High frequency of silent mutations in gyrA gene of Mycobacterium tuberculosis in Indian isolates
Anamika Gupta and Sudir K. Pal contributed equally to this work.
Accepted by: B. Gollapudi
Abstract
Reporting any uncommon or untapped changes in bacterial genetics or physiology would be of great importance to support the drug development process. We studied 120 Mycobacterium tuberculosis clinical isolates with different geographical origin within India and their resistance profile and found a significant number of isolates (109) harboring the polymorphism at nucleotide positions 61 and 284 of the gyrA gene. Bioinformatics analysis of these changes for drug binding suggested no significant change in the binding of the drug but have lower binding energies as compared with the wild-type proteins. Although functionally silent for the gyrA gene, these changes are indicating a silent geographical and evolutionary change that needs to be further studied for drug discovery and bacterial fitness.
1 INTRODUCTION
Drug resistance in Mycobacterium tuberculosis (M. tb) is the biggest obstacle in ending this disease. The problem in the high prevalence and low to middle-income countries is further exorbitant in terms of its management (Global tuberculosis report [World Health Organization, 2020]). Fluoroquinolones (FQ) which remain handy options after the first line of treatment fails are also getting resistant. The gene that serves as a very good marker for the diagnosis of resistance against FQ is gyrase A (gyrA) (Avalos et al., 2015). By looking at the mutations in this gene an assertive prediction can be made about FQ resistance. At the same time, new mutations keep emerging and may add to the list. Some of the mutations which emerge do not correlate with the emerging resistance and are termed silent ones (Gonzales et al., 2002). However, this change must have some effect, either from an evolutionary point of view or in terms of the overall physiology of the organism.
The study was part of a broader investigation in which multiple gene targets were sequenced to evaluate their conservation status. During this process, we identified two silent mutations in the gyrA gene that appeared in a significant number of isolates. Despite an extensive literature review, we were surprised to find that these mutations had been largely dismissed as inconsequential, with no further analysis in previous research. Driven by this gap in understanding, we utilized bioinformatics approaches to explore their potential impact, recognizing the importance of investigating even subtle genetic variations, especially when broader resources and studies were beyond our immediate reach.
Mutations with the change of amino acid but not having a drastic change in protein structure or those which are not inducing drug resistance are not given due consideration when drug resistance is a concern. However, accumulation of such mutation in a bigger population with high load of infection indicates an evolutionary change that can result in either better fitness of the pathogen or can support virulence in the long term. The present study focuses on one such observation and tried to check its impact using bioinformatics analysis. The outcomes indicate a non-significant change in drug binding with the change in sequences but definitely, they are showing lesser binding energies as compared to wild-type sequences. Their effect on the overall physiology of the pathogen is a subject of future studies.
2 MATERIALS AND METHODS
This study was approved by our institutional ethics committee wherein, we studied 120 M. tb isolates collected from various parts, that is, West, North, and South of India. The isolates were screened for primary drug resistance (SHRE—streptomycin [STR], isoniazid [INH], rifampin [RIF], and ethambutol [EMB]) along with ofloxacin (OFX) using BACTEC™ MGIT™ 960 Mycobacterial Detection System (BD, NJ) as well as with Nitrate reductase assay (NRA) (Gupta et al., 2020). The inoculum was prepared from fresh LJ medium. A growth control containing no antibiotic and a sterile control without inoculation were also used with every set of experiment (Gupta et al., 2017). We also sequenced the gyrA gene for mutations and submitted the representative sequences to GenBank (OQ348104–OQ348107). It was further explored if the mutations are translating into different amino acids and if so, if the change bringing drug resistance. The mutations at nucleotide position 61(Glu/Gln) and 284(Ser/Thr) were found to be translating into different amino acids but have no impact on phenotypic drug resistance status. Molecular docking was used for bioinformatics analysis as below.
Three-dimensional structure of the gyrA protein was downloaded from the RCSB PDB databank (Berman et al., 2002) and minimized using the YASARA minimization server (Krieger et al., 2009). The energy minimization experiment was completed after 1018 steps and the final energy was recorded as −46,944.379 kJ/mol. The structure so obtained was used for docking with the ligand Ofloxacin (obtained from PubChem database, https://pubchem.ncbi.nlm.nih.gov/compound/4583). The Docking server utilizing autodock was used for performing molecular docking (Bikadi & Hazai, 2009). The sampling mechanism of docking algorithm used was based on the Lamarckian genetic algorithm (LGA), the total and local search results of energy combined, and the semi-empirical function of the binding free energy used to rank the intermolecular energy scores between the receptor and the ligand. The box center was adjusted according to the active site of mechanism and built a 60 Å × 60 Å × 60 Å rectangular box with a grid space of 0.375 Å. One hundred conformations were collected in each docking run and 25 such runs were performed. The conformation of the lowest energy in the largest conformation cluster was defined as the near native conformation. Rest all parameters were set to default values and obtained results were recorded (Bikadi & Hazai, 2009).
3 RESULT AND DISCUSSION
Drug resistance studies using MGIT and NRA provided a mix of susceptible and resistant isolates for primary drugs as well as for the OFX. When confirmed with sequencing, the isolates were found to have mutations that could be correlated with drug resistance as per the published literate. To our surprise, all isolates were harboring mutations at nucleotide positions 61 and 284 as compared to Rv0006 except for 11 isolates of M. tb. Being present in drug-resistant as well as susceptible isolates, its correlation with drug resistance could not be established in the phenotypic assay.
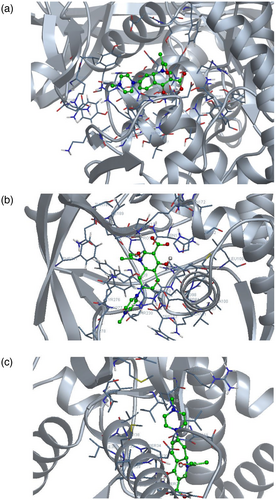
For bioinformatics analysis, the minimized structure of 3IFZ (gryA) was used to incorporate mutations and docked with OFX to see the impact of nucleotide change at 284 position (Ser95Thr). The H-bond pattern in Ser95 to Thr95 mutated gryA protein (3IFZ) where Thr95 was associated with three residues Ser91, Ile92, and Met99. Post mutation the H-bond interactions change (Figure 1a,b). Thr95 interacts with Ile92 with 2 H-bonds (distance 2.33 and 2.96 Å, respectively), Ser91 with 1 H-bond (distance of 3.16 Å) and Met99 with 2 H-bonds (distance 3.06 and 3.25 Å, respectively). However, this is not enough to drop the bound ligand (OFX) in our case as shown in docking work (Figure 1a,b).

The estimated free binding energy of docked structure of wild (3IFZ) gryA protein to the ligand ofloxacin is −5.58 kcal/mol (Figure 2a, Table 1) and the docked structure of mutated Ser95toThr95(3IFZ) gryA protein to the ligand ofloxacin has estimated free binding energy of −5.34 kcal/mol (Figure 2b, Table 2). This reveals that the interaction continues to happen and there would not be a fallback of the ligand. Hence proving, why this mutation does not result in drug resistance. Similar was the pattern for the mutation at nucleotide position 21 that resulted in a change of amino acid (E21Q) (Figure 2c). The mutated structure gave free energy of −3.74 which is higher than the wild type and hence would be a weaker binding of the ligand but not enough to resist ligand binding (Table 3).
| Rank | Est. free energy of binding | Est. inhibition constant, KI | vdW + H-bond + desolv energy | Electrostatic energy | Total intermolecular energy | Frequency | Interact. surface |
|---|---|---|---|---|---|---|---|
| 1. | −5.58 kcal/mol | 81.00 uM | −6.05 kcal/mol | −0.44 kcal/mol | −6.48 kcal/mol | 30% | 683.826 |
| Rank | Est. free energy of binding | Est. inhibition constant, KI | vdW + H-bond + desolv energy | Electrostatic energy | Total Intermolecular energy | Frequency | Interact. surface |
|---|---|---|---|---|---|---|---|
| 1. | −5.34 kcal/mol | 122.63 uM | −6.14 kcal/mol | −0.06 kcal/mol | −6.19 kcal/mol | 10% | 894.212 |
| Rank | Est. free energy of binding | Est. inhibition constant, KI | vdW + H-bond + desolv energy | Electrostatic energy | Total Intermolecular energy | Frequency | Interact. surface |
|---|---|---|---|---|---|---|---|
| 1. | −3.74 kcal/mol | 1.82 mM | −4.55 kcal/mol | −0.05 kcal/mol | −4.61 kcal/mol | 10% | 565.874 |
The overall results from phenotyping as well as bioinformatics analysis showed that there is no change in drug susceptibility or binding because of these two mutations. Also, in earlier drug resistance studies in M. tb, most of these polymorphisms were not taken into consideration as they were not found associated with FQ resistance and were regarded as nonfunctional or natural polymorphisms (Lau et al., 2011; Sandgren et al., 2009; Takiff et al., 1994). Several reports are available about this polymorphism as well as their co-relation with no phenotypic resistance (Chakravorty et al., 2011; Devasia et al., 2012). However, these two polymorphisms were not reported so frequently in previous studies which indicates a silent micro-evolution in these isolates with no predictive direction at this time. Agashe et al., have discussed a few such mutations in other microorganisms and have gathered evidence proving the non-negligible effect of silent mutations on altering mRNA structure and stability, binding sites of protein and DNA and overall fitness (Agashe et al., 2016). Other studies have seen such changes in light of evolution only and have used them to classify the isolates into various groups like the Beijing, Haarlem, and African M. tuberculosis clusters (Brosch et al., 2002) while other have used this information for classifying the M. tuberculosis strains into lineages using insertion sequence and spoligotype patterns, MIRU-VNTR typing, and SNP-based barcoding leading to their history of origin (Atavliyeva et al., 2024). Likewise, as observed in the present study, the codons are changing and there is a good possibility that the rate of the translation may get influenced due to codon bias. The rate of translation can be faster on the stretches wherein frequent codons are used as compared to those stretches where rare codons are being translated. Hence, this study opens the avenues for further research into the areas of expression analysis and physiological changes which could be induced due to such silent mutations in the case of gyrA gene in drug-susceptible and resistant strains of M. tb. In-depth studies in this area may further help understand the survival mechanism of M. tb under various stresses and may also impact drug and vaccine development.
AUTHORS CONTRIBUTIONS
Anamika Gupta: Review of Literature, Methodology and Writing – Original draft preparation. Sudhir K. Pal: Software. Bioinformatics, Figure preparation and Manuscript Writing. Vijay Nema: Conceptualization, Supervision Writing – Reviewing and Editing.
ACKNOWLEDGMENTS
The authors acknowledge the direct or indirect contribution of Sushama Jadhav, Sunita B Rathod, Divya Pandey and Najneen A Fakir ICMR-National AIDS Research Institute, Pune, India, during the execution of this work.
FUNDING INFORMATION
This work was supported by an ICMR extramural grant with grant no. as AMR/45/2011-ECD-I.
CONFLICT OF INTEREST STATEMENT
All the authors have no commercial, academic, or financial conflict of interest.
ETHICS STATEMENT
This study was approved by the institutional ethics committee with annual review and approval till the completion of the study.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in Genbank at https://submit.ncbi.nlm.nih.gov/about/bankit/, reference number OQ348104 - OQ348107.