Expression of genes involved in apoptosis is dysregulated in mucopolysaccharidoses as revealed by pilot transcriptomic analyses
Abstract
Mucopolysaccharidoses (MPS), a group of lysosomal storage diseases (LSD), are inherited disorders caused by mutations in genes coding for enzymes involved in the degradation of glycosaminoglycans (GAGs). Therefore, accumulated GAGs in lysosomes lead to severe symptoms in patients and significantly shortened life span. Although GAG accumulation in cells is the primary cellular defect in MPS, recent reports indicated that severe changes in cellular processes occur there as secondary or tertiary effects, which may contribute significantly to the disease pathomechanism. Apoptosis is one of such process, while mechanisms leading to dysregulation of this process in MPS remain largely unknown. To learn about these mechanisms, we have performed transcriptomic studies using cultures of fibroblasts derived from patients suffering from all types and subtypes of MPS, and assessed genes related to apoptosis. We found that there are significant changes in expression levels of many such genes relative to control fibroblasts (Human Dermal Fibroblasts-adult cell line), and the number of down- or up-regulated transcripts was between 19 and 73 in different MPS types. We have identified apoptosis-related genes, which were considerably dysregulated in many MPS types, as well as those in which expression was significantly changed in specific MPS types. BNIP3, C1D, CLU, GPER1, KREMEN1, and PRKCD genes displayed the most changed expression profiles in most MPS types relative to control cells. Caspase 3/7 activity was increased in MPS IVA and IX. These results indicate that changes in apoptosis, observed in MPS, may arise, at least partially, from dysregulation of genes coding for proteins involved in this process.
Abbreviations
-
- GAG
-
- glycosoaminoglycans
-
- LSD
-
- lysosomal storage diseases
-
- MPS
-
- mucopolysaccharidosis
Introduction
Mucopolysaccharidoses (MPS) are inherited metabolic diseases characterized by glycosaminoglycan (GAG) storage in lysosomes (Tomatsu et al., 2018). The storage results from deficiency in one of enzymes involved in GAG degradation, caused by mutations in corresponding genes. Progressive GAG accumulation in cells results in damage of the affected tissues, including bones, joints, respiratory system, heart, and central nervous system (CNS). The course of the disease is severe, resulting in average life span between one and two decades (Wraith, 2013). Depending on the kind of accumulated GAGs and clinical symptoms, seven types of MPS are distinguished, with subtypes in MPS type III and IV, in which defects of different enzymes cause the storage of the same kinds of GAGs. Following GAGs are accumulated in particular MPS types: heparan sulfate and dermatan sulfate in MPS I and MPS II (though degradation of these GAGs stop at different stages in each disease); heparan sulfate in MPS III (with four subtypes, designed A, B, C, and D, in which various enzymes are lacking or highly deficient); keratan sulfate and chondroitin sulfate in MPS IV (with two subtypes, designed A and B, in which various enzymes are lacking or highly deficient); dermatan sulfate in MPS VI; heparan sulfate, dermatan sulfate, and chondroitin sulfate in MPS VII; and hyaluronic acid in MPS IX (Tomatsu et al., 2018). MPS is considered a rare disease, with cumulative frequencies of all types between 1 per 40,000–50,000 live births (Muenzer, 2011).
Although GAG storage is undoubtedly the primary cause of MPS, secondary and tertiary effects can contribute significantly to the development of specific symptoms. Apoptosis is one of processes, which is affected in MPS cell although molecular mechanism leading to changes in this process (perhaps as a secondary or tertiary effect of GAG storage) remains unknown. This is despite the fact that disturbance of apoptosis was suggested as one of the crucial defects leading to cellular dysfunctions in lysosomal storage diseases, including MPS (Platt et al., 2012).
Generally, vast majority of studies on apoptosis in MPS demonstrated elevated levels of markers of this process. Noguti et al. (2012) demonstrated that apoptosis was stimulated in the mouse model of MPS I. Since Bcl-2 levels were decreased in tongue mucosa cells of this model, it was concluded that apoptosis might be induced due to down-regulation of the bcl-2 gene expression. In contrast, Viana et al. (2017) suggested that MPS I fibroblasts are more resistant to apoptotic induction than control cells as they responded less efficiently to staurosporine, one of the apoptosis stimulators. Studies on neural stem cells isolated from the MPS II mouse model indicated higher number of apoptotic cells relative to those isolated from wild-type animals (Fusar Poli et al., 2013). In MPS IIIA mouse model, enhanced expression of genes coding for tumor necrosis factor α (TNFα), TNF receptor 1 (TNFR1), caspase 3, and caspase 11 was observed, suggesting that apoptosis is induced (Arfi et al., 2011). When brains of MPS IIIB mice were analyzed for changes in expression of genes, using the microarray method, changes in messenger RNA (mRNA) levels of various apoptosis-related genes were found (Villani et al., 2007). Down-regulation of Bcl2, Bid, Birc2, Tnfrsf8, and Tnfrsf1b (Tnfr2) genes and up-regulation of Apaf1, Casp11 (Casp4), Tnfrsf1a (Tnfr1), and Traf3 genes were detected. Therefore, apoptosis disturbance could be suggested. In cultures of chondrocytes derived from MPS VI animals, the number of apoptotic cells was higher than in control experiments (Simonaro et al., 2001). Similar results, that is, increased number of apoptotic cells, were reported in studies on liver, spleen, and kidney of MPS VI rats (Tessitore et al., 2009). Levels of mRNAs of apoptosis-related genes were also estimated in brains of MPS VII mice, to find that genes coding for TRAIL R and caspase 2 were down-regulated, while genes coding for TNFα, TNF receptors 1 and 2, TNF RSF8, TLR-4, TRAIL, Fas, FADD, and caspases 3, 8, 9, and 11 were up-regulated (Richard et al., 2008). Moreover, extensive apoptosis has been reported in the Drosophila model of MPS VII (Bar et al., 2018).
As indicated above, there are several reports indicating enhanced apoptosis, however, mechanisms leading to this phenomenon are unclear, and all those studies were performed on models of only one MPS type at the time. In addition, different models were used, from cell cultures, through Drosophila to rodents. Therefore, in this work we aimed to perform complex transcriptomic studies, using fibroblasts of all MPS types and subtypes. Such experiments should provide a global picture of changes in expression of apoptosis-related genes in every MPS type/subtype.
Materials and methods
Cell cultures
Fibroblasts derived from patients suffering from all MPS types, and control fibroblast line Human Dermal Fibroblasts-adult (HDFa) were purchased from the NIGMS Human Genetic Cell Repository at the Coriell Institute for Medical Research (this institute possess all documents concerning bio-ethical issues). The genetic characteristics of MPS fibroblast lines is as follows: MPS I, affected gene—IDUA, mutations—p.Trp402Ter/p.Trp402Ter; MPS II, affected gene—IDS, mutation—p.His70ProfsTer29; MPS IIIA, affected gene—SGSH, mutations—p.Glu447Lys/p.Arg245His; MPS IIIB, affected gene—NAGLU, mutations—p.Arg626Ter/p.Arg626Ter; MPS IIIC, affected gene—HGSNAT, mutations—not determined; MPS IIID, affected gene—GNS, mutations—p.Arg355Ter/p.Arg355Ter; MPS IVA, affected gene—GALNS, mutations—not determined; MPS IVB, affected gene—GLB1, mutations—p.Trp273Leu/p.Trp509Cys; MPS VI, affected gene—ARSB, mutations—not determined; MPS VII, affected gene—GUSB, mutations—p.Trp627Cys/p.Arg356Ter; MPS IX, affected gene—HYAL1, mutations—not determined. Patients for which mutations were not determined, have been diagnosed on the basis of estimation of urinary GAG levels and activities of lysosomal enzymes in the plasma. Fibroblast was cultured in the Dulbecco's modified Eagle's medium (DMEM) supplemented with antibiotics and 10% fetal bovine serum (FBS), at 37°C, 95% humidity, and saturation with 5% CO2.
RNA isolation and purification
For each cell line, four biological repeats of the experiment (i.e., four independent cultures, each from different passage) were performed. In every single repeat, 5 × 105 cells were allowed to attach to 10-cm-diameter plate walls overnight and cultured on such plates. Cell lysis was performed using guanidine isothiocyanate, β-mercaptoethanol, and the QIAshredder column. RNeasy Mini kit (Qiagen) and Turbo DNase (Life Technologies) were used for RNA extraction, and the procedures were according to instructions of manufacturers. The quality of RNA samples was tested in the Agilent 2100 Bioanalyzer System with RNA Nano Chips (Agilent Technologies).
RNA-seq analysis
The mRNA libraries were prepared using Illumina TruSeq Stranded mRNA Library Prep Kit. After reverse transcription, complementary DNA (cDNA) libraries were sequenced using HiSeq4000 (Illumina, San Diego, CA, USA), employing the following parameters: 150 bp paired-end and minimum 40 × 106 raw reads, giving a minimum of 12 Gb of raw data for each sample. FastQC version v0.11.7 was used for quality assessment. RNA-seq data were deposited at NCBI Sequence Read Archive (SRA) (with the accession no. PRJNA562649). For mapping of raw readings to the GRCh38 human reference genome, the Hisat2 v. 2.1.0 program (from the Ensembl database) was used. Cuffquant and Cuffmerge programs (version 2.2.1) and the GTF Homo_sapiens.GRCh38.94.gtf file (the Ensembl database) were used to calculate the levels of transcripts. The Cuffmerge program was started with the library-norm-method classic-fpkm parameter normalizing the expression values by means of the FPKM algorithm. Statistical significance was assessed considering four biological repeats of each experiment (n = 4) and using one-way analysis of variance (ANOVA) on log2(1 + x) values, which have normal continuous distribution. To estimate the false discovery rate (FDR), the Benjamini–Hochberg method was used. For comparisons between two groups, post hoc Student's t test with Bonferroni's correction was employed. All statistical analyses were performed using R software v3.4.3, with significance assumed if P < 0.1. Transcript annotation and classification were performed using the BioMart interface for the Ensembl gene database. The raw RNA-seq data are available at NCBI Sequence Read Archive (accession no. PRJNA562649).
Measurement of caspase 3/7 activity in cells
For estimation of caspase 3/7 activity, 5 × 103 cells were seeded into each well of 96-well plate, and cultured for 24 h in the DMEM medium supplemented with antibiotics and 10% FBS, at 37°C, 95% humidity, and saturation with 5% CO2. Then, 0.1 μL of 2 mM CellEvent™ Caspase-3/7 Green Detection Reagent (Thermo Fisher Scientific) were added to 0.1 mL of the culture, and following 30 min incubation, the fluorescence was measured at 530 nm using EnSpire Multimode Plate Reader (Perkin Elmer). Simultaneously, the number of viable cells (cultured under exactly the same conditions are described above) was estimated by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) test as described previously (Pierzynowska et al., 2018). The results were calculated as caspase 3/7 activity per cell. Statistical analyses were performed with one-way ANOVA, assuming statistically significant differences when P < 0.05.
Results
For transcriptomic analyses, we have used total mRNA isolated from fibroblasts derived from all types and subtypes of MPS (types/subtypes: I, II, IIIA, IIIB, IIIC, IIID, IVA, IVB, VI, VII, and IX) and compared with the levels of mRNAs in control fibroblasts (HDFa cell line). RNA-seq technique was employed to obtain quantitative results of levels of particular transcripts. In this study, we have analyzed expression of genes listed in the QuickGO database under the term “apoptotic process” (GO:0006915), according to the Gene Ontology Consortium.
We have found that in each MPS type, numerous apoptosis-related transcripts were down- or up-regulated (Table 1). The highest number of affected transcripts occurred in MPS IVB (73 transcripts) while the lowest number in MPS VI (19 transcripts). In each type/subtype, both down- and up-regulated transcripts could be identified, though in almost all cases (except MPS II) the number of up-regulated transcripts was higher than that of down-regulated mRNA species (Table 1). These results indicate that expression of a high number of apoptosis-related genes is changed in MPS cells (all types/subtypes) relative to control cells, which can significantly influence this process.
| Number of significantly changed transcripts in MPS vs. HDFa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Transcripts | MPS I | MPS II | MPS IIIA | MPS IIIB | MPS IIIC | MPS IIID | MPS IVA | MPS IVB | MPS VI | MPS VII | MPS IX |
| Up-regulated | 43 | 13 | 38 | 40 | 32 | 32 | 19 | 41 | 10 | 46 | 40 |
| Down-regulated | 19 | 15 | 24 | 29 | 23 | 20 | 4 | 32 | 9 | 14 | 25 |
| Total | 62 | 28 | 62 | 69 | 55 | 52 | 23 | 73 | 19 | 60 | 65 |
To analyze the apoptotic genes changed in MPS in more detail, we have grouped them into categories according to the QuickGO database. This analysis indicated that particular sub-processes, defined by specific GO terms, are especially strongly affected in MPS as revealed by the number of genes in which expression is changed relative to HDFa cells (Table 2). The following sub-processes, characterized by changes in MPS cells, were identified: apoptotic mitochondrial changes (GO:0008637), apoptotic process involved in development (GO:1902742), apoptotic signaling pathway (GO:0097190), cysteine-type endopeptidase activity involved in apoptotic process (GO:0097153), execution phase of apoptosis (GO:0097194), negative regulation of apoptotic process (GO:0043066), negative regulation of mitochondrial membrane permeability involved in apoptotic process (GO:1902109), peptidase activator activity involved in apoptotic process (GO:0016505), positive regulation of apoptotic process (GO:0043065), positive regulation of mitochondrial membrane permeability involved in apoptotic process (GO:1902110), regulation of apoptotic process (GO:0042981), and regulation of mitochondrial membrane permeability involved in apoptotic process (GO:1902108). Among them, there are sub-processes in which changes in gene expression occurred in all MPS types/subtypes (Table 2). The highest number of genes with changed levels of transcripts in MPS could be classified into following terms: apoptotic signaling pathway, negative regulation of apoptosis, positive regulation of apoptosis, regulation of apoptotic process, and apoptotic mitochondrial changes. This indicates that disturbed regulation of expression of genes involved in the control of apoptosis may considerably contribute to changes occurring in this process in MPS cells.
| Number of significantly changed transcripts in MPS vs. HDFa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Subprocesses of apoptotic process | MPS I | MPS II | MPS IIIA | MPS IIIB | MPS IIIC | MPS IIID | MPS IVA | MPS IVB | MPS VI | MPS VII | MPS IX |
| Apoptotic mitochondrial changes | 11 | 6 | 6 | 8 | 10 | 5 | 3 | 9 | 6 | 10 | 9 |
| Apoptotic process involved in development | 1 | 0 | 0 | 0 | 3 | 1 | 0 | 0 | 0 | 0 | 1 |
| Apoptotic signaling pathway | 24 | 10 | 22 | 25 | 28 | 12 | 9 | 32 | 9 | 27 | 20 |
| Cysteine-type endopeptidase activity | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| Execution phase of apoptosis | 2 | 1 | 2 | 3 | 3 | 2 | 1 | 4 | 0 | 2 | 2 |
| Negative regulation of apoptosis | 12 | 5 | 8 | 15 | 12 | 5 | 4 | 11 | 5 | 17 | 10 |
| Negative regulation of mitochondrial membarne | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 1 | 0 | 1 |
| Peptidase activator activity | 0 | 0 | 0 | 3 | 0 | 0 | 1 | 2 | 0 | 1 | 0 |
| Positive regulation of apoptosis | 16 | 7 | 13 | 27 | 21 | 10 | 6 | 24 | 7 | 20 | 21 |
| Positive regulation of mitochondrial membrane | 1 | 1 | 2 | 0 | 1 | 1 | 1 | 2 | 2 | 0 | 3 |
| Regulation of apoptotic process | 5 | 1 | 9 | 10 | 8 | 7 | 5 | 10 | 3 | 7 | 7 |
| Regulation of mitochondrial membrane | 1 | 1 | 3 | 1 | 2 | 1 | 1 | 3 | 2 | 0 | 3 |
When considering individual transcripts, we could identify six genes in which expression was significantly changed in most (at least seven) MPS types/subtypes. This group of genes consists of BNIP3, C1D, CLU, GPER1, KREMEN1, and PRKCD (Table 3). Five of them were down-regulated in all MPS cell lines, and only CLU revealed enhanced expression in fibroblasts derived from MPS patients (Table 3).
| High-level changes in expression of selected genes in MPS vs. HDFa line (log2 FC) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | MPS I | MPS II | MPS IIIA | MPS IIIB | MPS IIIC | MPS IIID | MPS IVA | MPS IVB | MPS VI | MPS VII | MPS IX |
| BNIP3 | −0.8334 | −0.9357 | −0.8637 | −0.7140 | −0.7428 | −0.8010 | −0.6969 | ||||
| C1D | −1.2118 | −1.1290 | −1.4911 | −1.0865 | −1.4216 | −1.0683 | −1.3847 | −1.3832 | −1.0058 | −1.0096 | |
| CLU | 3.1058 | 3.3361 | 2.3442 | 3.4564 | 2.2600 | 3.8391 | 2.6958 | 2.2554 | 3.5694 | ||
| GPER1 | −1.4122 | −1.4569 | −1.8559 | −1.3795 | −0.9619 | −1.5090 | −1.5772 | ||||
| KREMEN1 | −1.3091 | −1.4533 | −3.0510 | −2.1959 | −2.0751 | −2.5427 | −1.3353 | −2.2162 | |||
| PRKCD | −1.2062 | −1.2300 | −0.7378 | −0.6532 | −0.9003 | −1.3367 | −0.8439 | ||||
We asked what genes are characterized by especially severe changes in expression in MPS cells relative to healthy fibroblasts. Therefore, we have identified transcripts with over fourfold change (log2FC > 2) in MPS versus HDFa fibroblasts (Table 4). Such genes occur in every MPS type/subtype. When considering sub-processes, the highest number of particularly significantly changed genes could be classified to the same terms as indicated for all genes, which revealed differences in expression in MPS cells (Table 5). This confirms that regulation of apoptosis is significantly disturbed in all types of MPS.
| Number of genes with high-level (log2 FC > 2.0) changes in expression in MPS vs. HDFa line | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | MPS I | MPS II | MPS IIIA | MPS IIIB | MPS IIIC | MPS IIID | MPS IVA | MPS IVB | MPS VI | MPS VII | MPS IX |
| Up-regulated | 3 | 2 | 2 | 4 | 4 | 2 | 1 | 2 | 1 | 2 | 2 |
| Down-regulated | 4 | 0 | 1 | 5 | 2 | 5 | 2 | 2 | 2 | 3 | 4 |
| Total | 7 | 2 | 3 | 9 | 6 | 7 | 3 | 4 | 3 | 5 | 6 |
| Genes with high-level (log2FC>2.0) changes in expression in MPS vs. HDFa line | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Subprocesses of apoptotic process | MPS I | MPS II | MPS IIIA | MPS IIIB | MPS IIIC | MPS IIID | MPS IVA | MPS IVB | MPS VI | MPS VII | MPS IX |
| Apoptotic mitochondrial changes | CLU↑ | CLU↑ | – | CLU↑ | CLU↑ | – | CLU↑ | CLU↑ | CLU↑ | CLU↑ | BBC3↑ |
| SOD2↓ | CLU↑ | ||||||||||
| Apoptotic signaling pathway | CLU↑ | ||||||||||
| CRIP1↑ | CLU↑ | CLU↑ | CLU↑ | ||||||||
| CAV1↓ | CLU↑ | CRIP1↑ | INHBA↑ | MKNK2↑ | CLU↑ | CRIP1↑ | CLU↑ | CLU↑ | BBC3↑ | ||
| CDKN1A↓ | CRIP1↑ | TNFRSF1B↓ | SOD2↓ | SOD2↓ | – | CDKN1A↓ | CAV1↓ | CDKN1A↓ | MLLT11↑ | CLU↑ | |
| SOD2↓ | |||||||||||
| Execution phase of apoptosis | – | – | – | – | – | – | – | – | – | – | BBC3↑ |
| Negative regulation of apoptosis | CLU↑ | CLU↑ | – | CLU↑ | CLU↑ | AKT1↓ | CLU↑ | CLU↑ | CLU↑ | CLU↑ | CLU↑ |
| CDKN1A↓ | TNFAIP3↑ | ||||||||||
| SOD2↓ | SOD2↓ | SOD2↓ | CDKN1A↓ | CDKN1A↓ | HSPD1↓ | ||||||
| Positive regulation of apoptosis | CLU↑ | CLU↑ | CLU↑ | ||||||||
| CAV1↓ | CLU↑ | TNFRSF1B↓ | INHBA↑ | IGFBP3↑ | IGFBP3↑ | CLU↑ | CLU↑ | CLU↑ | CLU↑ | CLU↑ | |
| SOD2↓ | SOD2↓ | SOD2↓ | AKT1↓ | IGFBP3↑ | CAV1↓ | IGFBP3↑ | MLLT11↑ | HSPD1↓ | |||
| Positive regulation of mitochondrial membrane | – | – | – | – | – | – | – | – | – | – | BBC3↑ |
| Regulation of apoptotic process | CDKN1A↓ | – | – | – | – | AKT1↓ | CDKN1A↓ | – | CDKN1A↓ | – | – |
| SGK1↓ | |||||||||||
| Regulation of mitochondrial membrane | – | – | – | – | – | – | – | – | – | – | BBC3↑ |
When considering individual genes with log2FC > 2 in MPS, it is interesting that most of them is either down- or up-regulated in all types of the disease (Table 6). This indicates that apoptotic changes may be similar in all or most MPS types/subtypes.
| High-level changes (log2FC > 2.0) in expression of selected genes in MPS vs. HDFa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | MPS I | MPS II | MPS IIIA | MPS IIIB | MPS IIIC | MPS IIID | MPS IVA | MPS IVB | MPS VI | MPS VII | MPS IX |
| AKT1 | – | – | – | – | – | ↓ | – | – | – | – | – |
| BBC3 | – | – | – | – | – | – | – | – | – | – | ↑ |
| CAV1 | ↓ | – | – | – | – | – | – | ↓ | – | – | – |
| CDKN1A | ↓ | – | – | – | – | – | ↓ | – | ↓ | – | – |
| CLU | ↑ | ↑ | – | ↑ | ↑ | – | ↑ | ↑ | ↑ | ↑ | ↑ |
| COMP | – | – | – | – | ↑ | ↑ | – | – | – | – | – |
| CRIP1 | ↑ | ↑ | ↑ | – | – | – | – | ↑ | – | – | – |
| DHCR24 | – | – | – | – | – | – | – | – | – | ↓ | – |
| DRAM1 | – | – | – | ↓ | – | – | – | – | – | ↓ | – |
| IGFBP3 | – | – | – | – | ↑ | ↑ | ↓ | – | ↓ | – | – |
| INHBA | – | – | – | ↑ | – | – | – | – | – | – | – |
| HSPD1 | – | – | – | – | – | – | – | – | – | – | ↓ |
| KPNB1 | – | – | – | – | – | ↓ | – | – | – | – | – |
| KREMEN1 | – | – | – | ↓ | ↓ | ↓ | – | ↓ | – | – | ↓ |
| MKNK2 | – | – | – | – | ↑ | – | – | – | – | – | – |
| MLLT11 | – | – | – | – | – | – | – | – | – | ↑ | – |
| PEG10 | – | – | – | – | – | ↓ | – | – | – | – | ↓ |
| SGK1 | – | – | – | – | – | ↓ | – | – | – | – | – |
| SOD2 | ↓ | – | – | ↓ | ↓ | – | – | – | – | – | – |
| SULF1 | ↑ | – | ↑ | – | – | – | – | – | – | – | – |
| TNFAIP3 | – | – | – | ↑ | – | – | – | – | – | – | – |
| TNFRSF11B | ↓ | – | ↓ | ↓ | – | – | – | – | – | ↓ | – |
| TMBIM4 | – | – | – | – | – | – | – | – | – | – | ↓ |
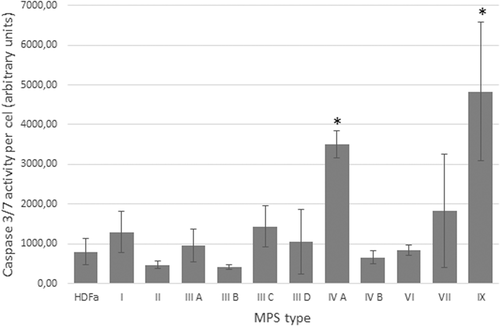
To test if the apoptosis process is actually affected in MPS cells relative to control cells in our experimental system, the activity of caspase 3/7 was measured in cultured fibroblasts. We found statistically significant increase in caspase 3/7 activity in MPS IVA and MPS IX, relative to HDFa (Figure 1). As these experiments were performed under non-stress conditions, it appears that apoptosis may be constantly stimulated in at least some MPS types, indeed.
Discussion
Although induction of apoptosis and dysregulation of this process in MPS have been reported previously by various authors (Simonaro et al., 2001; Villani et al., 2007; Richard et al., 2008; Tessitore et al., 2009; Arfi et al., 2011; Noguti et al., 2012; Fusar Poli et al., 2013; Viana et al., 2017; Bar et al., 2018), molecular mechanisms of these changes are not elucidated yet. Some authors demonstrated changes in the expression of apoptosis-related genes in MPS cells (Villani et al., 2007; Richard et al., 2008; Arfi et al., 2011). However, one type of MPS has been investigated in each of those works, and usually, only specific genes were tested. Therefore, in this work, we aimed to study cells of all types/subtypes of MPS and to analyze whole transcriptomes.
Results presented in this report indicated that expression of a battery of apoptosis-related genes is significantly changed in all MPS types. When considering sub-processes, according to the QuickGO database, it appeared that the most abundant changes occurred in genes classified in following terms apoptotic signaling pathway, negative regulation of apoptosis, positive regulation of apoptosis, regulation of apoptotic process, and apoptotic mitochondrial changes. This strongly suggests that regulation of apoptosis is significantly changed in MPS cells, which leads to overstimulation of this programmed cell death process. Although individual MPS types/subtypes differ in patterns of expressions of various apoptosis-related genes, general patterns might be considered similar, and we could find several genes in which expression is specifically dysregulated in most MPS types. This group of genes consists of BNIP3, C1D, CLU, GPER1, KREMEN1, and PRKCD. One might ask if these genes are responsible for similarity of apoptotic processes occurring in various MPS types.
The BNIP3 protein is closely related to another hypoxia-inducible protein NIX (for a review, see Rodger et al., 2018). Both are considered as mitophagy receptors in mammalian cells. BNIP3 can interact with the LC3/GABARAP complex, which is involved in the autophagy process (Novak et al., 2009; Rodger et al., 2018). In fact, a lack of BNIP causes impairment in mitophagy, leading to accumulation of dysfunctional mitochondria and enhanced production of reactive oxygen species (Chourasia et al., 2015). These changes can cause activation of apoptosis, thus, low-level expression of the BNIP3 gene, as observed in several MPS cell lines (Table 3) may promote apoptotic processes (see Burton and Gibson, 2009). This gene has been demonstrated to be regulated mostly at the transcription level, and in fact, its function has been demonstrated to be involved in the regulation of cell death (Ma et al., 2017). Therefore, one can assume that decreased levels of BNIP3 expression in MPS cells contributes significantly to apoptosis induction. Moreover, it was suggested that apoptotic process is stimulated in MPS I due to down-regulation of the blc-2 gene expression in mice (Noguti et al., 2012) while BNIP3 is known to be a BCL-2-interacting protein, and has been named after this feature (Vasagiri and Kutala, 2014).
Expression of the C1D gene is down-regulated in almost all MPS types (Table 3). This gene codes for a protein involved in maintenance of genomic stability (Jackson et al., 2016). The C1D coordinates actions of proteins involved in functions of exosomes and double-strand break (DSB) DNA repair system by recruiting regulatory factors (Jackson et al., 2016). Therefore, it is likely that low-level expression of the C1D gene may contribute to dysregulation of apoptosis in MPS.
The CLU gene is highly overexpressed in nine out of 11 MPS types/subtypes (Table 3). The product of this gene, clusterin, is a multifunctional glycoprotein occurring in two forms, nuclear and secretory (Garcia-Aranda et al., 2018). The nuclear form promotes apoptosis, most probably by sequestering the Bcl-XL protein, thus, releasing Bax and activating caspase 3 and cytochrome c release (Kim et al., 2012). Therefore, up-regulation of CLU in MPS cell lines can be considered as one of the crucial processes leading to apoptosis stimulation.
GPER1 is a G protein-coupled estrogen receptor (Hsu et al., 2019). Although cellular functions of this protein are still not clear, it was reported that it may have a pro-survival role, at least in some types of cells (Nilsson et al., 2011). Hence, decreased levels of mRNA of the GPER1 gene may contribute to enhanced apoptosis in MPS cells.
KREMEN1 codes for the Kremen 1 protein, which acts as a receptor of Dickkopf 1, the Wnt antagonist (Causeret et al., 2016). Kremen 1 has been proposed to be a pro-apoptotic protein (Causeret et al., 2016). Therefore, down-regulation of the gene coding for this protein in MPS cells (Table 3) might be considered hardly explainable in the light of apoptosis stimulation observed by others in this disease. However, recent studies indicated that Kremen 1 actually triggers apoptosis unless it is bound to Dickkopf 1, its ligand, because homodimerization of the receptor is required to transmit the apoptotic signal (Sumia et al., 2019). Binding of Dickkopf 1 results in inhibition of multimerization of Kremen 1, allowing its dimerization (Sumia et al., 2019). Therefore, we propose that decreased level of Kremen 1 in MPS cells, due to down-regulation of expression of its gene, may prevent its natural tendency to multimerization, allowing formation of dimers (like in the presence of Dickkopf 1), and resulting in stimulation of transduction of the signal causing apoptosis induction.
Levels of PRKCD transcripts are decreased in many MPS cell lines (Table 3), thus decreased amounts of the protein kinase Cδ, the gene product, can be assumed. Although it was demonstrated that overproduction and/or activation of this kinase causes apoptosis induction, there are also reports showing anti-apoptotic and pro-survival functions of this enzyme (discussed by Zhao et al., 2012). Apparently, the role of protein kinase Cδ in stimulation or inhibition of apoptosis depends on the cell type and physiological conditions; in fact, such conditions in MPS cells are definitely changed relative to healthy cells.
In conclusion, results presented in this study indicate that expression of many genes coding for proteins related to apoptosis is significantly changed in cells of all types of MPS. Some of these genes are dysregulated similarly in most of MPS types/subtypes, suggesting common mechanisms of apoptosis induction. In fact, analysis of expression of such genes (BNIP3, C1D, CLU, GPER1, KREMEN1, and PRKCD) provided a basis for proposal of putative molecular pathways, which may lead to stimulation of apoptosis in MPS cells, observed previously by others. This transcriptomic study might, therefore, open a new way to study molecular mechanisms of apoptotic dysregulation in MPS.
Acknowledgments and funding
This work was supported by National Science Center, Poland (project grant no. 2017/25/B/NZ2/00414 to G.W.).
Conflict of interest
The authors declare no conflict of interest.




