Stratifin (SFN) regulates lung cancer progression via nucleating the Vps34-BECN1-TRAF6 complex for autophagy induction
Lung cancer progression is regulated by various extrinsic factors derived from tumour microenvironment as well as intrinsic factors.1 Recent studies have shown that toll-like receptors (TLRs) are expressed in lung cancers, suggesting that TLRs may be implicated in lung cancer progression.2, 3 Although several studies have shown that stratifin (SFN, 14-3-3 sigma) facilitated lung cancer development and progression,4-6 the molecular and cellular mechanisms by which SFN is functionally involved in lung cancer progression, and the role of SFN in lung cancer progression in response to extrinsic stimulation, such as TLR agonist, are largely unknown. Here, we show that SFN expression is remarkably up-regulated in lung cancer tissues through The Cancer Genome Atlas (TCGA) data and primary non-small cell lung cancers (n = 31 of our cohort patients) analysis, and SFN positively regulates lung cancer progression through the autophagy induction by facilitating TRAF6- Vps34-BECN1 complex in response to an extrinsic TLR4 agonist.
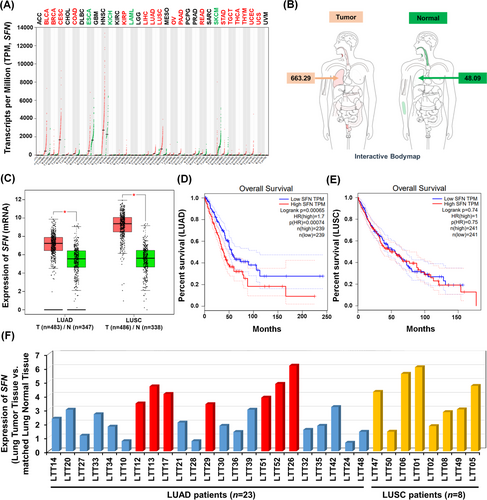
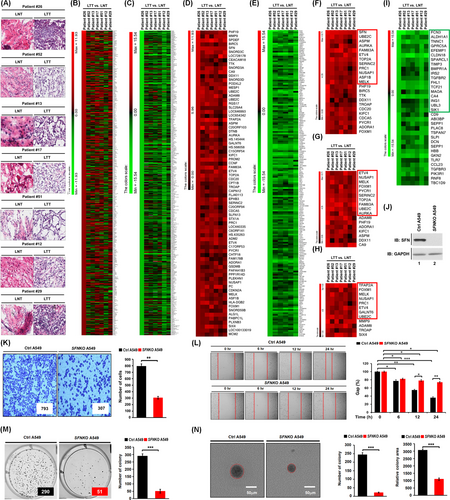
To get insight into the role of SFN in cancers, we first investigated whether the expression of SFN is associated with 33 different cancer types by analyzing TCGA datasets (http://gepia.cancer-pku.cn/detail.php?gene=SFN, Figure 1A). The gene expression of SFN was significantly up-regulated in 17 different tumour samples (Figure 1A, tumours are marked in red; Figure S1), whereas significantly down-regulation was observed in four different tumour samples (Figure 1A, tumours are marked in green; Figure S2). In lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) tumour samples, SFN expression was significantly increased compared to those of paired normal tissues (Figure 1B,C). Interestingly, the percentage of survival was significantly lower in LUAD patients with high expression of SFN (Figure 1D, red vs. blue; p = .00065), whereas there was no significant difference was observed in LUSC patients (Figure 1E, red vs. blue; p = .74). Notably, microarray data analysis showed that the expression of SFN was increased in primary 23 LUAD patients and eight LUSC patients (Figure 1F), indicating that SFN may be functionally associated with LUAD. Given the above results, we examined whether the expression of SFN is associated with gene expressions related to cancer progression in primary LUAD patient tissues. We selected seven primary LUAD patients with high expression of SFN (Figure 1F, red bars), and the pathological classification was confirmed by H&E staining in matched lung normal tissue and lung tumour tissue, patient #26,7 #52, #13, #17, #51, #12 and #29 (Figure 2A). To sort out genes related to cancer progression, 500 up-regulated genes or 500 down-regulated genes were arranged based on the data of patient #26 with the most up-regulation of SFN among the seven primary LUAD patients (Figure 2B, 500 up-regulated genes; Table S1: Figure 2C, 500 down-regulated genes; Table S2). Among the 500 down-regulated genes, commonly up-regulated genes (Figure 2D and Table S3) or down-regulated genes (Figure 2E and Table S4) were further sorted out. Interestingly, 23 genes related to cancer proliferation (Figure 2F and Table S5), 17 genes related to cancer migration or invasion (Figure 2G and Table S6) and 12 genes related to cancer metastasis (Figure 2H and Table S7) were up-regulated in the seven primary LUAD patient tumour tissues. Notably, 12, 10 and 8 genes were functionally associated with lung cancer proliferation, migration or invasion and metastasis, respectively (Figure 2F–H, red boxes; Table S5-S7, red letters). Although the effect of SFN on cell cycle progression is controversial, a previous report has demonstrated that SFN in lung adenocarcinoma cells might have tissue-specific functions and regulate cell cycle progression in a positive manner.4 Importantly, we found that 12 genes related to lung cancer proliferation have been reported to be involved in promoting cell cycle progression (Figure S3), indicating that SFN expression is associated with genes related to cell cycle progression in cancers. Moreover, 34 cancer suppressors genes were down-regulated in tumour tissues of the seven primary LUAD patients (Figure 2I, Table S8), and 18 genes were functionally associated with lung cancer (Figure 2I, green box; Table S8, green letters). To verify the function of SFN in lung cancer progression, we generated SFN-knockout (SFNKO) A549 human adenocarcinoma cells using the CRISPR cas9 gene editing method (Figure 2J, lane 2). The SFNKO A549 cells revealed marked inhibitions of cancer invasion and migration and colony formation and single-cell mobility and proliferation as compared to those of control (Ctrl) A549 cells (Figure 2K, invasion; Figure 2L, migration; Figure 2M, anchorage-dependent colony formation; Figure 2N, anchorage-independent colony formation; Figure S4A,B, single-cell mobility; Figure S5, cell proliferation: SFNKO A549 vs. Ctrl A549). Based on the TCGA and primary LUAD patient data analysis and the cancer progression assay of SFNKO lung cancer cells, we suggested that SFN might be functionally implicated in lung cancer progression.
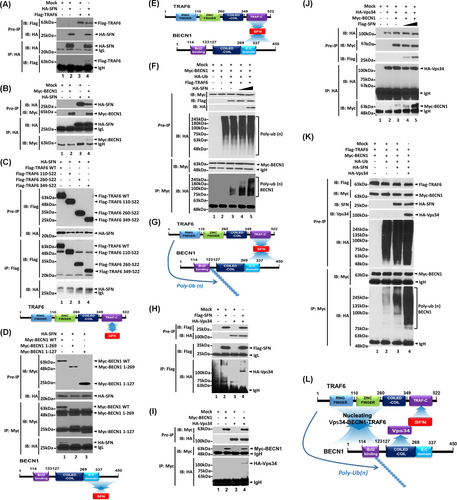
Further, we explored the molecular mechanism through which SFN is implicated in lung cancer progression. It has been reported that SFN accelerates lung cancer progression by regulating cell proliferation,4 and TLRs expressed in lung cancer promote the growth of cancer cells by inducing cell proliferation.3 Upon extrinsic TLR3/4 stimulations in lung cancer, additionally, autophagy was induced through the TRAF6-BECN1 signaling axis, leading to enhanced cancer migration and invasion.8 In this study, we hypothesized that SFN regulates lung cancer progression through the autophagy induced by extrinsic TLR stimulation. SFN interacted with TRAF6 (Figure 3A, lane 4) or BECN1 (Figure 3B, lane 4). SFN interacted with TRAF6 wild type (WT), and TRAF6 110-522 and TRAF6 260-522 and TRAF6 349-522 truncated mutants (Figure 3C). To verify the interaction, we performed an additional IP experiment with Flag-TRAF6 1-349 and Flag-TRAF6 349-522 truncated mutants. SFN interacted with Flag-TRAF6 349-522 but not with TRAF6 1-349 (Figure S6), indicating that SFN interacts with the tumor necrosis factor receptor-associated factor-C terminus (TRAF-C) domain of TRAF6 (Figure 3C, down). Additionally, SFN interacted with BECN1 WT, but not with BECN1 1-269 and BECN1 1-127 truncated mutants, indicating that SFN interacts with the C-terminal domain of BECN1 (Figure 3D). These results suggest that SFN can nucleate the association of TRAF6-BECN1 (Figure 3E). Importantly, the ubiquitination of BECN1 was markedly enhanced in the presence of SFN as compared to the absence of SFN (Figure 3F, lane 4 and 5 vs. lane 3), indicating that SFN nucleates the molecular association of TRAF6-BECN1 and enhances the ubiquitination of BECN1 (Figure 3G). Since Vps34 interacted with the coiled-coil domain of BECN1 and regulated autophagy,9 we further assessed whether SFN affects the association of Vps34-BECN1 complex. Our results showed that Vps34 interacted with SFN (Figure 3H, lane 4) or BECN1 (Figure 3I, lane 4). Furthermore, the interaction between Vps34 and BECN1 was markedly enhanced in the presence of SFN as compared to the absence of SFN (Figure 3J, lane 4 or 5 vs. lane 3). Consistently, the endogenous interaction between BECN1 and Vps34 was significantly decreased in SFNKO A549 cells in the presence or absence of lipopolysaccharide (LPS) as compared to those of Ctrl A549 cells (Figure S7, lane 3 and 4 in SFNKO A549 vs. lane 1 and 2 in Ctrl A549). Interestingly, the ubiquitination of BECN1 was significantly enhanced in the presence of SFN and Vps34 compared to their absence (Figure 3K, lane 4 vs. lane 3). These results suggest that SFN facilitates the molecular associations of the TRAF6-BECN1-Vps34 complex through the interaction with BECN1 (Figure 3D) or Vps34 (Figure 3H), leading to the enhancement of the BECN1 ubiquitination (Figure 3L).
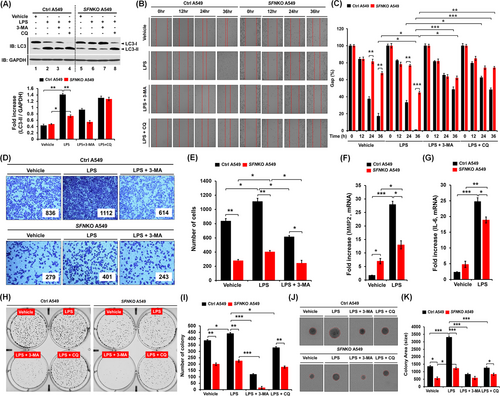
Autophagy regulated by the ubiquitination of BECN1 enhanced lung cancer migration and invasion in response to TLR3/4 stimulation.8 Upon TLR4 stimulation with LPS, the level of light chain 3-II (LC3-II) was significantly attenuated in SFNKO A549 cells as compared to Ctrl A549 (Figure 4A, lane 6 vs. lane 2). Moreover, cancer migration and invasion were markedly inhibited in SFNKO A549 cells treated with LPS, as compared to those of Ctrl A549 cells (Figure 4B,C, migration; Figure 4D,E, invasion: SFNKO A549 treated with LPS vs. Ctrl A549 treated with LPS). Consistently, single-cell mobility was significantly attenuated in SFNKO A549 cells treated with LPS (Figure S8A,B, SFNKO A549 vs. Ctrl A549). Matrix metalloproteinase-2 (MMP2) and interleukin-6 (IL-6), which are known to regulate cancer migration and invasion,8 were significantly down-regulated in SFNKO A549 cells treated with LPS as compared to Ctrl A549 cells treated with LPS (Figure 4F, MMP2; Figure 4G, IL-6). Furthermore, anchorage-dependent or -independent colony formation revealed a significant decrease in SFNKO A549 cells-treated LPS as compared to Ctrl A549 treated LPS (Figure 4H,I, anchorage-dependent; Figure 4J,K, anchorage-independent: SFNKO A549 cells treated LPS vs. Ctrl A549 cells treated LPS). Consistently, cell proliferation assay showed a significant decrease in SFNKO A549 cells in the presence of LPS (Figure S9, SFNKO A549 vs. Ctrl A549). In contrast, the co-treatment of autophagy inhibitors, 3-methyladenine (3-MA) and chloroquine (CQ), markedly attenuated cancer migration and invasion, and colony formation induced by LPS stimulation (Figure 4B–E,H–K, LPS vs. LPS + 3-MA or LPS + CQ). To verify these results, we performed rescue experiments with SFNKO A549 and SFNKO A549 transiently expressed with SFN (Figure S10). In SFNKO A549 cells transfected with hemagglutinin (HA)-SFN (Figure S10A, lane 2), cancer migration and invasion were significantly elevated as compared to those of SFNKO A549 cells transfected with mock vector (Figure S10B–C, HA-SFN-expressed SFNKO A549 vs. SFNKO A549). These results suggest that SFN positively regulates cancer migration and invasion, and colony formation through the autophagy induction by TLR4.
In summary, we demonstrated that SFN expression in lung cancer is clinically associated with poor patient survival and lung cancer progression, accompanying the up-regulation of genes related to cancer progression and the down-regulation of genes related to cancer suppression. Through the biochemical and cellular studies, we propose possible molecular and cellular mechanisms in which SFN is functionally implicated in lung cancer progression: (1) SFN nucleates the TRAF6-BECN1-Vps34 complex and enhances the ubiquitination of BECN1 and subsequently regulates autophagy, (2) upon extrinsic TLR4 stimulation, SFN enhances cancer progression including cancer migration and invasion, proliferation and colony formation through autophagy induction. Together, our clinically comparative results and functional investigations of SFN expression in lung cancer will potentially contribute to translational approaches for the development of lung cancer therapeutic agents.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
FUNDING INFORMATION
National Research Foundation of Korea (NRF) Grants, Korean Government, Grant Numbers: NRF-2021R1F1A1049324 and NRF-2021R1A2C1094478; Korea Basic Science Institute (National Research Facilities and Equipment Center) Grant, Ministry of Education, Grant Number: 2020R1A6C101A191; Ministry of Science ICT and Future Planning (MSIP), Korean Government, Grant Number: NRF-2016R1A5A2945889




