Object classification in analytical chemistry via data-driven discovery of partial differential equations
Funding information: National Institutes of Health, 1R01GM112490-04, 1R01GM130091-01, and 1U01CA225753; National Science Foundation, NSF 1903450
Abstract
Glycans are one of the most widely investigated biomolecules, due to their roles in numerous vital biological processes. However, few system-independent, LC-MS/MS (liquid chromatography tandem mass spectrometry) based studies have been developed with this particular goal. Standard approaches generally rely on normalized retention times as well as m/z-mass to charge ratios of ion values. Due to these limitations, there is need for quantitative characterization methods which can be used independently of m/z values, thus utilizing only normalized retention times. As such, the primary goal of this article is to construct an LC-MS/MS based classification of the glycans derived from standard glycoproteins and human blood serum using a glucose unit index as the reference frame in the space of compound parameters. For the reference frame, we develop a closed-form analytic formula via the Green's function of a relevant convection-diffusion-absorption equation used to model composite material transport. The aforementioned equation is derived from an Einstein–Brownian motion paradigm, which provides a physical interpretation of the time-dependence at the point of observation for molecular transport in the experiment. The necessary coefficients are determined via a data-driven learning procedure. The methodology is presented in an abstractly and validated via comparison with experimental mass spectrometer data.
1 INTRODUCTION
The biological significance of glycans is evident from the numerous studies demonstrating their roles in living systems. These molecules alone, as well as in conjunction with other biomolecules, participate in important biological functions. For example, glycosylation is one of the major post-translational modifications1, 2 and is known to mediate a broad range of biological processes such as cell recognition,3 cell signaling,3, 4 immune response,5 and protein stability.6 Furthermore, aberrations in glycosylation patterns are found to be related to various diseases, including cancers.3, 7-11
Glycans display high structural complexity owing to the presence of diverse monosaccharide composition, different linkages, and various branching options.12 Tandem mass spectrometry (MS) has emerged as an effective technique for glycan structural studies.1 This technique in conjunction with liquid chromatography (LC) provides a powerful tool for studying molecular structures.1, 13-16 Additionally, various derivatization techniques are employed to pursue sensitive and efficient investigation of the glycans. Some of the derivatization reagents include 2-aminobenzamide, procainamide, aminoxyTMT, RapiFluor-MS (RFMS) labeling, and iodomethane permethylation.17 Our particular method of choice for the current study is permethylation as it delivers several advantages over other derivatization techniques.13-16 In this technique, methyl groups replace the existing hydrogens, oxygen, and nitrogen atoms in a glycan structure. Permethylated glycans have increased hydrophobicity, which makes them ideal for reverse phase separation. It also prevents fucose (sugar) migration18, 19 and sialic acid loss.20 Also, due to increased positive ion efficiency of glycans, ionization efficiency is improved, thereby enhancing the sensitivity.21
Despite the availability of sensitive structural investigation techniques, inter-instrument, as well as inter-laboratory variations, in the data acquisition complicates the identification and characterization of glycans. This motivates the need for the development of universally applicable instrument, as well as laboratory, independent classification techniques. The glucose unit index (GUI) is one such method, as it relies only on the relative retention time of sample molecules with respect to the glucose units. Ashwood et al. recently reported the retention time normalization of native glycans based on GUI as well as m/z values.22
In this study, we develop a method which only utilizes GUI for characterizing permethylated glycans, independent of m/z values of the permethylated glycan structures. Dextrin, which is utilized as a reference frame in this study, consists of a mixture of oligosaccharides of D-glucose units which form linear chains consisting of either -(1 → 4) or -(1 → 6) glycosidic bonds. The retention times of these glucose units are used to calculate the normalized retention times of the reduced and permethylated N-glycans derived from samples. The use of Dextrin as an internal standard allows for the elimination of inter-injection variations and improves the accuracy of the measurements. This approach then allows for the development of a mathematical model employing the LC-MS/MS-based data for efficient identification of permethylated N-glycans.
In particular, we employ a so-called data-driven methodology for constructing an associated partial differential equation (PDE) which allows for a straightforward classification procedure. The use of data-driven PDEs and other data-driven methodologies have recently garnered much attention in the literature due to their ability to efficiently learn in relation to dynamical systems and physical processes (see, for instance, References 23-37 and the references therein). The coupling of such approaches with deep learning methods has allowed for the efficient and accurate handling of situations involving quite large data sets. Herein, we consider a modified approach which avoids standard regression methods in favor of a more mathematically informed process for determining the coefficients necessary for object classification. By modifying an approach employed by Einstein (e.g., Reference 38) we are able to deduce more clearly the form of the undetermined PDE—making our approach closer to a supervised learning method (with some distinct differences). Moreover, we demonstrate that performing a single learning procedure on a particular data set will allow for highly accurate classification of unknown data sets of a particular type. This, of course, motivates a wide array of novel questions related to learning procedures in both mathematics and the physical sciences.
This article is organized as follows. Section 2 provides a heuristic description of the algorithm in order to provide the reader clarity regarding our goal and general methodologies. In Section 3, we use a modification of the original Einstein argument for the classical development of standard Brownian motions (see Reference 38, for example) to derive the primary equation of interest. This section also includes a more generalized procedure for the derivation which reduces the necessary assumptions on the system of interest. Section 4 builds on the material in Section 3, allowing for the construction of a closed-form solution to our theoretical model—thus, bypassing the need for numerical approximations and complicated learning procedures. We then use this model in Section 5 to classify unknown samples via our proposed algorithm. This section also clearly outlines the parameters for the physical experiments carried out to produce the data set for classification. Finally, Section 6 provides some concluding remarks and also alludes to possible future endeavors related to the current work.
2 OUTLINE OF PROPOSED ALGORITHM
We briefly outline the ideas behind the newly proposed algorithm, below. Note that the algorithm will be more completely and rigorously described in Main Algorithm (see Section 5). For clarity, we allude to specific aspects of the experiment of interest, whose specific protocol is outlined in Section 5.2.
- (i)
Inject into the sample, A, simple chemicals which will serve as “markers” in the classification process. For our purposes, we use glucose molecules of different types and denote these types by Mi, i ∈ N∗ = {1, 2, … , n∗} (where ). Each molecule Mi, i ∈ N∗, has a linear structure and (possibly) different lengths.
- (ii)
We then “slowly” transport sample A through a short (approximately 10 cm) porous tube. We assume that the transport is one-dimensional and let this transport coincide with the positive x-axis (e.g., as in Figure 1). (Note that the speed of transport was selected through auxiliary experiments in order to maximize the high resolution of the signal-to-noise ratio, prior to our classification procedure.)
- (iii)
At the point of observation, which we denote by x = L (L < 10 cm), we record all signals which are obtained from the mass spectrometer (with a particular emphasis on the observed peaks in the signals).
- (iv)
In this situation it is assumed that for all i, j ∈ N∗, such that i ≠ j, it holds that Mi and Mj do not mix (i.e., do not undergo chemical bonding). We identify each Mi, i ∈ N∗, via the peaks in the signals of sample A obtained from spectrometer. These Mi, i ∈ N∗, will serve as our “marker” molecules.
Note that the family of “marker” molecules Mi, i ∈ N∗, will be a subset of all molecules in any other sample of interest: Mj, j ∈ N = {1, 2, … , n} (where ). In other words:
() - (v)
We then classify the peaks that are located between those of the “marker” molecules in the sample A via a so-called classification index. In the current article, this index will depend only on two parameters—which in turn depend only on the retrieval time (the time at which the signal has the largest peak).
- (vi)
Using the information obtained by completing (i)–(vii), we may then classify other samples of interest. That is, for all other samples which have been injected with the same “markers,” we extract data regarding the Mi, i ∈ N∗, in these samples by matching the spectrometer signal peaks which are closest to the “marker” molecules in sample A.
- (vii)
We can then classify the remaining signal peaks in these samples through associated so-called data-driven PDEs. This is accomplished by constructing appropriate diffusion and absorption coefficients, which will allow us to distinguish differences between the new samples and the original sample A (see the Main Algorithm). (Note that we use the terms diffusiont and absorption to mirror the description given in the thought experiment employing compound transport based on Einstein's paradigm of Brownian motion with absorption and drift—see Section 3 for more details.)
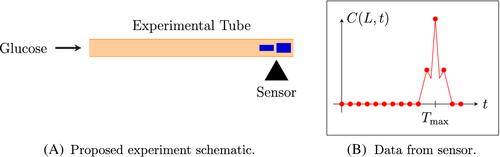
The generic description provided by (i)–(vii) in Proposed Algorithm are meant to provide a rough blueprint of the method we employ, herein. However, there are numerous mathematical details needed before we can rigorously formulate the final algorithm. A schematic depiction of Proposed Algorithm (and Main Algorithm) is provided in Figure 1, below.
As noted in (iv) of Proposed Algorithm, our method is based on the important assumption that for all i, j ∈ N∗, such that i ≠ j, the molecules Mi and Mj are not mixing throughout the experiment. This is formalized in the following assumption.
Assumption 1.For each i, j ∈ N∗ (cf. (i) of Proposed Algorithm), such that i ≠ j, it is assumed that Mi and Mj are not mixing. That is, they do not interact to create novel chemical compounds.
Assumption 1 is a vital assumption in our proposed method. However, it is worth noting that Assumption 1 may not be valid in all physical experiments of interest. For our particular situation, empirical evidence (obtained from experiments employing the guidelines outlined in Section 5.2) suggests that Assumption 1 does in fact hold. The process of molecular mixing requires further generalizations of Einstein's paradigm for Brownian motion and will be a focus of forthcoming research. More details on this process are provided in Section 3.
3 EINSTEIN'S PARADIGM FOR MOLECULAR TRANSPORT IN A TUBE
In this section we will reformulate Einstein's model for Brownian motion to the case where glucose molecules are being transported in the tube filled with porous material. Einstein derived his seminal mathematical framework based upon his visual observations of the random jumps of pollen grains of the plant Clarkia pulchella suspended in water (see References 38 and 39). He then provided a mathematical framework to describe the observed phenomena, which resulted in his model of classical Brownian motion. A less understood fact is that Einstein's approach can also be applied to many generic processes arising in physics, chemistry, and engineering. This is the key observation employed to derive our novel classification algorithm.
3.1 Assumptions for Einstein's paradigm
In order to employ Einstein's approach in novel situations, one must first understand the key principles which underpinned his work. These principles can be formulated into three main axioms, which we formulate into additional assumptions.
Assumption 2.For each molecule of interest, say M, we assume that there exists a time interval , which is small compared to the observable time intervals but large enough that the motions performed by M during two consecutive time intervals of length can be considered as mutually independent events.
Assumption 3.Let to be the set of all possible lengths of non-colliding jumps associated to M in the time interval (cf. Assumption 2). We will say that is a possible length of such “free jumps” if .
Assumption 4.All molecular interactions during such time intervals (cf. Assumption 2) are restricted to absorption of the surrounding media—which includes other molecules, the porous media, and all possible boundaries.
Remark 1.In general, the time interval of free jumps, (cf. Assumption 2), the expected value of the length of the free jumps associated to , which we denote by , and the frequency of the free jumps of length , which we denote by , are key parameters in our approach and may depend on underlying properties of the involved molecules (such as rates of change) and the surrounding environment (such as the components of the mixtures and the associated porous media) (see, for example, Reference 39 and the references therein).
We will use Remark 1 for the interpretation of the experimental data, herein.
3.2 Derivation of associated equation with drift and absorption
Remark 2.It is worth noting that Einstein's assumptions on φ are natural assumptions in the particular case involving Brownian motion. However, it is important to observe that this assumption may not be reasonable for all physical phenomena.
Next, let be the function which represent the number of particles per unit volume present at position at time t ∈ [0, ∞). With this Equations (3)–(5), we may now formulate a crucial axiomatic conservation law.
Axiom 1.There exists a continuous function such that the number of particles found at time (cf. Assumption 2) between two planes perpendicular to the x-axis with abscissas x and (where ) is given by
The second term in the right-hand-side of Equation (6) is a result of possible bonding and/or absorption with other molecules in the sample or with the porous media within the tube. In general, for Equation (6) to be well-defined, all one needs is that F is finite and measurable on the codomain of the function f. However, throughout this article, we will assume that there exists such that for all , t ∈ [0, ∞) this term is well-approximated by the linear function . The conservation law given by Equation (6) is depicted schematically in Figure 2, below.
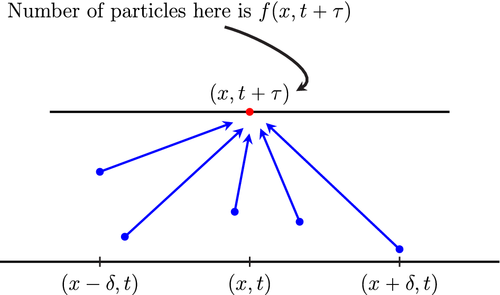
Remark 3.In Equation (7), we assume that φ has compact support. This assumption is physical in nature, as the ensuing theoretical work follows in a similar fashion if φ does not have compact support.
We conclude this subsection with some final remarks. In the arguments above, we have assumed sufficient smoothness conditions in order to allow for all claims to hold globally (i.e., for all t ∈ [0, ∞)). However, this can easily be circumvented by constructing Equation (13) locally (e.g., for some c ∈ (0, ∞) with t ∈ [0, c)) and then “gluing” the results together. This approach is avoided, herein, for simplicity and ease of exposition. Furthermore, the assumption that is not an inherently restrictive assumption. This assumption loosely can be thought of as removing the possibility of so-called long-range interactions within the experiment. One can allow for such interactions, but the resulting PDE may involve non-local operators such as the fractional Laplacian (see, for instance, References 41-44 and the references therein). Finally, we note that the assumptions in Equation (11) are purely for convenience, as we can obtain a result similar to Equation (13) through a simple re-scaling of the function f.
Remark 4.For the remainder of our study, we will assume that , φ, and (cf. Assumption 2) are independent of the function f. In general, these parameters can upon both the spatial and temporal variables, x and t, respectively, as well as the underlying porous media. Moreover, in more general situations, these parameters can further depend on the dependent variables' derivatives.
Remark 5.Herein, we assume that (cf. Assumption 2) is the same for each of the types of molecules of interest. That is, for each Mi, i ∈ {1, 2, … , n}, we assume that the associated . One can always find such a by simply choosing the minimum of the set of associated , i ∈ {1, 2, … , n}.
3.3 A remark on an alternative derivation
We conclude Section 3 with an alternative derivation of Equation (13). This alternative derivation is not simply an exercise in pure mathematics, but rather, it provides justification that the proposed algorithm may be applied to a wider class of problems than originally expected. First, this alternative derivation helps to resolve a slight inconsistency in the derivation presented in Section 3.2. That is, in Section 3.2 we simultaneously applied the Carathéodory theorem and Taylor's theorem to Equation (6). There is nothing inherently wrong with this approach—it simply seems strange. The approach presented below rectifies this concern. Next, this alternative derivation reduces the smoothness assumptions one imposes on the function f. In practice, it is often assumed that a function of interest is smooth enough to manipulate via expansions, but this assumption excludes numerous physically relevant situations. As such, we present a method for reducing the regularity assumptions needed to obtain Equation (13), which, in turn, increases the applicability of the proposed method.
Theorem 1.Let , t ∈ [0, ∞) and let be a function. Then f is twice differentiable with respect to the variable x at the point (c, t) if and only if there exist , i ∈ {1, 2}, such that it holds that
Proof of Theorem 1.We omit the details of this proof for brevity. However, the result follows from arguments analogous to those used to generate Equations (16)–(20). The proof of Theorem 1 is thus completed.
The alternative derivation of Equation (13) follows directly from Theorem 1 (combined with the previous results outlined in Section 3.2). It is worth noting that the alternative derivation presented above only requires the function f to be twice differentiable with respect to x and once differentiable with respect to t. In fact, Theorem 1 can be generalized to situations where f possesses even less regularity. However, we leave these considerations for future endeavors.
4 CLOSED-FORM SOLUTIONS TO THE EINSTEIN EQUATION IN APPLICATION TO GUI CLASSIFICATION VIA RETRIEVAL TIME
As should be clear from Section 3, we intend to use the Einstein model of random jumps to interpret the results of experimental observations. This approach should present a stark contrast to the traditional approach of employing Fick's law, flux conservation laws, and the thermodynamical law that density is proportional to the mass concentration function (see, for example, the classical work by Reference 45). The arguments in this section motivate the novel approach with the particular case of GUI classification via retrieval time in mind.
4.1 Development of closed-form solutions to the Einstein equation
4.2 Employing closed-form solutions to the Einstein equation to classify GUI via retrieval time
Note that Figure 3 presents a graph of the actual spectrometer signals, with associated peaks, which correspond to the molecules of GUI passing through the receiver of the mass spectrometer (obtained via the methods outlined in Section 5.2). As we can see, there are eight distinct GUIs and they each have different retrieval times. Figure 4(A) presents the calculated diffusion coefficients Di, i ∈ {1, 2, … , n}, for each retention time. Figure 4(A,B) together demonstrate the expected inverse proportional relationship between retention time and diffusivity.
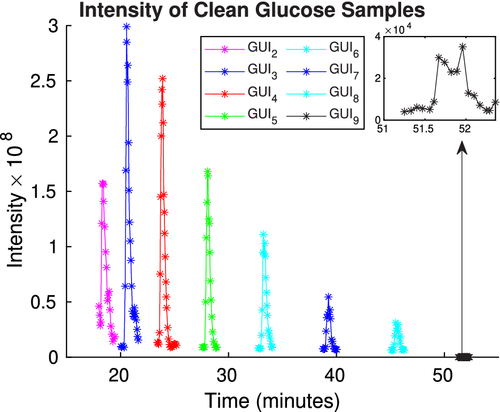
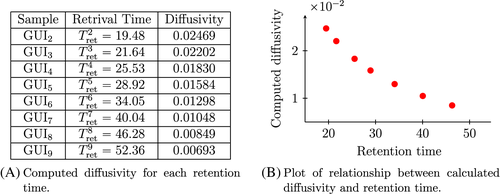
The intuition provided by the Einstein paradigm tells us that the molecule's retrieval time should depend on the molecule's time of free travel and that the value should increase proportionally with the molecules mass. This observation is clearly supported by our data (see Figure 4), as we have the mass of the GUI molecules increases with their index value. Furthermore, our post-processing—which can be performed using our analytical formula for pure samples—confirms this intuitive observation and Einstein's theory that the diffusion coefficients are inversely proportional to (cf. Assumption 2).
Note that the experimental observations presented in Figure 4 support precisely this claim. Further observe that the velocity of the filtration (drift) is so small that diffusion is the dominant transport property. Indeed, if drift had a larger influence than diffusion, then the decrease of the retrial time with respect to the GUI mass would be linear. Since Figure 4(B) clearly indicates an inversely propositional relationship, we conclude that diffusion is dominant. This is an important observation as drift mainly depends upon the boundary conditions of a problem whereas diffusion depends only on object versus tube structure properties. This crucial observation is what allows for the proposed classification method to work so well. Finally, we mention that in this preliminary result we have also ignored the associated absorption rates. However, classification methods for complex serum samples will consider these effects.
Remark 6.From the Einstein paradigm it follows that the calculated diffusion coefficients are inversely proportional to (the “free jump times”—cf. Equation 2) of each molecule. Equation (27) provides an explicit relationship between retrieval time and the “free jumps” for molecule Mi, i ∈ {1, 2, … , n}: smaller retrieval times correspond to larger .
5 SERUM CLASSIFICATION USING GUIS AS MARKERS
We will use the general ideas of Remark 6 as a basis for the development of our classification algorithm using GUIs as markers. The basic idea of the algorithm (which was outlined in the Proposed Algorithm) consists of determining the diffusivity coefficients of “marker” molecules in a base sample of interest (cf. Equation 13). The remaining molecules are classified by grouping them based on diffusivity coefficient ranges and computing the associated absorption coefficients, which serve as a correction term of sorts (cf. Equation 13). We assume throughout that drift coefficients are negligible (cf. Equation 13). This assumption is justified due to the intended use of post-processing in the actual physical experiment.
5.1 Serum classification algorithm
The proposed algorithm for the classification of a given unknown sample using two parameters, diffusivity and absorption, consists of five main steps. Note that throughout this section the primary focus in the classification of N-glycans.
- Step 1
Select a base sample, which without loss of generality we assume to be A0. Let represent the number of molecules in A0. Inject the “marker molecules,” or GUIs—which we designate as GUI1, GUI2, … , GUI8, into the sample A0. Without loss of generality, assume that the GUIs are indexed with respect to increasing mass. Collect all retention times , i ∈ {1, 2, … , n0}, from the mass spectrometer. Identify (manually) which retention times are associated to the GUIs.
- Step 2
Using the retention times from Step 1, calculate the diffusion coefficients for each GUI, which we designate as , i ∈ {1, 2, … , 8}, via Equation (27). Set the associated absorption coefficients, which we designate as , i ∈ {1, 2, … , 8}, to zero. This completes the baseline classification procedure (if desired, one can classify the remaining objects in A0).
- Step 3
Take a new (unknown) sample, which without loss of generality we assume to be A1, and again inject the GUI molecules from Step 1. Let represent the number of molecules in A1 (after injection with GUI molecules). Pass A1 through the mass spectrometer and again collect all retention times, , i ∈ {1, 2, … , n1}. Using Equation (27) compute all associated diffusivity coefficients, , i ∈ {1, 2, … , n1}.
- Step 4
Using the results from Step 1 find the coefficients i1, i2, … , in ∈ {1, 2, … , n1} which for each j ∈ {1, 2, … , 8} satisfy that
()Note that Equation (30) is well-defined for all j ∈ {1, 2, … , 8} by construction. The molecules associated with the indices i1, i2, … , i8 are the GUI markers GUI1, GUI2, … , GUI8. Let satisfy that()Finally, let , j ∈ {1, 2, … , 8}, satisfy for all j ∈ {1, 2, … , 8} that . - Step 5
We now classify the remaining molecules from A1 using Equation (31) to “classify” all remaining objects. Note that it is the case that . For all k ∈ {1, 2, … , n1} {i1, i2, … , i8} we compute the associated as
()(cf. Equation 26). - Step 6
Repeat Step 3, Step 4, and Step 5 for the remaining , i ∈ {2, 3, … , N}.
It is worth noting that only the diffusivity coefficients calculated from Step 1 and Step 2 need to be stored for future use. This data set serves as the baseline learning procedure for the data-driven classification algorithm. Thus, while the initial manual classification can be tedious, it results in an algorithm which can be used to classify a large number of other unknown samples (of appropriate type).
Remark 7.Main Algorithm can be significantly improved if we consider the classification index to be a time series instead. In this case, we may consider Equation (29) as the basis for classification. When considering a serum containing only the “marker” molecules, it follows that t is the retrieval time in Equation (29). An analogous (but improved) algorithm can then be obtained by approximating the integral implicitly (for numerical stability) and employing a Newton–Raphson-type method to solve for the desired parameters. As noted earlier, this will be a focus of forthcoming work.
5.2 Description of the experimental set-up and materials used
In this section, we briefly outline the materials and experimental protocols followed in order to obtain our experimental data, which is used for comparison.
5.2.1 Material
Standard glycoproteins, fetuin and ribonuclease B (RNase B), and pooled human blood serum (HBS) were purchased from Sigma Aldrich (St. Louis, MO). Formic acid (FA), borane-ammonia, dimethyl sulfoxide (DMSO), iodomethane and, sodium hydroxide beads were also obtained from the same vendor. HPLC grade water was obtained from Avantor Performance Materials (Center Valley, PA). HPLC grade acetonitrile (ACN), methanol, and ethyl alcohol were supplied by Fisher Scientific (Fair Lawn, NJ). PNGase F enzyme and 10XG7 buffer (0.5M phosphate buffer saline) were purchased from New England Biolabs.
5.2.2 Sample preparation
Model Glycoproteins and Dextrin: 20 g each of fetuin and RNase B were mixed with G7 buffer to get a final concentration of 20 mM for the buffer. The samples were then denatured at 90°C for 30 minutes. Samples were then cooled at room temperature and treated with 1.0 l of PNGase F, followed by incubation at 37°C for 18 hours. PNGase F digestion was followed by precipitation of de-N-glycosylated proteins with 90% ethanol at −20°C. Reduction of reducing ends of the purified glycans was done by addition of 10 of borane-ammonia complex (10 g/L) and incubating it at 60°C water bath for one hour. Methanol was later used to remove borane in the form of borate from reduced glycan samples. The methanol washing step was repeated three times to ensure the complete removal of borate from the samples. Reduction was then followed by permethylation of the samples, using a previously reported method.16 For this purpose, reduced and dried glycan samples were resuspended in 1.2 L and 30 L of DMSO. Later, 20 L of iodomethane was added to the samples and they were loaded on DMSO soaked sodium hydroxide beads packed in spin columns. The spin columns were washed with 200 L of DMSO, using a centrifuge at 1800 rpm for two minutes, prior to the loading of samples.
Once loaded, the samples were incubated at room temperature for 2 minutes. After 25 minutes, an additional 20 L of iodomethane was added and the samples were again incubated at the room temperature for 15 minutes. Permethylated glycans were then collected by centrifugation at 1800 rpm for two minutes. For complete elution of permethylated glycans, 30 L of ACN was added to the spin columns and again the eluants were collected by centrifugation. Permethylated glycans were further dried and resuspended in 20% ACN and 0.1% FA. Each of the samples were run in triplicates and 1 g of the samples were injected for each run. Dextrin standard was mixed with the samples prior to reduction and, therefore, was reduced and permethylated with each sample. 1 of sample was spiked with 100 ng of dextrin.
5.2.3 Human blood serum
10 L of human blood serum was mixed with 90 L of G7 buffer to get a final concentration of 20 mM for the buffer. Proteins from the samples were denatured in 90°C water bath for 30 minutes. After cooling at room temperature, 1.2 L of PNGase F was added to the samples. They were then incubated at 37°C for 18 hours. After the completion of the incubation, proteins were precipitated at −20°C for one hour. Reduction and permethylation were then performed as described previously for model glycoproteins. Resuspension was again done in 20% ACN and 0.1% FA. 1 L of the serum samples were then injected for each of the triplicate runs.
5.2.4 Liquid chromatography conditions
Chromatography was performed on UltiMate 3000 Nano UHPLC system using C18 column. Optimum temperature for the oven was kept at 55°C. A solution of 98% water, 0.2% ACN, and 0.1% FA was utilized as mobile phase A while, 100% ACN and 0.1% FA was mobile phase B. Initially, the gradient was set at 20% mobile phase B. It was then increased to 42% in 11 minutes. After 48 minutes, it was increased to 55% and then changed to 90% at 49 minutes. It remained at 90% for 54 minutes of total sample run and plummeted to 20% again for equilibration of the column for the final 6 minutes.
5.2.5 Mass spectrometry conditions
LTQ Orbitrap Velos (Thermo Scientific) was used to analyze the samples. The mass spectrometer was set to the positive ion mode with an ESI voltage of 1.6 kV. Full MS was performed at 100,000 resolution with 200–2000 m/z scan range. MS2 was acquired with collision induced dissociation (CID) and higher energy collision dissociation (HCD) with normalized dissociation energies of 30% and 45%, respectively. Activation Q (one of the parameters used in MS methods—the value controls the radio frequency applied to control fragmentation of ions during analysis) was 0.25. Injection time was 10 ms. Repeat count of dynamic exclusion and repeat duration were 2 and 30 s, respectively. The exclusion duration was 60 s. The four most intense ions were selected from the full MS for further CID and HCD based dissociation by applying data-dependent acquisition mode. The precursor ion selection window was 1.50. The MS2 intensity threshold was 5000 counts. Singly charged ions were excluded for MS2.
5.2.6 Data analysis
The extracted ion chromatograms (EIC) of full MS data were used to determine the glycan composition as well as retention times of reduced and permethylated glycans derived from model glycoproteins, and human blood serum, with a mass tolerance of 10 ppm. Retention times of reduced and permethylated glucose units were also determined using the EIC.
5.3 Data classification of an actual experiment using the proposed algorithm
We now demonstrate Main Algorithm and its efficacy through an experimental example. We will use data obtained via the methods and procedures outlined in Section 5.2. To obtain the data in Figures 5-7 below we implemented our algorithm for a particular set of experiments (obtained via the methods outlined in Section 5.2). In the first table (left table) in Figure 5, the GUIs were known (green rows). Then this data is then used according to Main Algorithm to identify the eight GUI markers in five other (unknown) experiments via the same instrumentation. We then later used precise verification methods to determine that Main Algorithm can distinguish GUIs with an error of no more than two percent.
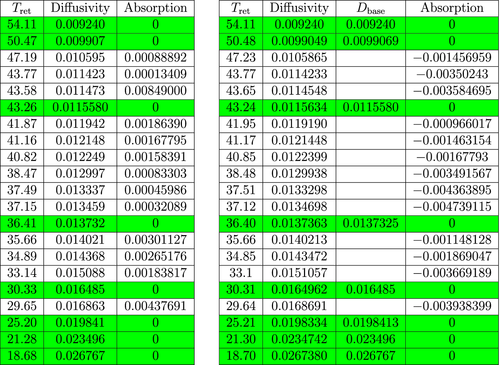
As noted above, the left table in Figure 5 consists of the data obtained from mass spectrometer readings for the sample A0 (cf. Main Algorithm) and the calculated absorption coefficients for that sample. The first column of the table contains the retention time data. The second column contains the corresponding calculated diffusion coefficients. The last column contains the calculated absorption coefficients. The green rows in each table represent the data corresponding to the injected GUI molecules. As mentioned above, for the sample A0, manual intervention to the mass spectrometer data is mandatory for finding the GUI retention times. See Section 5.2 for more details.
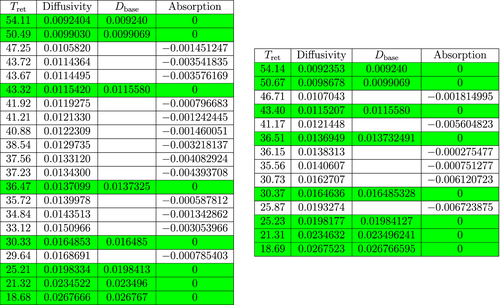
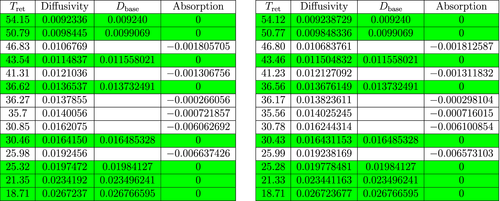
The second table in Figure 5 contains the data that was obtained from the mass spectrometer after the experiment for sample A1 (cf. Main Algorithm) and implementation of Main Algorithm for that sample. The first column of the table contains the retention times for the sample. The second column contains the corresponding calculated diffusion coefficients. The third column contains the coefficients of the GUI molecules from sample A0 (used for classification). The last column contains the calculated absorption coefficients. The tables in Figures 6 and 7 may be interpreted in a similar manner as the second table of Figure 5.
Remark 8.As is well understood, manual classification of molecular structures via the mass spectrometer is a time consuming and difficult task. However, Main Algorithm is demonstrated to be able to successfully identify GUI molecules in all experimental samples and also classify the remaining molecular structures using only retention times (after the initial classification of the “marker” molecules.
6 CONCLUSIONS AND FUTURE ENDEAVORS
In this article, we developed a novel mathematical method which allows for an efficient classification of experimental samples using a GUI as the reference frame. These interpretations were associated directly to the experimental spectrometer data in particular examples. In order to develop the novel method, we presented a data-driven partial differential equation model based on modified Einstein paradigm arguments. This extends Einstein's original study of Brownian motion to the situation of a more general conservation law. Once this data-driven model is obtained, we develop a closed-form solution of the model in order to avoid numerical approximations. A simple learning procedure is performed in order to determine the solutions coefficients on an initial sample. These coefficients are then used in additional learning procedures for computing the coefficients associated with unknown samples.
In order to further justify our method, we provide physical interpretations of the model coefficients. These coefficients are shown to be related to the experimental retrieval times, as well. This serves as the basis for the novel algorithm used for data classification (cf. Main Algorithm). Moreover, the proposed algorithm is successfully implemented and its efficacy is demonstrated via examples. Through the consideration of six independent samples we demonstrate that our method successfully classifies the unknown samples with errors which do not exceed two percent. Moreover, our novel method can be shown to be ten percent more accurate than the traditional method of using retrieval time only for classification.
It is important to mention that the retrieval time and shape of the peak in the spectrometer data depend on three main parameters: drift (velocity), variance (diffusion), and absorption. In this article we demonstrated that if the drift is fixed, due to reprocessing of the data, then the diffusion and absorption coefficients can accurately classify molecules in an unknown sample. Our future endeavors will focus on including all three characteristics for the N-glycan classification. This inclusion will make use of iterative deep learning algorithms of Kolmogorov-type (see, for instance, References 46-48).
Biographies

Joshua Lee Padgett is an Assistant Professor in the Department of Mathematical Sciences at the University of Arkansas. Prior to joining the University of Arkansas, Josh was a postdoc in the Department of Mathematics and Statistics at Texas Tech University. Josh earned his Ph.D. in mathematics at Baylor University under the supervision of Qin Sheng. Josh received his B.S. in Mathematics from Gardner-Webb University where his undergraduate research focused on the metabolic features of tumor cells and the occurrence of the so-called Warburg effect. While there, Josh was also a member of the Track and Field team (competing in the javelin and hammer throw). Josh is also an affiliated faculty member at the Center for Astrophysics, Space Physics, and Engineering Research and is a honorary adjunct faculty at Texas Tech University.
Josh Padgett's research lies at the intersection of pure, applied, and computational mathematics. Current research interests include applied mathematics, numerical analysis, geometric and Lie group integration methods, mathematics of deep learning, operator splitting methods, algebraic structures of numerical methods, fractional differential equations, stochastic differential equations, and the use of spectral and operator theory in theoretical and experimental physics.

Yusup Geldiyev has completed B.S. and M.S. degree in mathematics from TTU. Currently he is a first year phd student at UTD. He has participated in poster competition and given numerous talks in seminars. In short, his goal is to become a professional mathematician and enjoy the process along the way.

Sakshi Gautam is a PhD candidate in the Department of Chemistry & Biochemistry at Texas Tech University, working in the research group of Dr. Yehia Mechref. Her research focuses on the development of LC-MS based analytical methods for efficient separation and identification of isomeric glycans and glycopeptides from biological samples.

Wenjing Peng Dr. Wenjing Peng is currently working as a Research Assistant Professor in Chemistry and Biochemistry Department, Texas Tech University. Before working in Texas Tech University, Dr. Peng completed his undergraduate program and master's degree in Sichuan University, and Ph.D.'s degree in Chengdu Biological Institute, Chinese Academy of Sciences, China. Dr. Peng is focusing on artificial neural network based process optimization and LC-MS based glycomic and glycoproteomic studies for biomarker discovery of cancer diagnosis and progression. He also brings the expertise in design of experiments, microbiology, molecular biology, separation and purification techniques during more than seven years' experience as a Senior Researcher in the biological medicine developing group of Di'ao Pharmaceutical Company. His current projects include breast cancer brain metastasis biomarker discovery, multiple isotopic labeling technology development and anti-cancer drug development using liquid-chromatography-tandem mass spectrometry by integrated transcriptomics, glycomics, proteomics and glycoproteomics.
Research Interests: Glycomics, Proteomics, Glycoproteomics, Transcriptomics, Isotopic labeling, Separation, Purification, High Performance Liquid Chromatography (HPLC), Mass Spectrometry (MS), Design of Experiments (DOE), Biomarker Discovery, Microbiology, Fermentation.

Yehia Mechref Prof. Yehia Mechref is a Paul W. Horn Distinguished Professor in the department of Chemistry and Biochemistry at Texas Tech University. He is also the Chairman of the department of Chemistry and Biochemistry and the Director of the Center for Biotechnology and Genomics. He received a B.Sc. in chemistry from the American University of Beirut (Beirut, Lebanon) and a Ph.D. with an honorable mention from Oklahoma State University (Stillwater, OK, USA). Dr. Mechref is a world-renowned scientist.
He is a nationally and internationally recognized scientific leader in the development of sensitive biomolecular mass spectrometry methods enabling qualitative and quantitative assessments of the roles of proteins, glycoproteins, and glycans in biological systems. He has developed a fast, highly sensitive chromatographic method that efficiently separates and distinguishes among gangliosides in blood samples from patients with different diseases of the esophagus, including adenocarcinoma, high-grade dysplasia, and Barrett's Esophagus. His method for analyzing N-linked glycans, using multiple-reaction monitoring (MRM) on a triple quadrupole mass spectrometer, permits quantitative comparison of the expression of N-glycans in brain-targeting breast carcinoma cells and metastatic breast cancer cells. The permethylated N-glycan MRM method he developed yields rapid, reliable identification and relative abundances of glycans from varied, complex biological samples. He has further refined MRM to minimize the tissue needed and optimize the process, making it possible for the first time to profile glycans on the surfaces of tissues and cells, rather than whole-tissue samples, thus conserving limited tissue samples for other studies.
He pioneered a liquid chromatography-mass spectrometer (LC-MS) system to analyze glycomic samples in conventional proteomic laboratories, eliminating the need for more specialized facilities and making glycomic analysis more accessible to the broader scientific community. Dr. Mechref has, as well, completed a comprehensive characterization of bovine collagen glycosylation that may be foundational in developing the biochemical modifications necessary for its use in treating human diseases.
Over the past eight years, Dr. Mechref's research program has received over $9M in funding from the NIH, CPRIT, and CH Foundation, and he currently has five active NIH grants (3R01s, U01 and S10). Dr. Mechref is a prolific writer who has published 29 review articles, 14 book chapters, and 185 peer-reviewed research papers. He has guest-edited 7 journal special issues and he is a member of the editorial boards of numerous journals.
Currently, Dr. Mechref's Google Scholar h-index is 60, with 10,936 citations. He has 11 US patents. He has organized and co-organized numerous symposia and conferences, and is a standing member of the NIH EBIT study section. Dr. Mechref is the recipient of the 2019 TTU President's Academic Achievement Award, the 2019 TTU Nancy J. Bell Faculty Excellence in Mentoring Award, the 2016 Barnie E. Rushing, Jr. Faculty Distinguished Research Award, and the 2015 Barnie E. Rushing, Jr. Faculty Outstanding Research Award.

Akif Ibragimov is working as Assistant Professor in the Department of Mathematics, Atma Ram Sanatan Dharam College, University of Delhi. He has completed his Ph.D. in 2009 in Mathematics (celestial mechanics and space dynamics). His observations and comments in this field reflect his interests in nature, society, and nonlinear dynamics. He has more than 30 research articles in his name in reputed national as well as international journals in the field of celestial mechanics. He is also author of books related to his field. He is an individual member of International Astronomical Union (IAU obtained his Master's degree in Mathematics in 1975 from the Azerbaijan Oil Academy. He also interned at the Department of Computational Mathematics and Cybernetics (CMC) of Lomonosov Moscow State University (MSU) in 1974 and 1975 and one year later, in June 1976 Akif defended his PhD thesis at the CMC Department of MSU. During the next 10 years his research was focused on the Qualitative Theory of Partial Differential Equations and in 1985 he defended his dissertation at the Steklov Mathematical Institute of the Russian Academy of Sciences in Moscow, and obtained the degree Doctor of Science in Mathematics. Since then his research interests diversified; he worked at several academics and industrial positions in Russia, Azerbaijan and USA. From 2000-2004, he was a Visiting Professor at Texas A&M University, and Research Scientist at Knowledge Based Systems in College Station. Akif joined Texas Tech in September 2004. His research interests cover broad areas of applied mathematics including fluid flow in porous media, mathematical modeling in bio-medicine, fluid structure interaction, and image processing.
Research Interests: Applied Mathematics, Analysis, Industrial Mathematics, Mathematical Biology, Mathematical Physics, Ordinary Differential Equations, and Partial Differential Equations.




