DNA barcoding, identification, and validation of the pufferfish (Order: Tetraodontiformes) in China coastal waters
Abstract
The order Tetraodontiformes are one of the most unique groups of teleostean fish, exhibiting highly derived and greatly diversified phenotypes. It is a difficult task for both professionals and nonprofessionals to accurately identify these species only according to morphological characteristics. DNA barcoding can identify species at the molecular level to overcome the limitations of morphological classification. In this study, we collected 616 specimens of pufferfish from the coastal waters of China. According to the morphological characteristics, they were preliminarily identified as 50 species. Further analysis using DNA barcodes identified these specimens as 46 species, belonging to 23 genera, 6 families. According to the species classification results of DNA barcoding, the three species identified by morphology as Takifugu pseudommus, Takifugu chinensis, and Takifugu rubripes should be the same species. Similarly, Lagocephalus wheeleri is the synonym of Lagocephalus spadiceus. Another important discovery of DNA barcoding analysis is that there are closer interspecific genetic distances within the genus Takifugu. If T. rubripes, T. pseudommus, and T. chinensis are taken as one species, the average interspecific to intraspecific genetic distance ratio of Takifugu is only 6.21 times, which does not reach the DNA barcoding threshold of more than 10 times proposed previously. Although the interspecific genetic distance in the genus Takifugu is relatively small, each species can be clustered into independent clades in the NJ tree. In conclusion, this study not only found that there are synonymous phenomena in the order Tetraodontiformes but also provided molecular evidence for the valid species names of Takifugu rubripes and Lagocephalus Spadiceus. The results can provide reliable DNA barcoding information for the identification of pufferfish species, help solve the problem of classification confusion in this order, and provide technical support for the identification of the original components of related commodities on the aquatic product market.
1 INTRODUCTION
The latest record from Fishbase (https://www.fishbase.de/) shows that the order Tetraodontiformes comprises approximately 446 species overall, belonging to 106 genera and 10 families. According to the records of “Fauna Sinica,” there are 131 species in 61 genera of 10 families in China (Su & Li, 2002), of which only one species is freshwater pufferfish. Tetraodontiformes are widely distributed along the coast of China and occupy an important economic position in Chinese fishery (Su & Li, 2002). Among them, the pufferfish of three genera Thamnaconus, Takifugu, and Lagocephalus are with high economic value.
The pufferfish have a long history of consumption in China and Japan, and it is regarded as a precious aquatic product for its high protein content and delicious taste. Although pufferfish have a delicious taste and high economic value, many of them contain toxic tetrodotoxin (TTX) in the ovary, liver, kidney, eyes, and blood (Wu & Chen, 1981). If mishandled or ingested, people can become poisoned or even die. Therefore, it is urgent to screen the species of edible pufferfish to avoid poisoning caused by ingestion of toxic pufferfish. The taxonomy and identification of pufferfish is the key to solving the above problems.
In the traditional taxonomy, the classification and identification of Tetraodontiformes are mainly based on the morphological characteristics of stripes, spots, body color, mouth shape, etc. However, the morphology of closely related species of this order is very similar and difficult to distinguish (Chen & Zhang, 2015). Whether professionals or non-professionals, it is an extremely difficult task to identify pufferfish only according to their morphological characteristics. So far, the phenomenon of synonymous species in the order Tetraodontiformes is not uncommon (Cui et al., 2005; Liu et al., 1999; Matsuura, 2010; Reza et al., 2008; Sakai et al., 2021; Song et al., 2001), and the classification and identification of some species are relatively confused, which makes it difficult to accurately count the number of species in this order (Chen & Zhang, 2015; Su & Li, 2002). Moreover, most of the commodities of the pufferfish in the markets have been processed and lost their original morphological characteristics, which makes it impossible to identify species. It is evident that there is an urgent need for molecular identification of pufferfish species. Due to the ability of DNA barcoding, we can identify species at a molecular level, and the classification results are not affected by the developmental stage and individual morphological integrity of fish, as well as the experience of taxonomists (Hebert et al., 2003). If the morphological classification of species is combined with DNA barcoding technology for species identification, the results of species classification identification will be more objective and reliable.
At present, there have been many reports on the molecular taxonomy study of pufferfish. However, these studies only focused on a few genera or several species of the order Tetraodontiformes (Chen et al., 2019; Elmerot et al., 2002; Ishizaki et al., 2006; Reza et al., 2008; Santini et al., 2013; Song et al., 2001; Zhang & He, 2008), lacking systematic taxonomic study on the species of the entire order. Therefore, it is urgent to carry out research on DNA barcoding classification and identification of the pufferfish, to clarify the synonyms and cryptic species of this order.
In order to solve the difficulty of morphological classification of Tetraodontiformes, this study used DNA bar code technology and morphological methods to identify 616 samples of Tetraodontiformes collected from the coast of China, clarified the synonyms of some species of this order, and confirmed the effective species names of these species Building a perfect classification system of DNA barcode for the order Tetraodontiformes, will not only enrich the database of fish DNA barcode but also promote the rapid development of pufferfish taxonomy and systematics and provide technical support for aquatic product quality safety and trade supervision. The results of this study not only provide a basis for the taxonomic and phylogenetic research of Tetraodontiformes but also provide technical support for market supervision of pufferfish and their processed products.
2 MATERIALS AND METHODS
2.1 Sample collection
Since 2011, we have collected a total of 616 specimens of pufferfish from the coastal waters of China (Table 1). Samples collection and use comply with the relevant regulations of the Institutional Animal Care and Use Committee.
| Voucher no. | GenBank accession no. | Haplotype | Number of samples | Morphological identification | Molecular identification | BLAST result in NCBI | BLAST in BOLD | Family |
|---|---|---|---|---|---|---|---|---|
| YSFRI023–YSFRI026 | OQ700252–OQ700255 | Hap (13–15) | 4 | Balistoides conspicillum | B. conspicillum | B. conspicillum (99.23%–100%) | B. conspicillum (99.38%–100%) | Balistidae |
| YSFRI028 | OQ700257 | Hap17 | 1 | Canthidermis maculata | C. maculata | C. maculata (99.19%–100%) | C. maculata (98.95%–100%) | Balistidae |
| YSFRI236–YSFRI241 | OQ700465–OQ700470 | Hap (69–71) | 6 | Melichthys vidua | M. vidua | M. vidua (99.39%–99.85%), M. indicus (99.07%–99.69%) | M. vidua (99.39%–99.85%), M. indicus (99.08%–99.70%) | Balistidae |
| YSFRI279–YSFRI283 | OQ700508–OQ700511 | Hap79 | 4 | Rhinecanthus aculeatus | R. aculeatus | R. aculeatus (99.42%–100%) | R. aculeatus (99.38%–100%) | Balistidae |
| YSFRI308–YSFRI310 | OQ700537–OQ700539 | Hap (84–85) | 3 | Sufflamen chrysopterum | S. chrysopterum | S. chrysopterum (97.46%–99.85%) | S. chrysopterum (97.45%–99.85%) | Balistidae |
| YSFRI311–YSFRI313 | OQ700540–OQ700542 | Hap (86–87) | 3 | Sufflamen fraenatum | S. fraenatum | S. fraenatum (98.98%–99.82%) | S. fraenatum (98.98%–99.83%) | Balistidae |
| YSFRI616 | OQ700845 | Hap171 | 1 | Xanthichthys auromarginatus | X. auromarginatus | X. auromarginatus (98.61%–99.39%) | X. auromarginatus (98.6%–99.38%) | Balistidae |
| YSFRI075–YSFRI093 | OQ700304–OQ700322 | Hap (24–27) | 19 | Diodon holocanthus | D. holocanthus | D. holocanthus (99.05%–99.84%) | D. holocanthus (99.53%–99.84%) | Diodontidae |
| YSFRI094–YSFRI102 | OQ700323–OQ700332 | Hap (28–29) | 9 | Diodon hystrix | D. hystrix | D. hystrix (99.24%–99.85%) | D. hystrix (99.23%–100%) | Diodontidae |
| YSFRI103–YSFRI107 | OQ700332–OQ700336 | Hap (30–31) | 5 | Diodon liturosus | D. liturosus | D. liturosus (99.51%–100%) | D. liturosus (99.85%–100%) | Diodontidae |
| YSFRI001 | OQ700230 | Hap1 | 1 | Aluterus monoceros | A. monoceros | A. monoceros (98.93%–100%) | A. monoceros (97.41%–100%) | Monacanthidae |
| YSFRI002–YSFRI014 | OQ700231–OQ700243 | Hap (2–5) | 13 | Aluterus scriptus | A. scriptus | A. scriptus (99.24%–100%) | A. scriptus (99.08%–100%) | Monacanthidae |
| YSFRI027 | OQ700256 | Hap16 | 1 | Cantherhines pardalis | C. pardalis | C. pardalis (98.93%–100%), C. pullus (99.47%–100%), 2 C. sandwichiensis (99.69%–99.70%) | C. pardalis (98.8%–100%), C. pullus (99.07%–100%), 1 Stephanolepis setifer (99.85%) | Monacanthidae |
| YSFRI029–YSFRI063 | OQ700258–OQ700292 | Hap (18–19) | 35 | Chaetodermis penicilligerus | C. penicilligerus | C. penicilligerus (99.54%–100%) | C. penicilligerus (99.53%–100%) | Monacanthidae |
| YSFRI242–YSFRI248 | OQ700471–OQ700477 | Hap72 | 7 | Monacanthus chinensis | M. chinensis | M. chinensis (97.84%–100%) | M. chinensis (97.85%–100%) | Monacanthidae |
| YSFRI284–YSFRI307 | OQ700513–OQ700536 | Hap (81–83) | 24 | Stephanolepis cirrhifer | S. cirrhifer | S. cirrhifer (99.08%–100%), 1 S. diaspros (99.69%–100%) | S. cirrhifer (99.12%–100%), 1 S. diaspros (99.39%–99.69%) | Monacanthidae |
| YSFRI567–YSFRI582 | OQ700796–OQ700811 | Hap (153–158) | 16 | Thamnaconus hypargyreus | T. hypargyreus | T. hypargyreus (99.43%–100%), 2 T. septentrionalis (99.85%–100%) | T. hypargyreus (99.4%–100%), 4 T. septentrionalis (99.85%–100%) | Monacanthidae |
| YSFRI583–YSFRI605 | OQ700812–OQ700834 | Hap (159–167) | 23 | Thamnaconus modestus | T. modestus | T. modestus (99.36%–100%), 3 T. modestoides (99.1%–99.85%); 2 T. septentrionalis (99.84%–99.85%) | T. modestus (99.42%–100%), T. modestoides (99.69%–99.85%), 1 T. septentrionalis (99.19%–99.84%) | Monacanthidae |
| YSFRI108–YSFRI114 | OQ700337–OQ700344 | Hap (32–34) | 7 | Lactoria cornuta | L. cornuta | L. cornuta (99.81%–100%) | L. cornuta (99.35%–100%) | Ostraciidae |
| YSFRI249 | OQ700478 | Hap73 | 1 | Ostracion cubicus | O. cubicus | O. cubicus (98.01%–100%), 4 O. immaculatus (99.56%–100%) | O. cubicus (97.99%–100%), 5 O. immaculatus (99.42%–100%) | Ostraciidae |
| YSFRI250–YSFRI257 | OQ700479–OQ700486 | Hap (74–77) | 8 | Ostracion rhinorhynchos | O. rhinorhynchos | O. rhinorhynchos (99.39%–99.42%) | O. rhinorhynchos (98.76%–99.54%) | Ostraciidae |
| YSFRI015–YSFRI017 | OQ700244–OQ700246 | Hap (6–8) | 3 | Arothron hispidus | A. hispidus | A. hispidus (96.41%–100%) | A. hispidus (97.61%–100%) | Tetraodontidae |
| YSFRI018 | OQ700247 | Hap9 | 1 | Arothron mappa | A. mappa | 4 A. mappa (99.7%–99.85%), 1 A. stellatus (99.56%) | 2 A. mappa (99.7%–99.85%), 2 A. stellatus (99.56%)1 Tylerius sp. (100%) | Tetraodontidae |
| YSFRI019–YSFRI022 | OQ700248–OQ700251 | Hap (10–12) | 4 | Arothron stellatus | A. stellatus | A. stellatus (98.34%–99.67%), 1 A. multilineatus (99.56%–100%) | A. stellatus (98.23%–99.85%) | Tetraodontidae |
| YSFRI064–YSFRI074 | OQ700293–OQ700303 | Hap (20–23) | 11 | Dichotomyctere nigroviridis | D. nigroviridis | Tetraodon nigroviridis (98.22%–99.56%) | Tetraodon nigroviridis (98.16%–99.53%) | Tetraodontidae |
| YSFRI115–YSFRI131 | OQ700344–OQ700360 | Hap (35–43) | 17 | Lagocephalus gloveri | L. gloveri | L. gloveri (98.54%–100%); L. cheesemanii (99.42%–99.85%), 1 L. spadiceus (99.69%–99.85%) | L. gloveri (98.53%–100%); L. cheesemanii (99.42%–100%), 2 L. spadiceus (99.69%–100%) | Tetraodontidae |
| YSFRI132–YSFRI139 | OQ700361–OQ700368 | Hap (44–49) | 8 | Lagocephalus inermis | L. inermis | L. inermis (98.19%–99.82%) | L. inermis (98.01%–99.82%) | Tetraodontidae |
| YSFRI140–YSFRI153 | OQ700369–OQ700382 | Hap (50–53) | 14 | Lagocephalus spadiceus | Lagocephalus spadiceus | L. wheeleri (99.39%–100%), L. spadiceus (97.38%–100%); 2 L. gloveri (99.85%–100%) | L. spadiceus (97.17%–100%); 2 L. gloveri (100%), 1 L. inermis (99.81%–100%) | Tetraodontidae |
| YSFRI154–YSFRI235 | OQ700383–OQ700464 | Hap (50, 53–68) | 82 | Lagocephalus wheeleri | Lagocephalus spadiceus | L. wheeleri (99.39%–100%), L. spadiceus (97.38%–100%); 2 L. gloveri (99.85%–100%) | L. spadiceus (97.17%–100%); 2 L. gloveri (100%), 1 L. inermis (99.81%–100%) | Tetraodontidae |
| YSFRI258–YSFRI278 | OQ700487–OQ700507 | Hap78 | 21 | Pao leiurus | P. leiurus | P. sp. (99.84%–100%), 2 Monotrete leiurus (98.36%–99.56%), 1 P. abei (99.56%) | P. sp. (99.84%–100%), P. cambodgiensis (98.36%–100%), P. leiurus (98.11%–100%) | Tetraodontidae |
| YSFRI283 | OQ700512 | Hap80 | 1 | Sphoeroides pachygaster | S. pachygaster | S. pachygaster (97.48%–100%) | S. pachygaster (97.46%–100%), 1 Lagocephalus Lagocephalus. (99.85%) | Tetraodontidae |
| YSFRI314–YSFRI323 | OQ700543–OQ700552 | Hap (88–90, 108) | 10 | Takifugu alboplumbeus | T. alboplumbeus | T. alboplumbeus (99.13%–100%), T. niphobles (98.19%–100%) | T. alboplumbeus (98.33%–100%), T. niphobles (98.35%–100%), 2 T. porphyreus (99.64%–100%) | Tetraodontidae |
| YSFRI324–YSFRI356; YSFRI358 | OQ700553–OQ700585; OQ700587 | Hap (91–95,97) | 34 | Takifugu bimaculatus | T. bimaculatus | T. bimaculatus (99.71%–100%) | T. bimaculatus (99.32%–99.66%) | Tetraodontidae |
| YSFRI359–YSFRI366 | OQ700588–OQ700595 | Hap (98–99) | 8 | Takifugu chinensis | T. rubripes | T. chinensis (99.55%–100%), 2 T. pseudommus (99.56%–99.71%), T. rubripes (99.36%–99.84%), 1 T. flavidus (99.85%) | T. chinensis (99.51%–100%), 1 T. pseudommus (99.51%), T. rubripes (99.34%–99.85%), 2 T. flavidus (99.85%) | Tetraodontidae |
| YSFRI357;YSFRI367–YSFRI388 | OQ700586;–OQ700596–617 | Hap (96, 100–106) | 23 | Takifugu flavidus | T. flavidus | T. flavidus (99.54%–99.84%), 3 T. fasciatus (99.85%–100%) | T. flavidus (99.7%–99.84%), 4 T. fasciatus (99.85%–100%) | Tetraodontidae |
| YSFRI389–YSFRI409 | OQ700618–OQ700638 | Hap (88,107) | 21 | Takifugu niphobles | T. alboplumbeus | T. alboplumbeus (99.13%–100%), T. niphobles (98.19%–100%) | T. alboplumbeus (98.33%–100%), T. niphobles (98.35%–100%), 2 T. porphyreus (99.64%–100%) | Tetraodontidae |
| YSFRI410–YSFRI435 | OQ700639–OQ700664 | Hap (109–115) | 26 | Takifugu oblongus | T. oblongus | T. oblongus (98.45%–100%) | T. oblongus (98.26%–100%) | Tetraodontidae |
| YSFRI436–YSFRI449 | OQ700665–OQ700678 | Hap (116–119) | 14 | Takifugu obscurus | T. obscurus | T. obscurus (99.55%–99.85%), T. fasciatus (99.56%–99.69%) | T. obscurus (99.55%–99.85%), 2 T. fasciatus (99.56%) | Tetraodontidae |
| YSFRI450–YSFRI485 | OQ700679–OQ700714 | Hap (120–128) | 36 | Takifugu ocellatus | T. ocellatus | T. ocellatus (99.67%–100%) | T. ocellatus (99.55%–100%) | Tetraodontidae |
| YSFRI486–YSFRI487 | OQ700715–OQ700716 | Hap129 | 2 | Takifugu pardalis | T. pardalis | T. pardalis (99.42%–100%) | T. pardalis (99.7%–100%) | Tetraodontidae |
| YSFRI488–YSFRI492 | OQ700717–OQ700721 | Hap (130–132) | 5 | Takifugu poecilonotus | T. poecilonotus | T. poecilonotus (98.98%–100%), 1 T. exascurus (99.56%–99.71%), 1 T. alboplumbeus (99.26%–99.71%) | T. poecilonotus (99.4%–100%), 2 T. exascurus (99.56%–99.71%) | Tetraodontidae |
| YSFRI493–YSFRI495 | OQ700722–OQ700724 | Hap (133–135) | 3 | Takifugu porphyreus | T. porphyreus | T. porphyreus (99.41%–99.71%) | T. porphyreus (99.24%–99.71%) | Tetraodontidae |
| YSFRI496–YSFRI499 | OQ700725–OQ700728 | Hap (99, 136) | 4 | Takifugu pseudommus | T. rubripes | T. rubripes (99.52%–100%), 2 T. pseudommus (99.71%–99.85%), T. chinensis (99.54%–100%), 1 T. flavidus (99.71%–100%) | T. rubripes (99.5%–100%), 1 T. pseudommus (99.67%–99.84%), T. chinensis (99.35%–99.67%), 2 T. flavidus (99.71%–100%) | Tetraodontidae |
| YSFRI500–YSFRI526 | OQ700729–OQ700755 | Hap (99, 136–139) | 27 | Takifugu rubripes | T. rubripes | T. rubripes (99.52%–100%), 2 T. pseudommus (99.56%–99.85%), T. chinensis (99.54%–100%), 1 T. flavidus (99.71%–100%) | T. rubripes (99.5%–100%), 1 T. pseudommus (99.35%–99.67%), T. chinensis (99.35%–99.67%), 2 T. flavidus (99.71%–100%) | Tetraodontidae |
| YSFRI527–YSFRI528 | OQ700756–OQ700757 | Hap140 | 2 | Takifugu snyderi | T. snyderi | T. snyderi (100%), T. stictonotus (98.5%–100%) | T. snyderi (100%), T. stictonotus (98.5%–100%) | Tetraodontidae |
| YSFRI529 | OQ700758 | Hap141 | 1 | Takifugu stictonotus | T. stictonotus | T. snyderi (98.69%–99.85%) | T. snyderi (98.68%–99.85%) | Tetraodontidae |
| YSFRI530–YSFRI533 | OQ700759–OQ700762 | Hap (142–144) | 4 | Takifugu vermicularis | T. vermicularis | T. vermicularis (99.42%–99.71%) | T. vermicularis (99.38%–99.71%) | Tetraodontidae |
| YSFRI534–YSFRI566 | OQ700763–OQ700795 | Hap (145–152) | 33 | Takifugu xanthopterus | T. xanthopterus | T. xanthopterus (99.26%–99.85%) | T. xanthopterus (99.37%–100%) | Tetraodontidae |
| YSFRI606–YSFRI614 | OQ700835–OQ700843 | Hap (168–169) | 9 | Torquigener hypselogeneion | T. hypselogeneion | T. hypselogeneion (98.27%–100%), T. flavimaculosus (97.98%–99.85%), T. altipinnis (99.41%–99.55%) | T. hypselogeneion (97.77%–100%), T. flavimaculosus (96.59%–99.85%), 1 T. perlevis (99.7%–99.85%), 1 T. hicksi (99.53%99.69%), T. altipinnis (99.4%) | Tetraodontidae |
| YSFRI615 | OQ700844 | Hap170 | 1 | Triacanthus biaculeatus | T. biaculeatus | T. biaculeatus (99.68%–99.85%) | T. biaculeatus (99.69%–99.98%) | Triacanthidae |
| Total | 171 | 616 | 50 species | 47species | 6 |
- Note: The bold items in the table refer to samples with differences in morphology and molecular identification.
After preliminary classification according to morphological characteristics, they were preserved in 95% ethanol for subsequent DNA barcoding identification. All voucher specimens are deposited in the National Marine Fishery Biological Germplasm Resource Bank.
2.2 Morphological identification
Firstly, the body length, weight, and other measurable traits of the specimen were measured. Further species identification was conducted based on the morphological characteristics of pufferfish described by Chen and Zhang (2015) and Su and Li (2002), including body size, number of fins, stripes, spots, body color, mouth shape, etc.
2.3 DNA extraction and quality assessment
DNA was extracted from the muscle of fish by using TIANamp Marine Animals DNA Kit (Tiangen Biotech (Beijing) Co., Ltd.). For detailed steps of DNA extraction, refer to the kit manual. DNA samples were stored at −20°C.
Nano-300 micro-spectrophotometer (Allsheng) was used to detect the concentration and purity of DNA samples, and the quality of the DNA samples was detected by 1% agarose gel.
2.4 PCR amplification and DNA sequencing
- COIF: 5′-TCAACCAACCACAAAGACATTGGCAC-3′; COIR: 5′-TAGACTTCTGGGTGGCCAAAGAATCA-3′.
PCR amplification was in total volume of 25 μL, containing 12.5 μL Taq Mixture (2×, vazyme), 2 μL DNA template, 1 μL of each primer (10 μM), and 8.5 μL ddH2O. Thermo cycling was carried out as follows: 94°C for 5 min, 94°C for 30 s, 55°C for 30 s, 72°C for 1 min, cycle 35 times from step 2 to step 4, and finally 72°C for 10 min. PCR products were detected by 1% agarose gel, and the qualified samples were sent to BGI in Qingdao for direct bidirectional sequencing.
2.5 Data analysis
The raw sequences were assembled by seqman of Lasergene software package (Burland, 2000; Swindell & Plasterer, 1997) and aligned by Clustal W in BioEdit version 7.0.9 (Hall, 1999). After correction and alignment, the effective sequence length was 687 bp. The haplotypes were analyzed by DNASP version 5.10.01 (Librado & Rozas, 2009). Each haplotype sequence was compared for similarity in NCBI (https://www.ncbi.nlm.nih.gov/) and BOLD (http://www.barcodinglife.org/). The top matches showing ≥98% similarity were used as the preliminary identification results. Then based on Kimura-2-parameter (K2P) mode (Kimura, 1980), the genetic distance of different taxonomic levels of Tetraodontiformes was calculated.
Bothus myriaster (NC_030365) and Glyptocephalus stelleri (NC_060723) were selected as the outgroups, based on the haplotypes of COI gene of Tetraodontiformes, the Neighbor-joining tree (NJ tree) of Tetraodontiformes was constructed in Mega 7.0 (Kumar et al., 2016).
3 RESULTS
3.1 Morphological identification
According to the morphological characteristics, 616 samples were identified as 50 species, belonging to 23 genera, 6 families, and 4 Suborder (Table 1). Among them, 612 samples can be identified to species. The remaining four samples are larval and juvenile fish, which can only be identified to genus due to their atypical morphological characteristics.
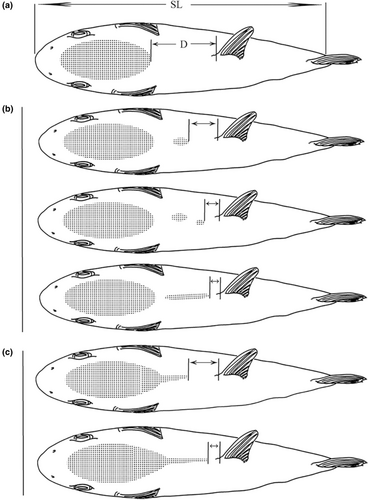
Among the 23 genera of Tetraodontiformes collected in this study, the various species of Takifugu genus have very similar morphologies and are difficult to distinguish with the naked eye. For example, the morphologic characteristics of T. alboplumbeus and T. niphobles are very similar. It is generally believed that T. niphobles has obvious chest spots. The chest spots of T. alboplumbeus are not obvious, but there are 4–6 dark bands on its back. Out of 616 specimens, 21 were identified as T. niphobles. And 10 specimens have the characteristics of T. alboplumbeus, but they are not exactly the same as T. alboplumbeus because they only present two dark bands. Furthermore, it is also difficult to distinguish T. rubripes, T. pseudommus, and T. chinensis, some studies suggest T. rubripes has irregular round black spots and white patterns in front of the caudal fin on the sides of the body, T. chinensis has no such black marks while T. pseudommus has white spots scattered on a black background on the dorsal and lateral sides of the body (Baek et al., 2018; Reza et al., 2008), as shown in Figure 1. Based on these morphological characteristics, 8 specimens were identified as T. chinensis, 4 as T. pseudommus, and 27 as T. rubripes.

Additionally, Lagocephalus wheeleri (Abe, Tabeta & Kitahama, 1984) and Lagocephalus spadiceus (Richardson, 1845) can also be distinguished by morphological characteristics. The former has elliptical dorsal spinule patch, while the latter has a rhomboidal patch with a posterior extension. In this study, 82 specimens were identified as L. wheeleri, 10 were identified as L. spadiceus, and 4 were found to have the intermediate characteristics of these two species, covering discontinuous patch. The above three morphological differences can be seen in Figure 2.
3.2 DNA barcoding of the Tetraodontiformes
3.2.1 Characteristics of DNA barcode sequences of Tetraodontiformes
The COI gene sequences of 616 pufferfish were retained with an effective length of 687 bp, consisting of 370 conserved sites, 62 transition sites (si), 43 transversion site (sv), and the si/sv ratio was 1.4. The results of base composition analysis showed: T 27.7%, C 29.9%, A 23.8%, G 18.6%, and the content of A + T was 51.5%, and C + G was 48.5%. Among them, the average G + C content of the first base of the codon was the highest, reaching 56.9%. The sequence analysis results also showed that the COI gene sequences of various species had high similarity with 561 conserved site in 253 sequences in Takifugu genus.
3.2.2 Haplotype analysis of DNA barcodes in Tetraodontiformes
Haplotype analysis showed that there were a total of 171 haplotypes in 616 COI gene sequences of Tetraodontiformes. Among them, there were 65 haplotypes in 253 COI gene sequences of genera Takifugu. Most noteworthy, the three species of T. pseudommus, T. rubripes, and T. chinensis share the same haplotype Hap 99; Hap 136 is the same haplotype shared by T.rubripes and T. pseudommus; and Hap 88 is the same haplotype shared by T. alboplumbeus and T. niphobles. In addition, among the 121 COI gene sequences of genera Lagocephalus, there were 36 haplotypes, with the shared haplotype of Hap 50 and Hap 53 for L. wheeleri and L. spadiceus.
3.2.3 Genetic distance of Tetraodontiformes
Using 50 species identified through morphological identification as taxonomic units, calculate the intra- and inter-species genetic distances of DNA barcodes. The results showed that the intraspecific genetic distance ranged from 0 to 0.009, with an average of 0.003. The interspecific genetic distance ranged from 0.001 to 0.298, with an average of 0.212. The ratio of the average intraspecific to interspecific genetic distance was 70.63. The average genetic distance within the genus was 0.016. The genetic distance between genera ranged from 0.12 to 0.298, with an average of 0.223. The average genetic distance within the family was 0.106. The genetic distance between families ranged from 0.192 to 0.247, with an average of 0.229.
We found that the interspecific genetic distance of several species was very low, far below the threshold of interspecific genetic distance for DNA barcoding. For example, the interspecific genetic distance between T. alboplumbeus and T. niphobles was only 0.002; between L. wheeleri and L. spadiceus was only 0.003. Additionally, the interspecific genetic distance of T. pseudommus, T. rubripes, and T. chinensis was 0.001 to 0.002.
Moreover, the results also showed that the species of Takifugu have very close genetic relationships. In this genus, the average interspecific genetic distance is only 6.21 times of the average intraspecific genetic distance, if T. rubripes, T. pseudommus, and T. chinensis are taken as one species, and T. alboplumbeus and T. niphobles are also considered as one species. It does not reach the DNA barcode threshold of more than 10 times proposed by Hebert et al. (2004). Among them, the genetic distance between T. oblongus and T. stictonotus is the largest, 0.045; and between T. bimaculatus and T. flavidus is the smallest, only 0.013. Meanwhile, there are several other species in this genus with interspecific genetic distance <0.02 (as shown in Table 2).
| Species 1 | Species 2 | Interspecific genetic distance |
|---|---|---|
| T. flavidus | T. bimaculatus | 0.0127 |
| T. flavidus | T. obscurus | 0.0196 |
| T. xanthopterus | T. obscurus | 0.0199 |
| T. xanthopterus | T. pardalis | 0.0159 |
| T. xanthopterus | T. poecilonotus | 0.0157 |
| T. stictonotus | T. snyderi | 0.0133 |
3.2.4 Phylogenetic relationships of Tetraodontiformes
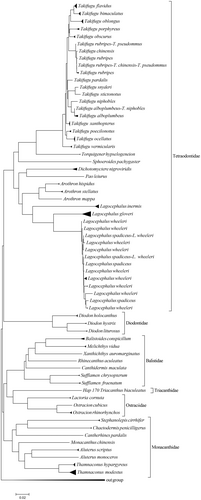
The Neighbor-joining tree was constructed for the order Tetraodontiformes, based on the haplotypes of COI gene. The results indicate that the six families of this order in this study are divided into two branches, namely the Diodontidae and Tetraodontidae families, which converge into one branch, while the Monacanthidae, Balistidae, Ostraciidae, and Triacanthidae families converge into another branch. As expected, in the Neighbor-joining tree, T. rubripes, T. pseudommus, and T. chinensis clustered into one clade, while T. alboplumbeus and T. niphobles clustered into one clade, and L. wheeleri and L. spadiceus also clustered into one clade. This once again confirms the previously proposed hypothesis of synonyms. In addition, we found that although the interspecific genetic distance of the genus Takifugu is small, some even <0.02, except for the few species that may be synonymous with the same species mentioned above, other species of this genus have clustered into different separate clades (as shown in Figure 3).
3.2.5 DNA barcode classification and identification of Tetraodontiformes
The classification and identification results based on DNA barcoding showed that 616 specimens were identified as 46 species, belonging to 23 genera, 6 families, and 4 Suborder (Table 1). Among them, although there are differences in morphological characteristics among the three groups of specimens of T. rubripes, T. pseudommus, and T. chinensis, they share the same haplotype, and their interspecific genetic distance ranges from 0.001 to 0.002 (average 0.002), far below the interspecific threshold of 0.02. In addition, in the NJ tree, they clustered into one clade. Therefore, T. rubripes, T. pseudommus, and T. chinensis should be synonymous with each other. According to the principle that the first published species name is the valid name, for the three species, T. rubripes should be their valid species name. Furthermore, this is also the case for L. wheeleri and L. spadiceus identified by morphological characteristics, which have the same haplotype and an interspecific genetic distance of 0.003, clustering into one clade in the NJ tree. Similarly, L. wheeleri and L. spadiceus should be synonymous, and L. spadiceus is the valid species name.
For T. alboplumbeus and T. niphobles, the DNA barcode identification results also showed that the two species may be synonymous. However, due to the fact that these 11 specimens identified as T. alboplumbeus do not completely have the morphological characteristics of T. alboplumbeus, it cannot be determined whether these specimens are hybrids of T. alboplumbeus and T. niphobles, or if T. alboplumbeus and T. niphobles are the same species, but they may undergo a morphological transformation process. More samples are needed to provide evidence for confirming the valid species names of these two species.
4 DISCUSSION
4.1 DNA barcoding threshold of Tetraodontiformes
DNA barcoding is used to identify species at molecular level, which is independent of time, region, and individual morphology (Hebert et al., 2003). This study shows that DNA barcoding can quickly and accurately identify the species of Tetraodontiformes, without being affected by key factors such as the integrity and development stage of the sample and the taxonomic expertise of the appraiser. For those closely related species, although the COI gene sequence may have high homology, they can still be distinguished by phylogenetic tree. For example, the sequence similarity between T. flavidus (Hap 100) and T. bimaculatus (Hap 91) has reached 98.7% (678 conserved sites in the 687 bp COI gene sequence), but the two species clustered into two independent clades in the NJ tree, thus enabling effective species identification.
This study indicates that species in Takifugu genus do have closer phylogenetic relationships than other reported species. The ratio of the average interspecific to intraspecific genetic distance is only 6.21 times. Among them, the largest interspecific genetic distance is between T. oblongus and T. stictonotus, with a value of 0.045; the smallest ones are T. bimaculatus and T. flavidus, with only 0.013; there are also multiple species with interspecific genetic distances <0.02 (as shown in Table 2). From this, it can be seen that the DNA barcode classification standard proposed by Hebert et al. (2004), with an interspecific genetic distance threshold of 0.02 and a ratio of interspecific to intra-specific genetic distances greater than 10, is not entirely applicable to the species of Takifugu genus.
4.2 Quality control of sequences downloaded from databases
NCBI's GenBank and BOLD are the two most abundant databases of DNA barcode resources. In recent years, the number of DNA barcode sequences submitted to NCBI and BOLD has increased explosively. When we use these two databases for DNA barcode sequence alignment analysis, we sometimes encounter inaccurate matching results. It has been reported that there are many errors in the sequences of the NCBI database (Liu et al., 2020; Shen et al., 2013). Some studies believe that the BOLD database has conducted a more rigorous review and screening on the submitted sequences, so the data in this database is more reliable (Macher et al., 2017; Wang et al., 2009). In fact, there are also wrong sequences in the BOLD database (Lis et al., 2016).
In this study, all sequences were run BLAST in NCBI and BOLD databases. The Blast results showed that most of the query sequences were able to match the target species with the highest identities in the database, which was consistent with the morphological identification results. But there were also some unexpected matching results of the query sequences, with multiple highly similar target species, some of which were consistent with morphological identification, while others were inconsistent. For instance, the query sequence of the T. porphyreus specimen was matched to the T. porphyreus, and in addition, two COI gene sequences of T. porphyreus (KT951818, KT951819) in the database were also matched, with identities of 100%. But apart from these two sequences, it could not match with other sequences of T. porphyreus in the database. Therefore, the information on those two sequences in the database is inaccurate. In addition, the query sequences of T. chinensis, T. pseudommus, and T. rubripes showed 99.56% identities to the two mitochondrial genome sequences (NC_024199, KJ562276) of T. flavidus in BOLD and NCBI. After checking, it was found that the information of these two sequences were also incorrect. Therefore, there are some incorrect sequences of Tetraodontiformes in the NCBI and BOLD databases.
The unreliable data in the databases will directly lead to the misidentification of species. To reduce the interference of the wrong sequence in the database on the analysis results, it is suggested that the relevant sequences in the database should be strictly screened when using DNA barcoding technology to identify species. For instance, when screening DNA barcode sequences, we can select sequences uploaded by at least three different authors, or select the sequences that have already been published and provide morphological characteristics of the species. If the information in NCBI and BOLD databases is found to be inaccurate when the voucher specimens with complete morphological characteristics are available, the database or data submitter shall be contacted in time to correct the sequence information. In this study, DNA barcoding technology and morphological methods were used synchronously to identify species of Tetraodontiformes, which could ensure the accuracy of species identification. Therefore, the DNA barcode sequences of Tetraodontiformes provided in this study can be used as reference sequences for the identification of the species in this order.
4.3 Synonym phenomenon in the order of Tetraodontiformes
Early taxonomic research was mainly based on morphological characteristics. Due to the fact that the morphological characteristics of fish are not only susceptible to subjective factors but also undergo significant change at different developmental stages and environmental conditions, coupled with delayed information exchange among taxonomists in different regions, the same species may have two or more different Latin names, which is also called synonym.
This study shows that there are a lot of synonyms in Tetraodontiformes. For example, M. leiurus is the synonym of P. leiurus, but the samples morphologically identified as P. leiurus were identified as M. leiurus in NCBI, and there is no information about P. leiurus can be found in NCBI. What's more, the samples which morphologically identified as D. nigroviridis, in NCBI and BOLD databases, were identified as T. nigroviridis. In fact, in the FishBase T. nigroviridis is the synonym of D. nigroviridis. The variety of species names in the database will affect the molecular identification results of species. We consider that the unsynchronized information in the database is also one of the reasons for the confusion of species identification of Tetraodontiformes.
In addition, in the FishBase, both Lagocephalus wheeleri (Abe, Tabeta & Kitahama, 1984) and Lagocephalus spadiceus (Richardson, 1845) are effective species, and they can be distinguished from each other in terms of morphological characteristics. For example, the former has elliptical dorsal spinule patch, while the latter has a rhomboidal patch with a posterior extension. However, some studies believe that these morphological features cannot be used to distinguish the two, and L. wheeleri may be a synonym for L. spadiceus (Matsuura, 2010; Sakai et al., 2021). In this study, the results of haplotype analysis showed that they had the same haplotype and clustered into the same clade in neighbor-joining tree. Additionally, their interspecific genetic distance was only 0.003. This shows, the molecular classification results support that L. wheeleri is synonymous with L. spadiceus.
Moreover, there are many reports believe that T. rubripes, T. chinensis and T. pseudommus may be different phenotypes of the same species, that is, T. chinensis and T. pseudommus are the synonyms of T. rubripes (Cui et al., 2005; Liu et al., 1999; Park et al., 2020; Reza et al., 2008; Song et al., 2001). However, all three species are valid in Fishbase and can be distinguished by morphological characteristics. T. rubripes has irregular round black spots and white patterns in front of the caudal fin on the sides of the body, T. chinensis has no such black marks, while T. pseudommus has white spots scattered on a black background on the dorsal and lateral sides of the body (Baek et al., 2018; Reza et al., 2008). In this study, the initial morphological identification results showed that eight samples were identified as T. chinensis, 4 as T. pseudommus, and 27 as T. rubripes. However, further research has found that samples identified by morphology as the three species had the same haplotype, and the interspecific genetic distance among them ranged from 0.001 to 0.002. In additional, in the NJ tree, they clustered into one clade. To sum up, the T. chinensis and T. pseudommus are the synonyms of T. rubripes, and T. rubripes is the valid name.
And the morphologic features of T. alboplumbeus and T. niphobles are so similar that they are difficult to distinguish. It is generally believed that T. niphobles have obvious chest spots, while the chest spots of T. alboplumbeus are not obvious, and there are several dark bands on the back of T. alboplumbeus. In this study, the DNA barcode identification results also showed that the two species may be synonyms. However, since the 11 specimens identified as T. alboplumbeus do not completely have the morphological characteristics of T. alboplumbeus, it is not certain that these specimens are hybrids of T. alboplumbeus and T. niphobles. Or that T. alboplumbeus and T. niphobles are the same species, but they will undergo a morphological transformation process. Matsuura (2017) regarded T. niphobles and T. alboplumbeus as synonyms based on the same color patterns and no differences in morphological characteristics between the type specimens. Some researchers support this view as well (Okabe et al., 2019), and these two species are also considered to be synonyms in NCBI. However, in the FishBase, it is considered that both T. alboplumbeus and T. niphobles are effective species. Zhang and He (2008) showed that 12 s and cytb genes could separate T. alboplumbeus and T. niphobles, but the study did not describe the morphological characteristics of the two species. Zhou et al. (2020) showed that the mitochondrial genome can separate T. alboplumbeus and T. niphobles effectively. However, the study did not describe the morphological characteristics of the two species as well. Therefore, whether T. alboplumbeus and T. niphobles are synonyms has not been determined.
4.4 Reconstruction of Tetraodontiformes phylogenetic relationships
Many researchers have studied the relationship between families of Tetraodontiformes based on different methods (Alfaro & Brock, 2007; Breder Jr & Clark, 1947; Holcroft, 2005; Leis, 1984; Rosen, 1984; Santini & Tyler, 2010; Tyler & Sorbini, 1996; Winterbottom, 1974; Yamanoue et al., 2007, 2008). In their study, there were more or less differences in the phylogenetic tree of Tetraodontiformes. The commonality is that they unanimously agreed that Balistidae and Monacanthidae were closely related, and Diodontidae and Tetraodontidae were closely related as well.
In this study, the Neighbor-joining tree based on the haplotype of COI gene of Tetraodontiformes shows that the species of six families are split into two branches. And Diodontidae and Tetraodontidae clustered into a branch, Monacanthidae, Balistidae, Ostraciidae, and Triacanthidae clustered into another branch, as shown in Figure 3. The result that Diodontidae and Tetraodontidae are clustered in a separate branch is consistent with the research results of others. However, unlike other studies, Balistidae and Monacanthidae did not cluster into a branch in this study. The reason for this result may be that there are not enough samples collected in this study. For example, only one sample of Triacanthidae was collected. It is also possible that the COI gene sequence is not suitable for the systematic relationship study of families and higher taxonomic levels.
In the Neighbor-joining tree based on the haplotype of COI gene of Tetraodontiformes, the species of the same genus clustered together, and the same species are clustered into a clade, which is basically consistent with the results of morphological identification. Although the interspecific genetic distance of some species of Takifugu is <0.02 (as shown in Table 2), in the Neighbor-joining tree, except for T. pseudommus, T. rubripes, and T. chinensis, each species can gather into a separate clade. It indicates that the COI gene sequence is suitable for the classification and identification of genera and lower taxonomic levels.
5 CONCLUSION
In this study, 616 samples of Tetraodontiformes were identified by using morphological characteristics and DNA barcoding technology. It revealed that DNA barcoding can be effectively used in the identification of Tetraodontiformes.
Moreover, it is suggested that DNA barcoding can provide molecular evidence for clarifying the problem of species synonyms and confirming valid species names. It provided molecular evidence for clarifying the valid species names of L. spadiceus and T. rubripes.
In addition, this study reconfirms that there are some incorrect sequences in both NCBI and BOLD databases. It is suggested that synonyms and unreliable data are also the reasons for the confusion of taxonomic identification of Tetraodontiformes. We suggest that when using DNA barcode technology to identify species, the sequence in the database needs to be strictly screened. When the sample has complete morphological characteristics, the final identification results of species should be determined by combining the morphological characteristics.
AUTHOR CONTRIBUTIONS
Kaiying Liu: Data curation (lead); formal analysis (lead); investigation (lead); methodology (lead); resources (equal); validation (lead); visualization (lead); writing – original draft (lead); writing – review and editing (equal). Hongyue Sun: Data curation (equal); investigation (equal); resources (equal); writing – original draft (equal). Xin Zhao: Investigation (equal). Chaoqun Wang: Investigation (equal); resources (equal). Changting An: Writing – review and editing (equal). Ang Li: Writing – review and editing (equal). Shufang Liu: Conceptualization (lead); data curation (equal); formal analysis (equal); funding acquisition (supporting); investigation (equal); methodology (equal); project administration (equal); resources (lead); supervision (lead); validation (equal); visualization (equal); writing – original draft (equal); writing – review and editing (equal). Zhimeng Zhuang: Conceptualization (equal); supervision (equal).
ACKNOWLEDGMENTS
This research was funded by the National Key Research and Development Program of China (2019YFC1604700), Central Public-interest Scientific Institution Basal Research Fund (NO.2021JC01 & 20603022022024). Meanwhile, we would like to express our sincere gratitude to all those who assisted us in the sampling and taxonomic research process.
CONFLICT OF INTEREST STATEMENT
We wish to draw the attention of the Editor to the following facts may be considered as potential conflicts of interest and significant financial contributions to this work. All authors agreed to this submission and the corresponding author has been authorized by co-authors. This manuscript has not been published before and is not concurrently being considered for publication elsewhere. This manuscript does not violate any copyright or other personal proprietary right of any person or entity and it contains no abusive, defamatory, obscene, or fraudulent statements, nor any other statements that are unlawful in any way.
Open Research
DATA AVAILABILITY STATEMENT
DNA sequences have been deposited in GenBank under Accession numbers OQ700230–OQ700845. Details regarding individual samples are available in Table 1.




