Statistical Genetic Analysis of Serological Measures of Common, Chronic Infections in Alaska Native Participants in the GOCADAN Study
ABSTRACT
This paper describes genetic investigations of seroreactivity to five common infectious pathogens in the Genetics of Coronary Artery Disease in Alaska Natives (GOCADAN) study. Antibody titers and seroprevalence were available for 495 to 782 (depending on the phenotype) family members at two time points, approximately 15 years apart, for Chlamydophila pneumoniae, Helicobacter pylori, cytomegalovirus (CMV), herpes simplex virus 1 (HSV-1), and herpes simplex virus 2 (HSV-2). Seroprevalence rates indicate that infections with most of these pathogens are common (≥20% for all of them, >80% for H. pylori, CMV, and HSV-1). Seropositive individuals typically remain seropositive over time, with seroreversion rates of <1% to 10% over ∼15 years. Antibody titers were significantly heritable for most pathogens, with the highest estimate being 0.61 for C. pneumoniae. Significant genome-wide linkage evidence was obtained for C. pneumoniae on chromosome 15 (logarithm of odds, LOD score of 3.13). These results demonstrate that individual host genetic differences influence antibody measures of common infections in this population, and further investigation may elucidate the underlying immunological processes and genes involved.
Introduction
The prevalence of cardiovascular disease (CVD) has increased among Alaska Natives in recent years, along with their adoption of a more “westernized” lifestyle including dietary changes, increase in smoking, and decreased physical activity [Howard et al., 2010; Kaufman et al., 2008]. It is possible that inflammation resulting from infection with common pathogens may also contribute to CVD prevalence in this population. Studies have reported associations between specific pathogens (e.g., Chlamydophila pneumoniae, Helicobacter pylori, influenza viruses, cytomegalovirus [CMV], Epstein-Barr virus, and herpes simplex virus 1 [HSV-1]) and CVD [Fraser et al., 2003; Guan et al., 2012; Ibarhim et al., 2005; Mendall et al., 1995; Simanek et al., 2011; Sorlie et al., 2000]. In addition, pathogen burden (i.e., the number of different infections in a given individual) has been implicated in the development of atherosclerosis, suggesting that cumulative infectious dose may increase CVD risk [Espinola-Klein et al., 2002; Rosenfeld and Campbell 2011; Rupprecht et al., 2001]. Individuals vary in their susceptibility to infectious diseases, which is attributable to various factors such as sex, age, nutritional status, socioeconomic status, and stress, etc. [Armstrong et al., 2001; Dowd and Aiello, 2009], and host genetic variation also plays a role [International HIV Controllers Study, 2010; Maran et al., 2012; McLaren et al., 2012; Timmann et al., 2012].
Participants in the Genetics of Coronary Artery Disease in Alaska Natives (GOCADAN) study were previously shown to have moderate-to-high seroprevalence rates for five pathogens (H. pylori, CMV, HSV-1, herpes simplex virus 2 [HSV-2], and C. pneumoniae) that are suspected to play a role in CVD [Zhu et al., 2006]. In this population, pathogen burden was positively correlated with C-reactive protein (CRP), which is a marker of chronic inflammation and is considered to be a CVD risk factor, and CRP in turn was positively correlated with CVD [Howard et al., 2008,]. Here, we report seroprevalence rates for a larger number of GOCADAN participants, as a follow-up to the results presented by Zhu et al., [2006], and we determine the genetic contribution to differences in antibody level and conduct genome-wide linkage analysis in order to localize genetic factors influencing these traits.
Methods
Study Population
Participants included 801 family members (Table 1) who participated in the GOCADAN study, which was designed to identify genetic risk factors for CVD [Ebbesson et al., 2006; Howard et al., 2005]. The study population resides in eight villages and the city of Nome in the Norton Sound region of Alaska, and is predominantly Inupiaq. The majority of individuals (678 out of 801) belong to a single, large, complex pedigree that was broken into smaller pedigrees in order to facilitate analysis. Phenotyped individuals in this sample belong to 128 pedigrees, the largest of which includes 162 family members, representing up to five generations. Most phenotyped individuals are members of extended pedigrees. Participants were recruited during the years 2000–2004, ranged from 18 to 91 years of age (with an average age of 45 years), and consisted of 463 women and 338 men. This study also utilized archived samples that were collected from the same participants 15 to 20 years earlier (referred to here as the “baseline” visit) by the Centers for Disease Control (CDC), prior to this study. The baseline age of participants ranges from >1 year to 74 years old. Permissions to conduct the study were granted by the Norton Sound Health Corporation and the institutional review boards of all participating institutions. Signed statements of consent were obtained from all study participants.
| Pedigree information | |
| Number of pedigrees | 128 |
| Maximum number of generations | 5 |
| Size of largest pedigree | 162 |
| Average sibship size (range) | 2.9 (2–8) |
| Familial relationships, observed pairs | |
| Parent-offspring | 283 |
| Full siblings | 270 |
| Half siblings | 193 |
| Grandparent-grandchild | 69 |
| Avuncular | 181 |
| Half avuncular | 277 |
| First cousins | 87 |
- Table values represent only the number of study participants (n = 801), excluding unphenotyped family members, required to establish pedigree relationships.
Serology
Serum samples were collected from participants during 2000 to 2003 and stored at –80°C. Archived serum samples were obtained from the CDC in Alaska, which were collected 15–20 years earlier, prior to the initiation of the GOCADAN study. Both the archived (or “baseline”) and “follow-up” serum samples were thawed immediately before testing, and assays were run at the same time for both sets of samples. Not all study participants were phenotyped for antibody levels against all pathogens for both visits, and the exact sample sizes (ranging from 495 to 782) are given in Table 2. Commercially available enzyme-linked immunosorbent assays (ELISAs) were used to quantify IgG antibody levels against the following pathogens: H. pylori and CMV (Wampole, Cranbury, NJ); and HSV-1 and HSV-2 (Focus, Cypress, CA). An IgG titer ≤0.9 was considered seronegative, 0.9 to 1.1 indeterminate, and ≥ 1.1 seropositive. IgG antibodies to C. pneumoniae were quantified using microimmunofluorescence. The antigen used in this test consisted of elementary bodies of the Finnish strain Kajaani 6 (Laboratory for Chlamydia and Respiratory Bacterial Infections, National Public Health Institute, Finland) and an IgG titer ≥1:32 was considered seropositive for C. pneumoniae, as previously described [Wang, 2000; Zhu et al., 2006]. Seroconversion was calculated as the proportion of seronegative individuals at the baseline visit who were seropositive at the follow-up visit. Seroreversion (i.e., a negative seroprevalence result at the follow-up visit for an individual previously found to be seropositive at the baseline visit) was also estimated for all pathogens. In addition, we summed up the number of seropositive reactions to the five targeted pathogens as an estimate of an individual's pathogen burden.
| Number of individuals | Seroprevalence | |||||
|---|---|---|---|---|---|---|
| Pathogen | Baseline visit | Follow-up visit | Baseline visit | Follow-up visit | Frequency of seroconversiona | Frequency of seroreversionb |
| C. pneumoniae | 495 | 484 | 39.6% | 44.0% | 15.6% | 10.8% |
| H. pylori | 781 | 766 | 76.8% | 77.5% | 50.0% | 6.0% |
| CMV | 782 | 767 | 86.7% | 94.1% | 67.1% | 0.9% |
| HSV-1 | 764 | 750 | 86.6% | 90.5% | 39.2% | 1.1% |
| HSV-2 | 764 | 749 | 33.2% | 47.3% | 17.1% | 4.9% |
- a Seroconversion rate was computed only on individuals who had information from both visits. It was defined here as the proportion of baseline seronegative individuals who are seropositive at the follow-up investigation. Changes to and from seroindeterminate status are ignored in this calculation.
- b Seroreversion rate was computed only on individuals who had information from both visits. It was defined here as the proportion of baseline seropositive individuals who are seronegative at the follow-up investigation. Changes to and from seroindeterminate status are ignored in this calculation.
Genotypic Data
DNA was extracted from buffy coats from study participants. Polymerase chain reaction was used to amplify 385 short tandem repeats, spaced at approximately 10 cM intervals across the 22 autosomes, using fluorescently labeled primer pairs (ABIPRISM Linkage Mapping Set MD 10 version 2; Applied Biosystems, Foster City, CA). Pedigree relationships were verified using PREST (pedigree relation statistical tests), and Mendelian inconsistencies in marker genotypes were identified and corrected using SIMWALK2 software [Sobel et al., 2002]. Multipoint identity-by-descent matrices were calculated at 1 cM intervals throughout the genome using LOKI [Heath, 1997]. The chromosomal map used in the analysis was based on marker locations reported by deCode genetics [Kong et al., 2002].
Statistical Methodology
The IgG quantitative antibody titer and pathogen burden traits were analyzed. Quantitative traits were transformed by inverse normalization prior to analysis, and sex, age, and their interactions were included as covariates in all analyses. Additive genetic heritability (h2) was estimated using a variance components pedigree analysis with the SOLAR computer program [Almasy and Blangero, 1998]. Multipoint genome-wide linkage analysis was performed in order to identify regions of the genome that may harbor genetic variants influencing the antibody trait phenotypes. A GAUSS script, implementing the methodology of Feingold et al. [1993], was used to estimate the logarithm of odds (LOD) score corresponding to a genome-wide adjusted significance level of 0.05. The method takes into account the complete pedigree structure as well as the chromosomal marker density. An LOD score of 2.86 was identified as the approximate 0.05 genome-wide significance level.
Results
Seroprevalence, Seroconversion, and Antibody Persistence
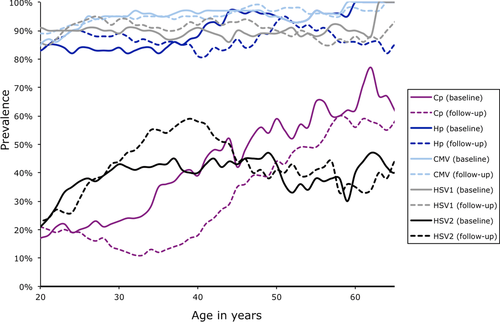
Seroprevalence rates were high (>20% in all age groups, except for C. pneumoniae among some younger age brackets) for all five examined pathogens. Seroprevalence was particularly high (>80%) for H. pylori, CMV, and HSV-1 (Fig. 1). Seroprevalence typically increased over the examined age range, though the seroprevalence plateaus out for most pathogens beyond a certain age. There were no significant differences in sex- and age-adjusted seroprevalence between the two visits (based on likelihood ratio tests comparing sample means of antibody titers after adjusting for covariates while taking pedigree relationships into account), which took place approximately 15 years apart. The proportion of baseline seronegative individuals who then converted to seropositive status by the follow-up visit was fairly low for C. pneumoniae and HSV-2 (16% and 17%, respectively) and quite high for HSV-1, H. pylori, and CMV (seroconversion rates ranged from 40% to 67%), reflecting the higher seroprevalence of these pathogens (Table 2). As primary infection with many of these pathogens occurs in childhood, it is not surprising that some individuals seroconverted during the 15- to 20-year time period given that a number of individuals were children at the time that the baseline samples were collected by the CDC. However, these numbers represent only 5–10% of the total sample, as the majority of individuals were already seropositive for HSV-1, H. pylori, and CMV at the baseline visit. The frequency of seroreversion (seronegative results for individuals previously determined to be seropositive) was low for these pathogens over the examined time period, ranging from 0.9% for CMV to 10.8% for C. pneumoniae, which suggests that the dichotomized antibody traits are fairly stable over an extended period of time. On average, individuals were seropositive to three out of the five pathogens examined (Fig. 2). Mean IgG antibody levels for the different categories of serostatus are provided in Supplementary Table S1. Values for C. pneumoniae IgG levels are higher than for the other pathogens due to different methods used for antibody determination (i.e., microimmunofluorescence vs. ELISA).

Heritability
We then investigated whether genetic factors of the human host in aggregate influence antibody levels. Heritability estimates, based on quantitative antibody titer, were highly significant except for HSV-2 and the follow-up measurement of H. pylori (Table 3). Estimates range from 0.14 for HSV-2 to 0.61 for C. pneumoniae. Heritability of pathogen burden (i.e., the number of seropositive reactions to the pathogens examined) was not significant for either the baseline or follow-up visits.
Linkage Results
Given the convincing evidence for host genetic effects, we conducted a genome-wide linkage scan to search for major loci influencing pathogen-specific antibody levels. We estimated that for our study sample, a LOD score of 2.86 corresponds with a genome-wide P-value of 0.05, based on theory developed by Feingold et al. [1993], which takes pedigree relationships and chromosomal distribution of markers into account. Genome-wide linkage plots for baseline and follow-up visits for each pathogen are provided as Supporting Information (Supplementary Fig. S1–S5). We obtained significant linkage evidence for C. pneumoniae, with a maximum LOD score of 3.13 on chromosome 15q22.31, near marker D15S153 (Fig. 3, Table 4). The 1 LOD unit support interval spans a large genomic region between markers D15S117 and D15S130 (approximately 56–92 Mb q terminus) containing ∼547 genes. In addition, results suggestive of linkage (LOD score ≥ 2.0) were obtained for C. pneumoniae on chromosomes 1 and 19, and for H. pylori, CMV, and HSV-1 on other chromosomes (Table 4). We also ran genome-wide bivariate linkage analysis in which we simultaneously analyzed both baseline and follow-up antibody measures for each pathogen, based on the rationale that the two phenotypic measurements jointly may provide more accurate phenotypic information and thus potentially permit mapping of additional loci (although power can also be decreased owing to the increase in degrees of freedom). The sample sizes and results are given in Supplementary Table S2, and genome-wide linkage plots are included as Supplementary Fig. S6–S10. We replicated the suggestive LOD score for the baseline visit for HSV-1 on chromosome 17q23.2 near marker D17S944, and obtained additional suggestive linkages results for C. pneumoniae on chromosome 15 and CMV on chromosome 13. The lowering of the significant LOD score obtained for the follow-up visit for C. pneumoniae may be related to the reduced sample size and increased degrees of freedom for the bivariate analysis. No additional genome-wide significant loci were identified.
| Heritability estimate of quantitative antibody | ||
|---|---|---|
| traits (P-value) | ||
| Pathogen | Baseline visit | Follow-up visit |
| C. pneumoniae | 0.47 ± 0.11 (5.9 × 10−6) | 0.61 ± 0.11 (3.1 × 10−9) |
| H. pylori | 0.33 ± 0.11 (5.9 × 10−4) | 0.13 ± 0.11 (0.12) |
| CMV | 0.42 ± 0.13 (3.4 × 10−4) | 0.50 ± 0.12 (2.2 × 10−6) |
| HSV-1 | 0.59 ± 0.09 (2.1 × 10−13) | 0.55 ± 0.10 (5.5 × 10−10) |
| HSV-2 | 0.18 ± 0.10 (0.02) | 0.14 ± 0.09 (0.05) |
| Pathogen burdena | 0.18 ± 0.15 (0.10) | 0.14 ± 0.14 (0.14) |
- a Number of seropositive reactions to pathogens examined in this study.
| Closest | Chromosomal | Maximum | ||
|---|---|---|---|---|
| Pathogen (visit)a | marker | cM location | location | LOD score |
| C. pneumoniae (f) | D1S450 | 21 | 1p36.22 | 2.22 |
| D15S153 | 62 | 15q22.31 | 3.13 | |
| D19S220 | 62 | 19q13.13 | 2.76 | |
| H. pylori (b) | D8S264 | 1 | 8p23.2-p23.3 | 2.07 |
| CMV (f) | D10S249 | 2 | 10p15.3 | 2.72 |
| D13S171 | 25 | 13q13.1 | 2.43 | |
| HSV-1 (b) | D17S944 | 83 | 17q23.3 | 2.75 |
- a b, baseline visit; f, follow-up visit.

Discussion
This research confirms that there is a high level of seroprevalence and long-term antibody persistence among the study participants. Our results are similar to those of an earlier investigation of the same five infectious pathogens in a smaller subset of 610 participants [Zhu et al., 2006], with only slight differences due to our exclusion of indeterminate values (i.e., samples with antibody titers falling within the 0.9 to 1.1 range) and an increased sample size used here. Both studies report a substantial level of chronic infection in this Alaska Native population, which suffers from CVD. Chronic infection, such as seen in this population, may lead to prolonged systemic and/or localized inflammation caused by the host's immune system that may ultimately result in atherosclerosis and other ageing-related diseases, as suggested by other studies [Espinola-Klein et al., 2002; Fraser et al., 2003; Guan et al., 2012; Ibarhim et al., 2005; Mendall et al., 1995; Rosenfeld and Campbell, 2011; Rupprecht et al., 2001; Simanek et al., 2011; Sorlie et al., 2000]. Indeed, previous research involving GOCADAN study participants demonstrated a significant, positive correlation between pathogen burden and CRP, a risk factor for CVD [Howard et al., 2008].
The heritability estimates presented here show that individual genetic differences do contribute to the majority of these serological phenotypes in this sample of Alaska Natives, suggesting that pathogen exposure alone is not enough to determine infection status, at least if one assumes that genetic factors do not primarily modify behavior that influences exposure risk, frequency and intensity. Many of these pathogens are common and are transmitted via the oral/respiratory route, suggesting that every individual is exposed repeatedly throughout life. For this reason, a seronegative status is informative for genetics investigations, as this status is unlikely to simply reflect the absence of exposure. HSV-2 is an exception. It is the only primarily sexually transmitted pathogen examined here, and exposure to it therefore may be limited. This may also explain the small heritability estimates for this pathogen, as seronegative individuals may provide little information on host genetic effects. In an attempt to address this, we analyzed the quantitative antibody levels of HSV-2 seropositive individuals only (n = 254 for baseline visit, and n = 354 for follow-up visit), as these individuals must have been exposed (assuming absence of false positives). The resulting heritability estimates were 0.00 (P = 0.5) for both visits. This may indicate that the low heritability levels obtained with HSV-2 cannot be explained entirely by nonexposure, although the reduced sample size complicates interpretation. The biological interpretation of our results is complicated by the nature of serological phenotypes. The presence of antibodies indicates a past or present infection, but it is not clear whether, for example, a high antibody titer represents success in warding off infection, or whether the immune system was less efficient at dealing with the invading pathogen, or if it is related to the timing of infection (that the infection was more recent).
Significant heritability estimates justify further investigation in order to localize underlying genetic factors that may influence these traits. Our genome-wide linkage analysis for C. pneumoniae IgG antibody titer trait gave a significant LOD score of 3.13 at 15q22.31. Several potential candidate genes lie within this large region (the 1-LOD linkage region spans 15q22.1–15q26.2), including SMAD6 (located at 15q22.31), which is involved in endocytosis and negative regulation of apoptosis by inhibiting signaling by the TGF-β superfamily [Lee et al., 2011; Park, 2005]. C. pneumoniae, a common cause of acute respiratory infection, is an obligate intracellular bacterium that induces apoptosis resistance in host cells in order to prevent eradication of infected epithelial cells during the early stage of infection [Fischer et al., 2001; Rajalingam et al., 2001]. Other genes in this region that may be involved in genetic control of antibody levels to C. pneumoniae include: SEMA7A (at 15q24.1), which stimulates cytokine production, chemotaxis, and superoxide release by monocytes; IL-16 (at 15q25.1), which encodes a proinflammatory cytokine that influences CD4+ T lymphocytes, eosinophils, and monocytes and upregulates IL-2 receptors; and AEN (apoptosis-enhancing nulease, at 15q26.1). We did not find significant evidence of linkage, however, for the remaining four pathogens. This may have to do with antibody titers being an imprecise measure of current or past infection, as they are not a direct measure of the pathogens themselves. It is also possible that the size of the effect is small and/or that our modest sample size (by genome-wide analysis standards) makes us unable to detect potential susceptibility loci for these other pathogens at this time. However, a comparison of the genomic regions of interest identified in our study with previously published genome-wide association studies (Supplementary Table S3) points to a number of genes involved in immune response (including FREM2, which may be involved in antibody response to infection with CMV), coronary heart disease, and other age-related diseases (e.g., type 2 diabetes and Alzheimer's disease) that are potentially of interest.
We compared the heritability estimates of this study with those of another minority population, Mexican Americans from San Antonio, Texas, for whom data were also available for the same five pathogens (Rubicz et al., 2011a). It should be noted that these studies are not directly comparable because different serological assays were used in quantifying the antibody titers (Rubicz et al., 2011b), in addition to obvious differences in ethnicity of study participants and environments. Nevertheless, both studies demonstrated a significant contribution of host genetic factors to antibody titer levels for the majority of the pathogens examined. The smallest heritability estimates were obtained for the only sexually transmitted pathogen that was examined, HSV-2 (h2 = 0.18 for both studies). For the remaining pathogens, estimates ranged from h2 = 0.26–0.39 for the Mexican Americans, compared to h2 = 0.13–0.61 for Alaska Natives. Several differences were noted between the two study populations, including the seroprevalence rates for some pathogens. For example, the C. pneumoniae seroprevalence rate was 40–44% in GOCADAN, vs. 86% in the Mexican Americans, which may be due in part to low population density in the region of Alaska inhabited by the Inupiaq study population [Zhu et al., 2006]. In addition, our heritability estimates for HSV-1 were higher for the GOCADAN study (h2 = 0.55–0.59) than for the Mexican Americans (h2 = 0.26). Although the reason for these observed differences is unknown, it is possible that the genetic factors influencing susceptibility or resistance to these infectious diseases may be population specific, possibly resulting in part from their adaptation to the local environment and differential exposure to pathogens.
In conclusion, our study provides further evidence that individual genetic variation influences infectious disease serological phenotypes, and that these variants can be localized to particular regions of the genome. Further investigation may point to novel methods for treating or even preventing infection with common pathogens, thereby eliminating subsequent disease progression including chronic inflammation and its contribution to the development of CVD.
Acknowledgments
We thank the Norton Sound Health Corporation (NSHC) and the participants in this study. The study was funded by NIH grants HL064244, MH059490, HL082458, and HL080149. The NIH LRP health disparities division provided additional support. Parts of this investigation were conducted in facilities constructed with support from the Research Facilities Improvement Program Grant Number C06 RR013556 from the National Center for Research Resources, National Institutes of Health. The AT&T Genomics Computing Center supercomputing facilities used for this work were supported in part by a gift from the AT&T Foundation and with support from the National Center for Research Resources Grant Number S10 RR029392. The authors have no conflict of interest to declare.




