; (E) primary Gunn rat hepatocytes, transfected with FPL loaded with pT2-pCAAGS-hUGT1A1//eIF-SB10.
Intravenous Injection of FPL-DNA Results in Transgene Expression Exclusively in the Liver.
To determine short-term tissue distribution of transgene expression, FPL loaded with the plasmid expressing firefly luciferase (pT2-pCAAGS-Luc//eIF-SB10, 4 μg DNA) was injected intravenously. Three days after injection, luciferase activity was detected in the liver (1,200,000 RLU/mg tissue) but not in the kidney, spleen, brain, and lungs. When the firefly luciferase gene was injected without entrapment in FPL, no luciferase activity above the background (noise) was detectable in the liver or any other tissue, 72 hours after the injection.
Integration of the hUGT1A1 Transgene into the Genomic DNA of Gunn Rat Liver.
To determine whether the expression of the hUGT1A1 was from integrated transgene or from episomally carried copies of the SB-Tn, total liver DNA was PCR-amplified to determine the presence of the HSB coding sequence. No amplification products were found in treated Gunn rats (Fig. 3A). In contrast, when liver DNA from an untreated Gunn rat was spiked with ≈15 ng of plasmid containing the HSB3 CDS, an amplicon of the predicted size (136 base pairs [bp]) was readily detectable. Using 2 μg of the isolated liver DNA from the treated and control Gunn rats as template, inverted nested PCR was used to recover genomic insertion sites. Four sequences were retrieved, one located within an intron of a Reference Sequence-identified gene RGD1310066 and the location of the other three were not identified using the megablast search against the existing genomic rat data base. This may be explained by the Gunn rat being on a Wistar-RHA (Roman high avoidance) background, whereas the rat genomic sequences compiled to date are from outbred Harlan-Sprague Dawley rats. Moreover, intergenic regions of the rat genome have not been completely sequenced and may also contribute to the lack of genomic assignment as ≈2/3 of SB-mediated insertions occur in nontranscribed regions of the genome. Together, the results indicated that, whereas the hUGT1A1 transcription units are integrated into the rat genome, the SB coding region is lost as a result of the transposase activity, which cleaves the transposon plasmid, at the IR/DR sequences, resulting in its degradation.
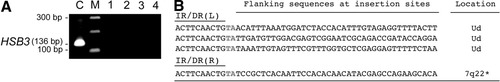
Sleeping Beauty-mediated genomic insertion. (A) Total liver DNA isolated from Gunn rats was subjected to PCR to detect the presence of HSB3 coding sequences. C, control untreated Gunn rat DNA spiked with the pT2-pCAAGS-hUGT1A1//eIF-SB10 plasmid. Amplification of the plasmid yielded the expected 136 bp amplicon. M, DNA ladder in 100-bp increments. Positions of the 300 bp and 100 bp markers are indicated. Lane 1, water control; lane 2, untreated Gunn rat; lanes 3 and 4, livers from two Gunn rats treated with four daily intravenous injections of pT2-pCAAGS-hUGT1A1//eIF-SB10 (4 μg DNA per dose). (B) Insertion sites determined by inverted nested PCR using the genomic DNA isolated from the treated animals. The first 10 nucleotides of the IR/DR are shown with the bold gray TA being the duplicated TA genomic dinucleotide. The first 40 nucleotides of the flanking genomic sequence is shown with the location determined by megablast against the National Center for Biotechnology Information (NCBI) rat genome assembly indicated at far right. Ud, genomic location using megablast search on rat genome sequence did not provide a location. *Located in the intron of RGD1310066 reference sequence gene on chromosome 7.
Long-Term Studies In Vivo.
In initial studies we tested the dose response by injecting 2 to 32 μg of FPL-entrapped pT2-pCAAGS-hUGT1A1//eIF-SB10 into Gunn rat tail veins. Two weeks later, Western blot analysis of the liver showed a maximum hUGT1A1 expression at the 8 μg dose, above which the signal did not increase. Thereafter, we injected 4 μg FPL-entrapped DNA daily for 1, 2, 3, or 4 days, which showed a progressive increase in the Western blot signal intensity. For initial determination of the effect of incorporating histidine lipid into FPL, we gave Gunn rats four daily injections of 4 μg of pT2-pCAAGS-hUGT1A1//eIF-SB10 entrapped into FPL with or without histidine lipid. After 14 days, Western blot of the liver homogenates showed a 40%-50% increase in hUGT1A1 expression when histidine lipid was incorporated into FPL (Supporting Fig. 2). Therefore, in subsequent long-term experiments we used daily injection of 4 μg or 8 μg DNA for 4 days. Five groups of Gunn rats, each consisting of six rats, received daily injections for 4 days as follows: (1) Age-matched controls receiving intravenous saline, (2) 4 μg DNA in FPL without histidine lipid, (3) 4 μg DNA in FPL with histidine lipid, (4) 8 μg DNA in FPL with histidine lipid, (5) 8 μg naked DNA without FPL. Serum bilirubin levels were followed for 28 weeks, when the bile was collected for bile pigment analysis and the rats were sacrificed. Tissues were analyzed by DNA PCR, Western blot, liver histology and immunohistochemical staining, and UGT1A1 assay as described below.
Liver-Specific Persistence of the Transposition-Competent Transgene Delivered by FPL.
Human UGT1A1 coding sequences were amplified by PCR from total DNA extracted from various tissues 28 weeks after injections of FPL-entrapped pT2-pCAAGS-hUGT1A1//eIF-SB10 once daily for 4 days. As shown in Fig. 4A, DNA extracted from the livers, but not lungs, kidney, and spleen, yielded amplicons of the expected size.
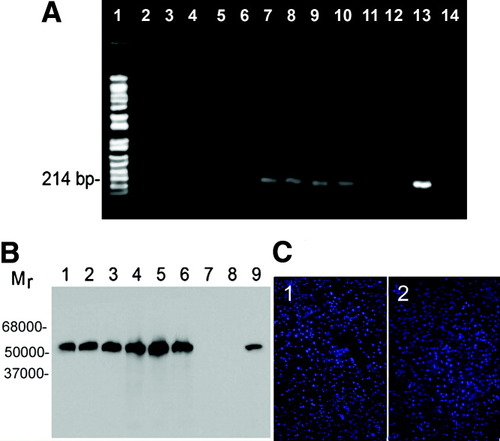
Tissue distribution of human UGT1A1 transgene and its expression. Gunn rats were given intravenous injections of FPL loaded with pT2-pCAAGS-hUGT1A1//eIF-SB10 (4 μg DNA) or saline (control) once a day for four doses as described in Materials and Methods. The recipients were sacrificed after 28 weeks and tissues were harvested. (A) PCR amplification of DNA extracted from various tissues. The data represent results from individual mice representative of the various experimental groups. Lane 1, molecular weight markers; lane 2, lungs; lane 3, kidney; lane 4, spleen; lane 5, brain; lane 6, testis; lanes, 7-12, DNA from livers of six different Gunn rats treated with FPL loaded with 4 μg of pT2-pCAAGS-hUGT1A1//eIF-SB10; lane 13, pT2-pCAAGS-hUGT1A1//eIF-SB10; lane 14, liver from saline injected control. (B) Western blot analysis of human UGT1A1 expression. Lanes 1-6, Gunn rats treated with FPL loaded with pT2-pCAAGS-hUGT1A1//eIF-SB10: lanes 1-3, 4 μg DNA; lanes 4-6, 8 μg DNA; lanes 7 and 8, untreated Gunn rat liver; lane 9, Gunn rat skin fibroblast, stably transfected with human UGT1A1. (C) Immunofluorescent staining of liver cryostat sections. 1, Untreated Gunn rat; 2, a Gunn rat treated with FPL loaded with pT2-pCAAGS-hUGT1A1//eIF-SB10 (4 μg DNA). The DAPI-stained nuclei exhibit blue fluorescence, whereas human UGT1A1 exhibits red fluorescence.
Western Blot Analysis.
Twenty-eight weeks after treatment, expression of human UGT1A1 was demonstrated in liver homogenates of the Gunn rats (Fig. 4B). The 52 kiloDalton (kDa) transgene product comigrated with human UGT1A1 expressed in stably transduced Gunn rat fibroblasts.18 As expected, saline-treated Gunn rat livers did not show any immunoreactive bands. To evaluate the requirement of the SB transposase in the persistence of transgene expression, we injected an FPL-entrapped plasmid expressing hUGT1A1, but not HSB (pT2-pCAAGS-hUGT1A1) (8 μg DNA/day for 4 days). Western blot of liver homogenates showed the expression of hUGT1A1 1 week after the injections, but not the 28-week time point (data not shown in Fig. 4B).
Immunohistochemistry.
Immunofluorescent staining of frozen liver sections showed hUGT1A1 expression in livers of Gunn rats treated with FPL-entrapped pT2-pCAAGS-hUGT1A1//eIF-SB10, but not in the saline-treated age-matched controls (Fig. 4C, left panel). Clumpy cytoplasmic distribution and linear distribution in the nuclear envelope were consistent with the endoplasmic reticulum localization of UGT1A1 (Fig. 4C, right panel).19
UGT1A1 Activity Toward Bilirubin.
UGT1A1 activity toward bilirubin in liver homogenates from wildtype Wistar-RHA rats (congeneic with the Gunn rats used in this study) was 2.2 ± 0.4 μmol/g wet weight per hour. As expected, there was no detectable UGT1A1 activity in the livers of saline-treated control Gunn rats. Twenty-eight weeks after injections of FPL-pT2-pCAAGS-hUGT1A1//eIF-SB10 in Gunn rats, UGT1A1 activity in the liver homogenates of the recipients was 0.31 ± 0.11 μmol/g wet weight per hour.
Serum Bilirubin Levels.
As serum bilirubin levels change with age in Gunn rats, we expressed serum bilirubin concentrations both as percentage of the levels in age-matched controls (Fig. 5A) and in absolute values (Fig. 5B). After four injections 4 μg of pT2-pCAAGS-hUGT1A1//eIF-SB10 entrapped in FPL without histidine lipid, serum bilirubin levels declined slowly to ≈75% of the levels in saline-treated age-matched controls in 8 weeks (P < 0.01) (Fig. 5A,B). Reduction of serum bilirubin was more pronounced when histidine lipid was incorporated in the FPL bilayer, so that serum bilirubin levels were significantly lower at all timepoints beyond 2 weeks, and stabilized at 42% to 50% below the control levels (P < 0.01). Mean serum bilirubin levels in the group receiving 8 μg DNA/day entrapped in FPL containing histidine lipids were slightly lower than those in the 4 μg DNA/day group at timepoints after 8 weeks, but the difference was not statistically significant. When pT2-pCAAGS-hUGT1A1//eIF-SB10 was administered without FPL, there was no significant reduction of serum bilirubin at any timepoint, indicating the effectiveness of FPL in gene delivery (Fig. 5A,B).
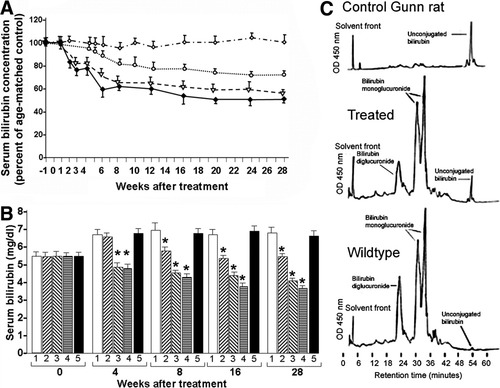
Metabolic effect of FPL-mediated gene transfer. (A) Serum bilirubin levels relative to age-matched controls. Gunn rats were given daily intravenous injections of FPL loaded with pT2-pCAAGS-hUGT1A1//eIF-SB10 for 4 days or saline (untreated control). (A) Serum bilirubin levels are shown as percent of mean bilirubin concentration in age-matched, saline-treated controls. Values are means ± standard deviation; n = 6 in each group. Mean values for the group receiving 4 μg plasmid DNA per dose in FPL without histidine lipid (open circles) were significantly lower (P < 0.01) than in age-matched controls from the 8-week timepoint onward. In groups receiving either 4 μg (triangles) or 8 μg DNA (solid diamonds) per dose in FPL containing histidine lipids the mean values were significantly lower (P < 0.01) than in controls at all time 4 weeks and beyond. In contrast, injection of the plasmid without incorporation into FPL did not result in significant change in mean serum bilirubin levels (open diamond, P > 0.5). (B) The experimental groups are identical to those in (A), but serum bilirubin levels shown as absolute concentrations. 1, untreated age-matched controls; 2, 4 μg plasmid DNA per dose in FPL without histidine lipid; 3, 4 μg DNA per dose in FPL containing histidine lipids; 4, 8 μg DNA per dose in FPL containing histidine lipids; 5, 8 μg naked DNA per dose without FPL. Asterisks indicate values that are significantly lower (P < 0.01) than in age-matched controls. (C) High-pressure liquid chromatographic analysis of pigments excreted in bile. A single representative sample from the control (saline-treated) group (n = 6), the treated group that received FPL with histidine lipid plus pT2-pCAAGS-hUGT1A1//eIF-SB10 (4 μg) (n = 6) and a congeneic wild-type Wistar-RHA rat are shown.
Excretion of Bilirubin Glucuronides in Bile.
Chromatographic analysis of bile pigments from control Gunn rats showed no bilirubin glucuronides and unconjugated bilirubin (UCB) was the major peak (Fig. 5A), whereas bile of Gunn rats that had received injections of FPL-entrapped pT2-pCAAGS-hUGT1A1//eIF-SB10 contained both bilirubin diglucuronide (BDG) and bilirubin monoglucuronide (BMG), in addition to a small amount of UCB (Fig. 5B). BDG and BMG were the major pigments in bile from wildtype Wistar-RHA rats (Fig. 5C).
Serum Albumin and ALT Levels.
Serum albumin and ALT levels were determined before injections and 1, 2, 4, 8, and 28 weeks after the series of FPL-DNA injections to evaluate hepatic protein synthetic function and liver injury, respectively. Serum albumin levels remained between 3.8 and 4.3 g/dL in both control and treated groups, and serum ALT levels remained between 35 and 45 IU. At no time was there a significant difference between the values in the control and the treated groups.
Evaluation of Humoral Immune Response Against FPL and Human UGT1A1.
Western blot analysis was performed to determine whether there was an antibody response in the treated Gunn rats against the F-protein or human UGT1A1. Sera from FPL-treated rats did not yield a positive band against the F-protein at the range of dilutions tested (Fig. 6A, lanes 1-3). An immunoreactive band of expected size for the F-protein was observed when a rabbit anti-Sendai virus envelope antiserum was used (positive control, Fig. 6A, lane 4).
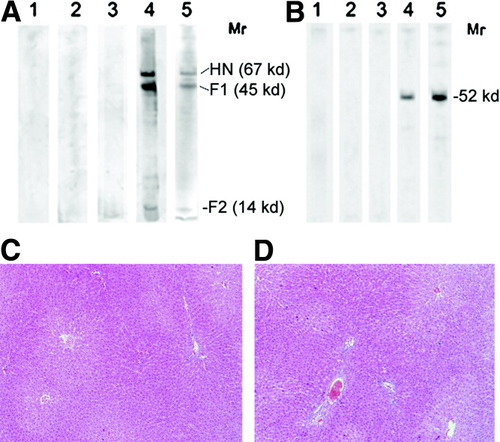
Evaluation of antibody response and liver histology. (A) Western blot using sera from recipient Gunn rats against proteins of the Sendai virus envelope. Gunn rats were injected with FPL loaded with pT2-pCAAGS-hUGT1A1//eIF-SB10 once a day for 4 days (lanes 1 and 2, 4 μg DNA daily; lane 3, 8 μg DNA daily). Sera were collected 28 weeks after the injections. Western blot analysis was performed against Sendai virus envelope proteins resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Various dilutions of the sera were tested. The figure shows results with 1:400 dilution. Lane 4, serum from a Gunn rat, which had been injected with both HN and F protein of the Sendai virus envelope, prepared as described in the text. Lane 5, Sendai virus envelope proteins were resolved by SDS-PAGE and proteins were visualized by Coomassie blue staining. HN, hemagglutinin; F1 and F2, the two subunits of the F-protein. (B) Western blot using sera from recipient Gunn rats against human UGT1A1. Gunn rats were treated as in (A). The sera were tested by Western blots against an extract of Gunn rat skin fibroblasts stably transduced with human UGT1A1. Lanes 1-3 are from rats as in the corresponding lanes in (A); lane 4, serum from a Gunn rat that was injected a lentiviral vector expressing human UGT1A1; and lane 5, a monoclonal antibody that recognizes human UGT1A isoforms at a 1:4,000 dilution. (C) Hematoxylin-eosin stained paraffin-embedded liver section from a Gunn rat 7 days after four injections of FPL loaded with pT2-pCAAGS-hUGT1A1//eIF-SB10 (8 μg). (D) Liver section 28 weeks after the injection.
Sera from the treated Gunn rats did not yield an immunoreactive band against human UGT1A1 expressed in Gunn rat fibroblasts (Fig. 6B, lanes 1-3). In contrast, sera from rats treated with a lentiviral vector expressing human UGT1A1 from the ubiquitous PGK promoter showed a single immunoreactive band of the size expected for human UGT1A1 (Fig. 6B, lane 4).
Histological Examination of the Liver.
Hematoxylin-eosin staining of liver sections from the treated and control Gunn rats showed completely normal architecture and cellular morphology. There was no evidence of infiltration by inflammatory cells and no increase in the number of apoptotic bodies (Fig. 6C,D).
Discussion
We have shown efficient transgene integration specifically into the genome of hepatocytes, mediated by nonviral systemic delivery. To our knowledge, this is the first report of long-term amelioration of a liver-based metabolic disease by intravenous administration of a plasmid entrapped in liposomes. The success of this strategy depended on two components of the gene transfer system. FPL permitted gene delivery targeted to hepatocytes after systemic administration, which may explain the observed lack of host immune response toward the gene delivery vehicle and the transgene product. The second component was the plasmid construct, which was engineered to coexpress a hyperactive SB transposase that promoted integration of the human UGT1A1 into the host genome.
For hepatocyte-specific gene delivery, we generated liposomes with a single glycoprotein, F, derived from the Sendai virus envelope, incorporated in the lipid bilayer. Other investigators have demonstrated that efficient gene transfer can be achieved in vitro and in vivo using liposomes containing both of the two major proteins of the Sendai virus, HN and F.8 However, in the presence of HN the F-protein binds ubiquitously to all cell types, which is a potential source for toxicity and immunogenicity against the proteins contained in the gene transfer vehicle, as well as the transgene product, which may be recognized as a foreign protein in the recipient. We reported previously that in the absence of HN the F-protein loses its ubiquitous cell surface binding property and its binding becomes dependent on the interaction of its carbohydrate component to ASGPR, which is expressed specifically on the hepatocyte cell surface.6
The fusogenic property of the F-protein is partly retained after removal of HN, so that following binding to ASGPR-positive cells, the contents of the liposome are deposited directly into the cytosol, bypassing the classical endocytotic pathway that directs the ligand to lysosomes. However, the full complement of fusogenic activity of the F-protein requires interaction with histidine groups of HN. Therefore, incorporation of histidine lipids in the FPL increases the fusogenic activity and macromolecule transferring ability of FPL.7, 20 Based on these observations, we hypothesized that FPL-loaded transgenes would be delivered and expressed specifically in hepatocytes in vitro and in vivo. In a cell culture system, ASGPR+ Hepa-1 cells but not ASGPR− 293 cells expressed the transgene after FPL-mediated delivery of a plasmid expressing human UGT1A1, which was consistent with the requirement of ASGPR for gene transfer.
To examine whether transgenes delivered in vivo by way of systemic administration of FPL are expressed specifically in the liver, we performed both short-term and long-term studies. In both cases we used ubiquitous promoters to avoid transcription restriction of transgene expression. Three days after FPL-mediated administration of a luciferase-expressing plasmid, luciferase activity was detectable in the liver, but not in the other tissues tested. Similarly, in long-term experiments, 28 weeks after gene transfer, the transgene was detectable by PCR only in the liver. Consistent with our previous observation that histidine lipids increased its fusogenic activity of the F-protein,6 there was a more pronounced hypobilirubinemic response when histidine lipids were incorporated in the FPL. The liver-specificity of transgene delivery after intravenous injection of FPL was in sharp contrast to the reported results using liposomes containing both HN and F, which resulted in transgene expression mainly in the spleen, but not in the liver.8 For gene delivery to the liver, other investigators have used “hydrodynamic” injection, which consists of very rapid intravenous injection of the DNA in a large volume (1.5 × blood volume) to cause transient cardiac failure and consequent hepatic congestion resulting in gene transfer.21 Using FPL, we obtained comparable levels of hepatic transgene expression using one-hundredth the dose of DNA and at an injection volume, which has no hemodynamic consequence. Therefore, in terms of clinical translatability, FPL is superior to existing methods of liver targeted gene delivery by systemic administration.
Our second hypothesis was that the highly efficient ASGPR-mediated internalization of FPL and its DNA cargo by hepatocytes would minimize the exposure of the F-protein and the transgene product (human UGT1A1) to antigen-presenting cells, thereby circumventing host immune response. This is relevant to gene therapy in general because the lack of immune response would permit repeated administration of the vehicle-DNA complex and would spare the recipient tissues from immune-mediated injury. In addition, in many cases of inherited metabolic diseases, the mutant had never been exposed to the missing protein. Therefore, the therapeutic gene product may be treated as a foreign protein, resulting in host immune response. Expression of human UGT1A1 in Gunn rats by systemic administration of lentiviral vectors has been reported to result in the development of anti-human UGT1A1 antibodies and coincident reduction of hepatic UGT1A1 activity.22 Here we have confirmed the development of antibodies against human UGT1A1 after lentiviral gene transfer in Gunn rats. In contrast, FPL-mediated expression of human UGT1A1 did not result in an antibody response. Although we did not directly examine cell-mediated immune response against the transgene-expressing cells, normal morphology of the hepatocytes, the lack of infiltration by inflammatory cells, and the absence of transaminase elevation indicate the absence of any cell-mediated immune reaction. For both the lentiviral vector and our transposition-competent plasmid, human UGT1A1, expression was driven by a ubiquitous promoter, so that there was no translational restriction to specific tissues. This suggests that the lack of immune response in the case of FPL-mediated gene transfer was due to restriction of gene delivery to hepatocytes, which is consistent with our finding of liver-specific distribution of hUGT1A1 sequences.
As inherited metabolic disorders are lifelong conditions, gene therapy for these conditions may be facilitated by integration of the transgene into the host genome, so that the gene would be transmitted to the progeny of the transduced cells after mitosis. The plasmid utilized in our study expresses the SB transposase and human UGT1A1 from two different transcription units. The transcription unit expressing human UGT1A1 is flanked by IR/DR DNA sequences that are substrates for the transposase. When the SB transposase is expressed, it catalyzes the excision of the DNA segment located between the IR/DRs, followed by insertion of DNA segment at AT dinucleotide sites in the cellular chromosomes. The mechanism and characteristics of SB-mediated gene transcription have been described.10, 11 In this study we used a partially sequence modified version of SB, which was generated by Yant and associates and was reported to be more efficient in mediating DNA transposition than previous versions of the enzyme. Notably, excision of the target DNA unit by the transposase results in cleavage and destruction of the plasmid, minimizing the chance of repeated transposition. We determined the flanking genomic sequence at several sites of insertion of the hUGT1A1 transcription units in the rat genome by inverse PCR. The presence of the TA dinucleotides at the insertion sites provided evidence that the integration was mediated by SB transposase. Because the map of rat genome is not as complete as the human or mouse genome, we were able to identify the location of the integrations on rat chromosomes in some but not all cases.
In summary, intravenous injection of FPL, loaded with a therapeutic plasmid, resulted in liver-specific delivery and expression of the transgene. In vivo transduction of hepatocytes was efficient enough to achieve reduction of hyperbilirubinemia in the Gunn rat model of CN-1. The SB transposition system enabled integration of the therapeutic gene into the host genome, permitting long-term metabolic effect. FPL-based gene delivery obviates the use of recombinant viral vectors and did not result in an immune response and detectable toxic effect. SB-mediated transposition sites are scattered throughout the genome, without preference to sites close to expressed genes. This reduces the probability of insertional mutagenesis. Several laboratories are engaged in engineering additional safety features, such as inserting insulator sequences flanking the transcription units to prevent accidental activation of promoters of neighboring genes23 and adding DNA targeting elements to the transposase to promote site-preference of integration.24, 25 Clinical trials are being proposed for gene therapy based on the SB transposition system.26 Furthermore, liver-targeted delivery of a plasmid expressing a short-hairpin RNA directed against the internal ribosomal entry site (IRES) of the hepatitis C virus resulted in selective inhibition of HCV IRES-mediated translation.27 Thus, our combined strategy of FPL-based hepatocytes targeted delivery of transposition competent DNA has the potential to be developed for gene therapy for inherited liver-based metabolic disorders.