Downregulation of Rab23 inhibits hepatocellular carcinoma by repressing SHH signaling pathway
Abstract
Hepatocellular carcinoma (HCC) is the sixth most common malignant tumors and the third leading cause of cancer-related death worldwide. As an oncogene, Rab23 has been shown to be significantly related to the growth and migration of hepatocellular carcinoma in both in vitro and in vivo studies, but its underlying mechanism remains obscure. In the present study, we examined the effect of inhibiting Rab23 expression on the pathological progression of HCC. The correlation between liver Rab23 gene expression and survival probability in human HCC patients was analyzed using the TCGA database and CPTAC database. Rab23 knockdown hepatocellular carcinoma cell line was generated through lentiviral transduction, then we established a nude HCC xenograft model by subcutaneously implanting the transfected cells. The analysis of gene and protein expression was carried out using Western blot or RT-qPCR, respectively. Flow cytometry analysis was used to detect the level of apoptosis. The expression levels of key proteins involved in the Sonic Hedgehog (SHH) signaling pathway were assessed. The results showed that HCC patients with low levels of hepatic Rab23 mRNA and protein had a better survival tendency than those with higher levels of Rab23. Cell proliferations were reduced and apoptosis levels were increased after Knocking down Rab23 in HCC cell lines. Furthermore, in vivo studies have demonstrated that suppression of the Rab23 gene results in decreased tumor size, proliferation rate, and reduced levels of SHH-related proteins Smoothened and GLI-1. The above results suggest that Rab23 is involved in the pathological progression of HCC as an important regulator of the SHH signaling pathway, which also provides an important research basis for new therapeutic strategies for HCC.
1 BACKGROUND
The incidence of hepatocellular carcinoma (HCC) has increased worldwide, not only in East Asia but also in western Europe and the United States. HCC currently ranked sixth in incidence rate and third in mortality rate among all malignant tumors.1 It is estimated by The World Health Organization that over 1 million patients will die from HCC by 2030.2 Unfortunately, the current diagnosis rate of HCC is very low, and effective clinical treatments are lacking.3 HCC is a complex process involving multiple risk factors, including hepatitis B virus (HBV), Hepatitis C virus (HCV), cirrhosis, and alcohol use, which further promote abnormal activation of specific molecular signaling pathways, leading to disruption of the balance of oncogenes and tumor suppressor genes in cells, respectively.4 Currently, various oncogenes such as TP53, CTNNB1, AXIN1, and ARID1A have been identified to promote the initiation and progression of HCC by inducing abnormalities in cell proliferation, apoptosis, and signal transduction.5, 6 In addition, providing a clear understanding of the relationship between these oncogenic genes and HCC, as well as comprehending their essential mechanisms in tumor progression, will contribute to the development of targeted treatments and preventive measures for HCC.
Rab23 is a conserved member of the Ras-related small GTPase family, which is crucial for membrane trafficking and protein transport.7-9 In cancer, Rab23 has been found to be highly expressed in carcinoma cells and is closely related to tumor cell proliferation, migration and apoptosis as an oncogene with tumor-promoting activity, which makes it a useful cancer biomarker and potential therapeutic target.9-14 Furthermore, studies have demonstrated that Rab23 represses hedgehog (Hh) activities Via GLI-2 and promotes the proteolytic cleavage of GLI-3 into its cleaved repressor form. In addition, Rab23 also appeared to regulate Hh pathway activity through Smoothened.15 Both Rab23 gene and SHH pathway have also been implicated in tumor invasion and migration of HCC through their regulation of cell growth.16, 17 However, the potential mechanisms of Rab23 in HCC are not yet clear.
In previous studies, we have demonstrated that Rab23 exists in both GDP- and GTP-bound forms. Disrupting this binding or reducing the level of Rab23 has a significant impact on the expression and nuclear localization of Gli1 in human liver cancer cell line Hep3B and HepG2, ultimately leading to suppression of cell growth. Therefore, we conducted this study to investigate the role and in vivo molecular mechanism of Rab23 silencing in HCC.
2 MATERIALS AND METHODS
2.1 Datasets collecting and preprocessing
The TCGA_LIHC cohort survival analysis for this study used Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/) online analysis, with the data from The Cancer Genome Atlas (TCGA, https://portal.gdc.Cancer.gov/). At the same time, the protein data obtained in this study is downloaded from the The Clinical Proteomic Tumor Analysis Consortium (CPTAC, https://proteomics.cancer.gov/programs/) database.
2.2 Survival analysis
First, HCC patients were divided into Rab23 high group and Rab23 low group according to the median Rab23 as a cut-off value. Load the R package “survival”, then create the Surv object, and use Kaplan–Meier to build an overall survival (OS) analysis model to plot the survival distribution curve between the Rab23 high group and the Rab23 low group.
2.3 Cell culture
Both the liver cancer cell lines Hep3B and HCCLM3 (iCell Bioscience Inc, Shanghai, China) were used in this study. Hep3B cells were grown in high-glucose DMEM (Corning, China) medium, while HCCLM3 cells were cultured in RPMI-1640 (Corning, China) medium. Both types of cells were cultured in a medium supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin at a temperature of 37°C and 5% CO2.
2.4 Lentivirus transfection
Three specific shRNAs (Table 1) were designed to target the Rab23 gene, and the lentivirus was purchased from BioSCI Res Company (China). Before transfection, approximately 4 × 105 Hep3B and HCCLM3 cells were plated in a 6-well plated. The lentivirus particles were added to the cells at a multiplicity of infection (MOI) of 10 overnight. Afterward, the medium was replaced with 2 mL of fresh complete medium and incubated for an additional 48 or 72 h. Stable cell lines were obtained after 48 or 72 h of screening using 2 μg/mL puromycin (BeyoTime, China). Infection efficiency was calculated by observing GFP fluorescence under a fluorescence microscope. Quantitative real-time PCR or Western blotting was used to measure Rab23 expression in each stable cells line.
| shRNA | Sequences |
|---|---|
| shRab23-1 | AGGGAATGGAGCAGTTGGAAA |
| shRab23-2 | ACAAACAAAGGACCAAGAA |
| shRab23-3 | CAGAACTAACGCATTCAAGTA |
2.5 Cell proliferation assay
The Cell Counting Kit-8 (CCK-8, Gongsheng, China) was used for cell viability analysis following the manufacturer's instructions. In detail, a total of 10 × 103 cells/well of stable cells were plated onto a 96-well plate for 8 h and then treated with 10 μL of Cell Counting Kit-8 (CCK-8) solution (5 mg/mL) for 4 h. After that, the absorbance is measured at 450 nm in a microplate reader (FLx800, BioTek, Winooski, VT, USA).
2.6 Flow cytometric analysis
The apoptosis level was evaluated using an Annexin V-APC Cell Apoptosis detection kit (BeyoTime, China) on the Guava EasyCyte flow cytometer (Millipore Merck, USA). The cells were dissociated with 0.25% trypsin and washed twice with PBS after transfection. The cell pellets were resuspended in binding buffer and then mixed successively with 200 μL of 1× apoptosis buffer. Annexin V-APC (5 μL) and PI (5 μL) were added, and the cells were stained for 15 min. Then, the cells were resuspended in 800 μL of 1× apoptosis buffer and the final volume was adjusted to 1 mL. Next, 200 μL of the cell suspension was transferred to a 96-well plate (with three duplicate wells in each group) and subsequently analyzed using a flow cytometer.
2.7 Western blot assay
Total protein was isolated from cultured cells using lysis buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 10% Glycerol, 1% Triton X-100, 1.5 mM MgCl2, 1 mM EGTA, 10 mM Sodium Pyrophosphate, 100 mM Sodium Fluoride) with a mixture of protease inhibitors including 1 mM PMSF and 10 μg/mL complete protease inhibitor mixture (Roche). The protein concentration was determined using a bicinchoninic acid protein assay kit (TaKaRa, Japan). The protein samples were run on 10% SDS-PAGE, transferred to PVDF membranes according to standard procedures.
The membranes were blocked with 5% nonfat milk for 1 h at RT and then incubated overnight at 4°C with primary antibodies, including Rab23 (11101-1-AP, Proteintech, USA), GLI-1 (SC-515751, Santa Cruz, USA), alpha Smoothened muscle actin (Ab32575, Abcam, USA) and GAPDH antibody (60004-1-lg, Proteintech, USA). Subsequently, the corresponding secondary antibodies were added to the membranes after washed by Tris-buffered saline/Tween-20 (TBST). Bands were visualized using the ChemiDocTM MP Imaging System (Bio-Rad, USA). Densitometric analysis was performed by Image J software.
2.8 Quantitative real-time PCR
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, USA), and cDNA was synthesized by Hiscript III All-in-one RT SuperMix Perfect for qPCR (Vazyme, China) according to the manufacturers' instruction. The specific primers used are listed as follows: Rab23 F1 (5′-GCTTGTGTGCTCGTGTTCTCTA-3′) and R1 (5′-CCACTTCGGCTACTACTTTCTCTC-3′); GAPDH F1 (5′-CCTGGAGAAACCTGCCAAGTA-3′) and R1 (5′-TCATACCAGGAAATGAGCTTGAC-3′). Quantitative real-time PCR (qRT-PCR) was preformed to quantify the RNA of interest using the 2−ΔΔCT method and SYBR Green PCR reagents (Vazyme, China).
2.9 In vitro studies
2.9.1 Human HCC xenograft model
Eight-week-old BALB/c nude mice (GemPharmatech, China) were used for in vivo experiments and divided into two groups (shCtrl group and shRab23 group). The mouse had free access to standard feed and water in a temperature-controlled facility, which maintained a 12-hour light and 12-hour dark cycle. Tumorigenesis was induced by subcutaneous injection of mice with 4 × 106 transfected HCCLM3 cells. The size of the tumor is regularly measured using a caliper at the same time point each week. Optical imaging was captured via the IVIS spectral imaging system (PerkinElmer, USA), and image analysis was conducted using the Living Image version 3.2 software. The nude mice were euthanized by injection of 3.5% aqueous solution of carbachol (0.1 mL/g) when the tumors reached an appropriate size. The tumor size is measured to further calculate the tumor volume using the formula length × width × 0.5. Each tumor tissue will be collected and weighed. All animal experiments were carried out in accordance with the U.K. Animals (Scientific Procedures) Act, 1986 and associated guidelines, EU Directive 2010/63/EU for animal experiments, or the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978).
2.10 Histological staining
Collected tumor tissues were paraffin embedded after fixed with 10% formalin fixation, and cut into appropriate thin slices. Hematoxylin and eosin (H&E) and Ki67 staining were performed according to a standardized procedure. Immunohistochemical analysis was used to detect the expression of Rab23 in tumor tissues using Rab23 antibody (Proteintech, USA).
2.11 Statistical analysis
Descriptive data were presented as means ± standard deviations (SD)s. The professional statistical analyses were performed using GraphPad Prism software version 5.01. Student t tests, one-way or two-way analysis of variance (ANOVA) were used to analyze the differences between the groups, respectively. A p value of <0.05 is considered statistically significant.
3 RESULTS
3.1 Correlation of hepatic Rab23 gene expression with survival probability in human liver cancer patients
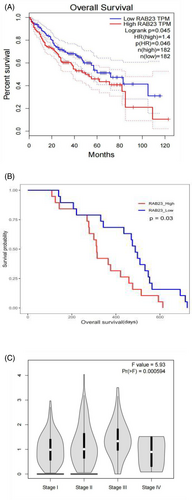
As the role of Rab23 in HCC is not well-understood, we conducted an analysis to examine the correlation between hepatic Rab23 gene expression and survival probability in human HCC patients using the TCGA dataset and CPTAC dataset. Our analysis revealed that patients with lower levels of hepatic Rab23 mRNA and protein had better survival rates compared to those with higher levels of Rab23 (p < 0.05) (Figure 1A,B). Moreover, the expression level of Rab23 was also shown to be significantly and positively correlated with the stage of HCC (Figure 1C).
3.2 Expression of Rab23 in liver cancer cell lines
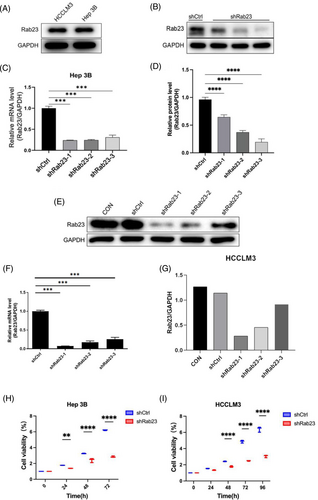
We first examined the expression of Rab23 protein in wild and knockdown cell lines by Western blot analysis, respectively. The results showed that both Hep3B and HCCLM3 cells had similar levels of endogenous Rab23 protein expression (Figure 2A). We generated Rab23 knockdown cells in Hep3B cell lines using three different shRNA sequences targeting Rab23 (shRab23-1, shRab23-2 and shRab23-3). The gene expression of Rab23 significantly decreased in these knockdown cell lines compared to shCtrl (p < 0.001), and the knockdown efficiency is a maximally of 70% (Figure 2C). Moreover, all three shRNAs reduced the protein levels of Rab23 in cells with different knockdown efficiencies (22% for shRab23-1, 61% for shRab23-2, and 80% for shRab23-3) (Figure 2B,D). We established a Rab23 knockdown cell line in HCCLM3 cells using the aforementioned shRNA sequences. Compared to the control group, both the gene level (Figure 2F) and protein level (Figure 2E,G) of Rab23 were significantly decreased. Among them, the knockdown efficiency of shRab23-1 was the highest, reaching nearly 90%.
3.3 Rab23 knockdown decreases cell proliferation and induces cell apoptosis in liver cancer cells
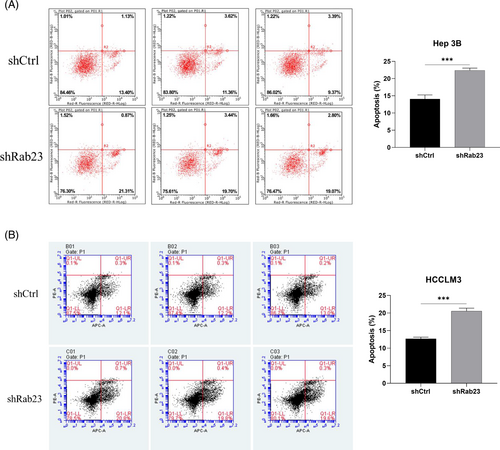
We next examined cell proliferation in these Rab23 knockdown cell lines. The cell proliferation levels of Hep3B cells at 72 h and HCCLM3 cells at 96 h were significantly lower than those of the shCtrl group (p < 0.001) (Figure 2H,I). Furthermore, we investigated the impact of Rab23 knockdown on apoptosis levels in hepatocellular carcinoma cell lines using flow cytometry. The results showed that the apoptosis rate within both Hep3B and HCCLM3 cells was significantly increased in the shRab23 knockdown group compared to the shCtrl group (p < 0.001) (Figure 3A,B).
3.4 Rab23 knockdown suppresses GLI-1 expression
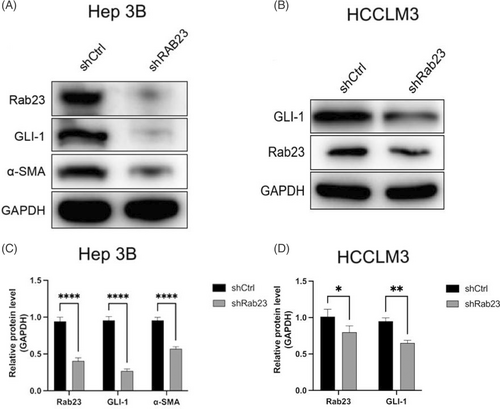
To evaluate the effect of Rab23 knockdown on SHH signaling pathway, the expressions of GLI-1 and cancer-associated fibroblast markers α-SMA in the shRab23 group were further detected. We found that when compared to WT control, both Rab23, GLI-1 and α-SMA protein levels were all significantly downregulated in shRab23 cells l (p < 0.001 in Hep3B cells; p < 0.05 (GLI-1) or p < 0.01 (Rab23) in HCCLM3 cells) (Figure 4A–D).
3.5 Proliferation inhibition of BALB/c xenograft tumor with shRab23 in vivo
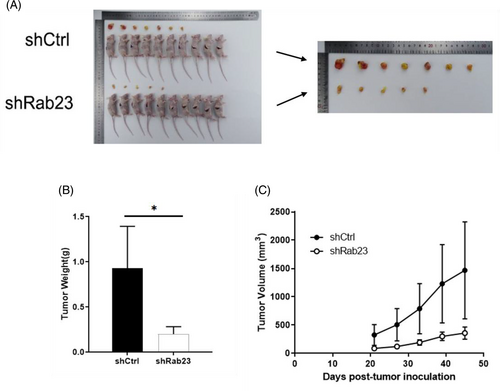
To further validate the suppressive influence of Rab23 on tumorigenesis in vivo, we implanted Rab23 knockout HCCLM3 cells into nude mice and examined the enlargement of tumor cells. Compared with the shCrtl group, we found that tumors from shRab23 knockdown cell lines grew more slowly characterized by lower tumor tissue weight (p < 0.05) and smaller volume (Figure 5A–C). H&E staining also indicated that tumor cells in the Rab23 knockout group had significantly reduced irregularly shaped tumor cell nucleosomes, incomplete cytoplasmic membranes, and heterogeneous chromatin structures compared to the control group (Figure 6D). However, according to photon flux detection, there were no significant differences of the metastasis of HCC between two groups (Figure 6A–C).
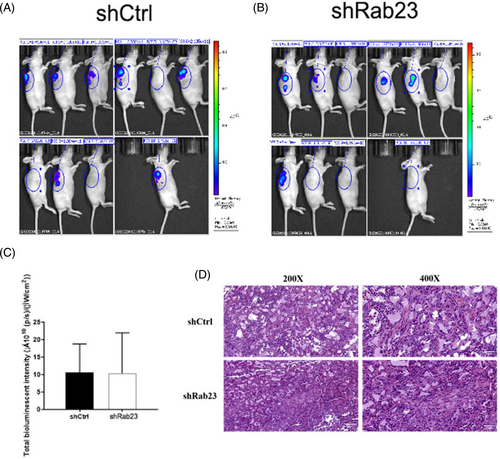
3.6 Silencing of Rab23 suppress the progression of HCC in vivo via inhibiting the SHH signaling pathway
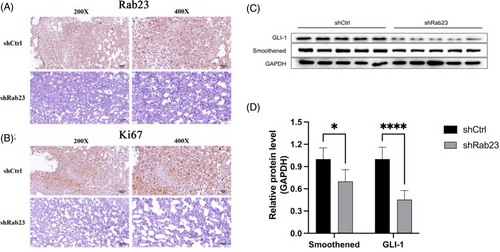
Immunohistochemical (IHC) staining showed reduced Rab23 expression in knockout liver cancer cells (Figure 7A), along with decreased levels of KI-67 (Figure 7B), a proliferation marker for human tumors that has been strongly linked to advanced HCC.18 Furthermore, the expression levels of the major SHH pathway proteins Smoothened (SMO) and GLI-1 were also significantly reduced in shRab23 liver cancer cells compared to the control (Figure 7C,D). Therefore, these findings indicated that Silencing of Rab23 may suppress the progression of HCC via inhibiting the SHH signaling pathway in vivo.
4 DISCUSSION
HCC is a prevalent malignant tumor worldwide and is the primary cause of cancer-related deaths. In Asia such as China, 5-year survival rates are reported to be as low as 12%.19 Conventional treatments, including resection and anti-tumor medication, are not yet optimal due to the high diagnosis rate at advanced stages, leading to insignificant improvements in survival rates.
Rab23 is a small GTPase belonging to the Rab superfamily, whose gene is located on human chromosome 6p12.1.8 Over the past few years, it has been confirmed by several studies that Rab23 is an oncogene that plays a critical role in promoting cancer development.10 Furthermore, it has been found to be involved in promoting invasion of various types of cancer including breast cancer, gastric cancer, lung cancer, and so forth.20-22 Sicklick's study revealed Rab23 had elevated levels in human liver cancer tissues or liver cancer cells and was closely associated with tumor size.23 Zhang et al.11 found that overexpression of Rab23 promoted the migration of Hep3B cells, and this process could be reversed by silencing Rab23. Currently, only a limited number of papers have reported on the role of Rab23 in HCC, and the specific molecular mechanisms by which Rab23 promotes the progression of liver cancer are far from being elucidated.
In previous study, we found that exhibited a positive expression rate of 53.5% in patients with HCC and was significantly associated with tumor size.14 The downregulation of Rab23 mRNA in Hep3B cells has also been shown to promote apoptosis and inhibit cell growth.14 Furthermore, we also demonstrated that the nuclear localization of Rab23 depends on its GDP/GTP binding state, and that the biological function of Rab23 is to control GTP hydrolysis as a molecular switch.16 In addition, Rab23 exhibited inhibitory effects on the transcriptional activity of Gli1 in HepG2 cells, although it was not directly regulated by the SHH signaling pathway.16 However, the above studies were all performed in vitro, and there is still a lack of evidence to support the function of Rab23 in vivo. Therefore, we conducted this present study to gain a deeper understanding of the molecular mechanisms underlying Rab23 by utilizing Xenograft mouse models.
In our research, we generated a nude mouse xenograft model of HCC by subcutaneously implanting a HCCLM3 cell line with Rab23 knockdown. The findings demonstrated that Rab23 knockdown suppressed the growth of cancer cells and increased apoptosis in vivo. Importantly, we found that the deficiency of Rab23 resulted in a significant decrease in protein levels of both SMO and GLI-1. This suggests that Rab23 plays an essential role in liver cancer development and the SHH signaling pathway, which is consistent with our previous studies.
The SHH pathway is a well-known signal transduction pathway comprising of main components such as hedgehog (Hh), the patched receptor(Ptc), a 12-domain transmembrane receptor, the Smoothened receptor(SMO), a 7-domain transmembrane receptor coupled to G protein-coupled receptor(GPCR), zinc finger transcription factor cubitusinterruptus (Ci), and other components that regulate embryonic development.24 Numerous studies have indicated a close correlation between the aberrant activation of the SHH signaling pathway and the occurrence of various types of tumors such as cancers of skin, brain, liver, gallbladder, pancreas, stomach, colon, breast, lung, prostate, and hematological malignancies.20-22 Sicklick et al. found the SHH signaling pathway plays an important role in the pathogenesis of HCC because of high expression levels of Ptc, SMO, and GLI-1 in liver cancer.23 SMO is a major component involved in SHH signaling transduction, which can be coupled with G protein-coupled receptors (GPCRs) to inhibit the activity of suppressor of fused protein (SUFU) and GLI family of transcription factors. When the SHH signaling pathway is activated, PTCH-1, which destabilizes SMO and inhibits its activity, is degraded, leading to the activation of G-protein coupled receptor (GPCR)/SMO activity.25 This promotes the dissociation of GLI from its inhibitor protein SUFU, resulting in the release and transport of GLI into the nucleus, triggering downstream target gene expression and activating cellular functions.26 However, our results revealed a significant decrease in SMO and GLI-1 protein levels in tumor tissues, suggesting that Rab23 plays a negative regulatory role in the SHH signaling pathway and can suppress tumor growth in HCC.
Overall, our study indicate that reducing Rab23 gene expression inhibit tumor cell growth and promoted apoptosis in vivo by downregulating the SHH signaling pathway. These experimental results present new evidence for the mechanisms of Rab23 on HCC, which will help to improve the development of therapeutic strategies for patients with hepatocellular carcinoma.
AUTHOR CONTRIBUTIONS
Yun-Jian Liu conceived and designed the present study. Si-Jia Liu analyzed and interpreted data. Yu-Wei Zang and Cui-Jun Huang performed experiments, wrote the manuscript. All authors approved the final version to be submitted.
ACKNOWLEDGMENT
None.
FUNDING INFORMATION
This study was supported by the Key Science Foundation of Jiangxi Province Department of Education, China (Grant No. GJJ161064) and Natural Science Foundation of Jiangxi Province (No. 20202BABL206092).
CONFLICT OF INTEREST STATEMENT
The authors have stated explicitly that there are no conflicts of interest in connection with this article.
ETHICS STATEMENT
All procedures performed in studies involving animal experiments were accordance with the ethical standards of the Research Ethics Committee of the affiliated hospital of Jiujiang University (jjuhmer-b-20220503) and with the 1964 Helsinki declaration and its later amendments.




