lncRNA KIAA0125 functions as a tumor suppressor modulating growth and metastasis of colorectal cancer via Wnt/β-catenin pathway
Abstract
Colorectal cancer (CRC) is the third most common cancer in all races worldwide in recent years. The survival of the CRC patients is mostly affected by the stage of the disease at the time of diagnosis. Thus, the current challenge is to find sensitive and reliable biomarkers in early screening of CRC. Recently, emerging evidence has shown that long non-coding RNAs (lncRNAs) may play crucial roles in tumorigenesis. In this study, we found that lncRNA KIAA0125 was downregulated in colorectal tissues and cells. The functional study demonstrated that overexpression of KIAA0125 suppressed cell proliferation, migration, and invasion whereas the reversal effects were seen in silencing experiment. Besides, KIAA0125 inhibited epithelial–mesenchymal transition through Wnt/β-catenin signaling in CRC. Our findings suggested that KIAA0125 may act as an oncosupressor gene and could be considered as a potential diagnosis biomarker in CRC.
Abbreviations
-
- AML
-
- acute myeloid leukemia
-
- APC
-
- adenomatous polyposis coli
-
- CCK-8
-
- cell counting kit-8
-
- CeRNA
-
- competing endogenous RNAs
-
- CRC
-
- colorectal cancer
-
- EMT
-
- epithelial–mesenchymal transition
-
- FBS
-
- fetal bovine serum
-
- LncRNA
-
- long non-coding RNA
-
- PBS
-
- phosphate-buffered saline
-
- qRT-PCR
-
- quantitative real-time polymerase chain reaction
-
- SEM
-
- standard error mean
Introduction
In recent years, colorectal cancer (CRC) becomes the third most common cancer in all races worldwide. The incidence rates decreased since 1999 but became stable after 2012 (Cronin et al., 2018). Most cases are sporadic and less than 10% of cases are hereditary autosomal dominant disorders (non-polyposis colon cancer and familial adenomatous polyposis coli [APC]) (Brenner et al., 2014). The survival of CRC patients is mostly affected by the stage of the disease at the time of diagnosis (Vatandoost et al., 2016). Thus, the early screening is quite necessary for improving the clinical outcome of patients. In the current clinical diagnosis, colonoscopy remains the gold-standard diagnostic test to recognize colonic pathology, however, it is not prevalent due to its invasive and expensive attributes (Heiss and Brenner, 2017; Januszewicz and Fitzgerald, 2019).
The molecular biomarkers of CRC are starting to play an important role in the detection of colorectal malignancies these years by the easy practice and low cost of sequencing-based technologies (Januszewicz and Fitzgerald, 2019). Researchers previously focused on gene mutations and proteins that function as oncoproteins or tumor suppressors, such as p53, KRAS, and APC (Huh et al., 2013; Coppedè et al., 2014; Wang et al., 2015). Most recently, due to the development of bioinformatics methods, many non-coding transcripts have been identified and attracted an increasing attention because of their biological significance in cancers (Mercer et al., 2009; Ferracin et al., 2016). These studies provided new insights into the opportunity for diagnosis and therapy of cancers.
Long non-coding RNAs (lncRNAs), one kind of transcripts containing no less than 200 nucleotides, have been found to be differentially expressed in CRC tissues (Xue et al., 2015). H19, the first CRC-related lncRNA was identified, acted as an oncogenic lncRNA that abundantly expressed in CRC (Hibi et al., 1996). Since then, many other lncRNA have been discovered to be involved in all stages of CRC (Yang et al., 2017).
The lncRNA KIAA0125 (on chromosome 14q32.33) was found aberrantly expressed and functionally involved in different cancers and diseases (Yao et al., 2017; Diniz et al., 2018; Hornung et al., 2018). However, its expression and function was never explored in CRC. In our present study, we observed that KIAA0125 was downregulated in CRC tissues and cells. The overexpression of KIAA0125 suppressed cell proliferation, migration, and invasion whereas KIAA0125 silencing promoted cell proliferation, migration, and invasion in HCT116 and SW480 cells. In the meanwhile, the functions of KIAA0125 in epithelial–mesenchymal transition (EMT) were through wnt/β-catenin signaling. Our results suggested that KIAA0125 acted as an oncosuppressor gene in CRC and might provide a new insight into CRC clinical diagnosis and treatment.
Materials and methods
Patient samples
Twenty-five pairs of human colorectal carcinoma tissues and adjacent normal colon tissues were collected from China to Japan union hospital of Jilin University. For each patient, at least three samples were collected from different parts of cancer tissues. The patients were diagnosed with CRC based on the histopathological detection and divided into early stage (I/II) group (n = 13) and late stage (III/IV) group (n = 12). All the patients did not undergo preoperative therapy prior to the surgical resection and volunteered to participate in the research by written consent form. This study was approved by the ethic committee of Jilin University and all the procedures were performed according to the approved protocol.
Real-time quantitative polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the CRC tissues or cells with TRIzol reagent (Invitrogen, USA) according to the manufacturer's protocols. After concentration measurement, the RNA was reverse transcribed into complementary DNA (cDNA) and analyzed by qRT-PCR. The targeted gene expression was normalized with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and calculated using 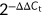 method. The primer sequences were used as follows: KIAA0125: 5′-TGGCAAAGGCAAGTGAC-3′ and 5′-GGCAGAAGGATGAACCC-3′; GAPDH: 5′-TGACAACTTTGGTATCGTGGAAGG-3′ and 5’-AGGCAGGGATGATGTTCTGGAGAG-3′; β-catenin 5′-GGCAGCAACAGTCTTACCT-3′ and β-catenin 5′-CATACAGGACTTGGGAGGT-3′.
method. The primer sequences were used as follows: KIAA0125: 5′-TGGCAAAGGCAAGTGAC-3′ and 5′-GGCAGAAGGATGAACCC-3′; GAPDH: 5′-TGACAACTTTGGTATCGTGGAAGG-3′ and 5’-AGGCAGGGATGATGTTCTGGAGAG-3′; β-catenin 5′-GGCAGCAACAGTCTTACCT-3′ and β-catenin 5′-CATACAGGACTTGGGAGGT-3′.
Cell culture and cell transfection
FHC, HCT116, LS174T, SW480, SW620, and LOVO cell lines were obtained from the American Type Culture Collection (ATCC). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotics and maintained at 37°C in a humidified incubator with 5% CO2.
si-KIAA0125 (5′-GGAGCAAAUGGAGAGGGAA-3′) or scramble small interfering RNA (siRNA) (5′-GGAAAUAGGGAGAGGCGAA-3′) was transiently transfected into cells using Lipofectamine RNAi Max (Invitrogen) according to the manufacturer's instructions. For the lncRNA-KIAA0125 overexpressing construct, the full-length lncRNA-KIAA0125 was cloned into pcDNA3.1 vector (Invitrogen) between HindIII and BamHI sites. The primer sequences are as follows: forward, 5′-CCCAAGCTTAGGGTTGTCTGGATGGGCAG-3′ and reverse, 5′-CGCGGATCCTTAGCCTACAAATTTAATGTAA-3′. pcDNA3.1-KIAA0125 or pcDNA3.1 empty vector was delivered into cells using Lipofectamine 2000 (Invitrogen) transfection reagent according to manufacturer's protocol.
Cell counting kit-8 (CCK-8) cell proliferation assay
The CCK-8 assay was used to define HCT116 or SW480 cells viability under KIAA0125 knockdown or overexpression. The cells were seeded into 96-well plates and cultured for the indicated time points. Ten microliters of CCK-8 reagent was added into each well and incubated for another 2 h. The optical density of solution was detected at 450 nm.
The scratch assay
HCT116 and SW480 cells were seeded into a 24-well plate and reached 80–90% density after 24 h of growth. A linear wound was scratched across the center of the well using a pipette tip. The wound closure rate was measured after 36 h.
Transwell assay
The invasion and migration of HCT116 and SW480 cells was detected using matrigel-coated or non-coated chambers with a pore size of 0.8 μM. The cells were seeded into upper chamber in DMEM with 1% FBS and DMEM with 10% FBS in the lower chamber was added as chemoattractant. After 36 h incubation, cells in the upper chamber were removed and cells in the lower side were fixed with 4% paraformaldehyde and stained by 1% crystal violet in phosphate-buffered saline.
Western blotting
The cellular proteins were extracted in 2× Laemmli buffer supplemented with protease inhibitor. Nuclear and cytoplasmic proteins were separated using cytoplasmic extract buffer (10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], 60 mM KCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 0.075% NP40, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1 mM dithiothreitol, pH 7.6) and nuclear extract buffer (20 mM Tris-HCl, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM PMSF, and 25% glycerol, pH 8.0). The proteins were analyzed by gel electrophoresis system and transferred to the polyvinylidene fluoride membrane. After blocking in 3% bovine serum albumin, the membrane was incubated with primary antibodies at 4° overnight followed by incubation with peroxidase-conjugated secondary antibody for one hour at room temperature. The protein signals were detected and the intensity of the bands was quantified using Image Lab Software (Bio Rad, China).
Statistical analysis
All data are expressed as mean ± standard error mean (SEM). The difference between two groups was compared using Student's t test and the differences among multiple groups were analyzed using one-way analysis of variance. A P < 0.05 was considered as statistically significant (*P < 0.05 and **P < 0.01).
Results
The expression of KIAA0125 is downregulated in CRC tissues and cells
LncRNA KIAA0125 was found differentially expressed in many cancers (Yao et al., 2017; Diniz et al., 2018; Hornung et al., 2018). From our previous RNA-sequencing bioinformatic analysis, lncRNA-KIAA0125 was differentially expressed in several carcinomas (data unpublished). To understand the role of KIAA0125 in the CRC, we examined the expression of KIAA0125 in both CRC tissues and CRC cells. The 25 pairs of tumor and paratumor tissues were analyzed by qRT-PCR and the expression level of KIAA0125 was normalized by GAPDH. As shown in Figure 1A, the expression of KIAA0125 was significantly downregulated in CRC tissues when compared with adjacent normal tissues. Besides, the relative expression of KIAA0125 in each patient showed different levels of decrease as indicated in Figure 1B. Moreover, we classified the CRC tissues into two groups (Stage I/II as early stage and Stage III/IV as late stage). The expression levels of KIAA0125 in Stage III/IV (n = 12) were much lower than in Stage I/II (n = 13), which suggested that the expression of KIAA0125 correlated with the clinical cancer development (Figure 1C). Next, we analyzed the expression of KIAA0125 in CRC cell lines (HCT116, LS174T, SW480, SW620, and LOVO) and human normal colon epithelial cell line fetal human cell (FHC). The expression levels of KIAA0125 in CRC cells were also lower than in FHC cells (Figure 1D). Collectively, the above results demonstrated that KIAA0125 was significantly downregulated in CRC tissues and cells.
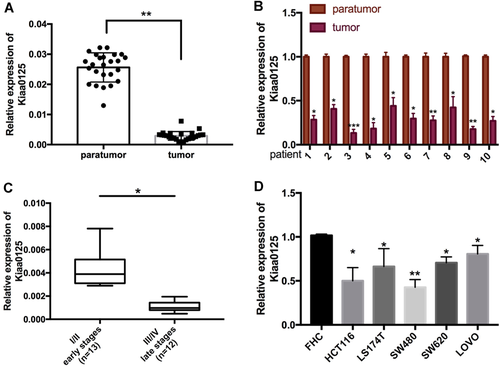
KIAA0125 is downregulated in colorectal cancer (CRC) tissues and cells. (A) The expression of KIAA0125 was determined by quantitative real-time polymerase chain reaction (qRT-PCR) in CRC tissues and adjacent normal tissues (n = 25). (B) The individual expression level of KIAA0125 was examined by qRT-PCR in 10 pairs of CRC samples and adjacent normal tissues. (C) The expression of KIAA0125 was compared between Stage I/II (n = 13) group and Stage III/IV (n = 12) group. (D) The expression levels of KIAA0125 were determined by qRT-PCR in human normal colon epithelial cell line (FHC) and CRC cell lines (HCT116, LS174T, SW480, SW620, and LOVO). *P < 0.05, **P < 0.01.
Overexpression of KIAA0125 suppresses cell proliferation, migration, and invasion in CRC
To elucidate the mechanism of KIAA0125 in cell proliferation, migration, and invasion, we transfected KIAA0125 expressing plasmid or empty vector into HCT116 and SW480 cells, which were chosen for further study because of their relatively low expression levels of KIAA0125 (Figure 2A). For cell proliferation, KIAA0125 overexpression led to growth retardation of HCT116 and SW480 cells, which was confirmed by CCK-8 assays (Figure 2B). Since cell motility and consequent invasion have been recognized as the major features during cancer progression (Reya et al., 2001), we further investigated the functions of KIAA0125 in cell migration and invasion. In wound healing assays, the wound closure rates in HCT116 and SW480 cells were delayed by KIAA0125 overexpression (Figures 2C and 2D). Moreover, the property of invasion was determined by transwell assays. The results indicated that KIAA0125 overexpression suppressed their invasive potential (Figures 2E and 2F). Overall, these data indicated that KIAA0125 acted as a tumor suppressor gene that suppressed cell proliferation, migration, and invasion in CRC cells.
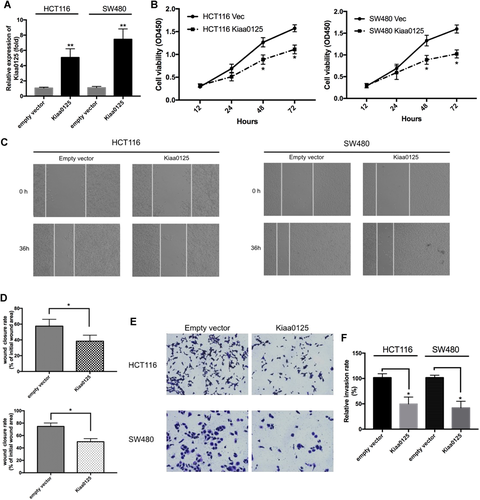
Overexpression of KIAA0125 suppresses cell proliferation, migration, and invasion in colorectal cancer (CRC). (A) quantitative real-time polymerase chain reaction (qRT-PCR) analysis of KIAA0125 in HCT116 and SW480 cells transfected with empty vector or KIAA0125 expressing plasmids. *P < 0.05, n = 3. (B) The effect of KIAA0125 overexpression on cell proliferation at the indicated time points was evaluated in HCT116 (left) and SW480 (right) cells by cell counting kit-8 (CCK-8) assays. *P < 0.05, n = 3. (C and D) Wound healing assay was performed to evaluate the migration of HCT116 and SW480 cells after overexpressing empty vector or KIAA0125 plasmids. Cell migration was determined by the wound closure rate. *P < 0.05, n = 3. (E and F) The effect of KIAA0125 overexpression on cell invasion was assessed by Transwell assay with matrigel-coated chambers. The relative invasion rate was calculated from five random microscopic fields in each experiment. *P < 0.05, n = 3.
Knockdown of KIAA0125 promotes cell proliferation, migration, and invasion in CRC
To further confirm the functions of KIAA0125 in cell proliferation, migration, and invasion, we used siRNA to knockdown endogenous KIAA0125. The pre-designed siRNA effectively silenced KIAA0125 in HCT116 and SW480 cells (Figure 3A). After siRNA transfection for 72 h, the cell viability was increased in HCT116 and SW480 cells when compared with scramble siRNA transfection groups, which suggested that KIAA0125 silencing promoted cell proliferation (Figure 3B). Next, we examined the potential of cell migration and invasion under KIAA0125 knockdown using transwell assays with or without matrigel coating, respectively. As shown in Figures 3C, 3D, and 3E, KIAA0125 silencing increased the migratory and invasive ability of HCT116 and SW480 cells. In summary, KIAA0125 was directly involved in the regulation of cell proliferation, migration, and invasion in CRC cells.
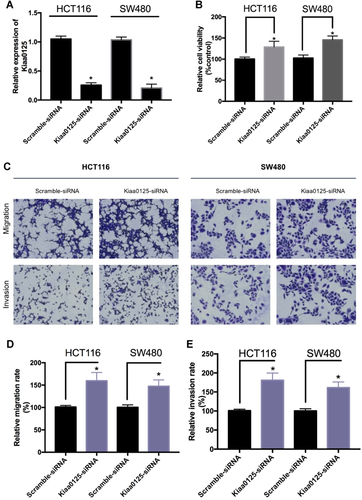
Knockdown of KIAA0125 promotes cell proliferation, migration, and invasion in colorectal cancer (CRC). (A) quantitative real-time polymerase chain reaction (qRT-PCR) analysis of KIAA0125 in HCT116 and SW480 cells transfected with scramble small interfering RNA (siRNA) or KIAA0125 siRNA. *P < 0.05, n = 3. (B) The effect of KIAA0125 knockdown on cell proliferation was evaluated in HCT116 and SW480 cells by cell counting kit-8 (CCK-8) assays. *P < 0.05, n = 3. (C, D, and E) The effect of KIAA0125 knockdown on the migration and invasion in HCT116 and SW480 cells was assessed by Transwell migration and invasion assay, respectively. The relative migration (D) and invasion (E) rate was calculated from five random microscopic fields in each experiment. *P < 0.05, n = 3.
KIAA0125 inhibits EMT through Wnt/β-catenin signaling in CRC
EMT has been demonstrated as a key program and been activated during cancer development (Mani et al., 2008). Among several signaling pathways, Wnt signaling pathway has been shown to directly contribute to EMT (Gasior et al., 2017). Thus, we first examined the expression of several EMT-related proteins under the condition of ectopic KIAA0125 expression in HCT116 and SW480 cells. As shown in Figures 4A and 4B, the epithelial marker, E-cadherin, was increased and mesenchymal markers, N-cadherin and vimentin, were decreased after KIAA0125 overexpression. In addition, the expression of matrix metalloproteinase-2, which represented the cell invasive potential was decreased after KIAA0125 overexpression. Next, we explored the effects of KIAA0125 overexpression on β-catenin, which is the key regulator in Wnt canonical pathway (MacDonald et al., 2009). The overexpression of KIAA0125 suppressed β-catenin at both messenger RNA (mRNA) level (Figure 4C) and protein level (Figures 4D and 4E). We then analyzed myc and cyclin D1, which are the targets of β-catenin. The levels of protein expression were also decreased after KIAA0125 overexpression. Taken together, the above data suggested that KIAA0125 exerted functions by regulating Wnt/β-catenin signaling in CRC cells.
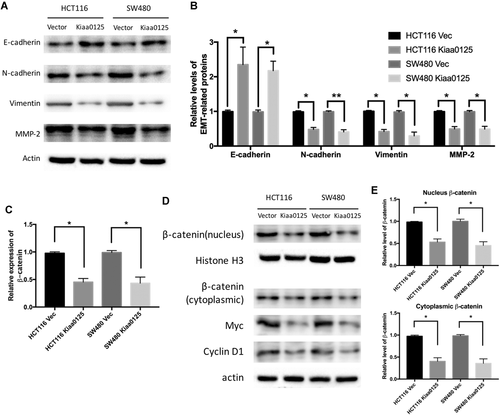
KIAA0125 inhibits epithelial–mesenchymal transition (EMT) through Wnt/β-catenin signaling in colorectal cancer (CRC). (A and B) Western blot analysis of EMT-related proteins (E-cadherin, N-cadherin, vimentin, and matrix metalloproteinase-2 [MMP-2]) in HCT116 and SW480 cells after empty vector or KIAA0125 plasmids transfection. The relative protein level was normalized by actin. *P < 0.05, **P < 0.01. n = 3. (C) quantitative real-time polymerase chain reaction (qRT-PCR) analysis of β-catenin in HCT116 and SW480 cells transfected with empty vector or KIAA0125 expressing plasmids. (D and E) Representative immunoblots of nuclear and cytoplasmic β-catenin, total myc, and cyclin D1 protein expression in empty vector and KIAA0125 overexpression group in HCT116 and SW480 cells. The relative nuclear protein level was normalized by Histone H3 and cytoplasmic protein level was normalized by actin. *P < 0.05, n = 3.
Discussion
With the advances of sequencing technologies, many differentially expressed non-coding transcripts have been identified in cancers. In the further functional studies, many of them were demonstrated to be involved in tumorigenesis, cancer progression and metastasis (Li et al., 2017). Most recently, numerous studies working on lncRNAs show that they play critical roles in different cancers acting as oncogenes or tumor suppressors (Dey et al., 2014). In our study, we found lncRNA KIAA0125 was significantly downregulated in CRC tissues and associated with stages of CRC. The relative expression in CRC cells was also decreased, which is consistent with the results in CRC samples.
lncRNA KIAA0125 has been studied in different diseases. It exerts a regulatory effect in neurogenesis and may be responsible for Abeta ratio in Alzheimer disease (Uhrig et al., 2009). Some studies have shown that KIAA0125 as a mediator gene correlated with the survival of acute myeloid leukemia (Hornung et al., 2018). In the meanwhile, the expression pattern of KIAA0125 in tumors also seems inconsistent. It is upregulated in ameloblastomas but the function has not been characterized (Diniz et al., 2018). The role of KIAA0125 in gallbladder cancer is not certainly defined because of the original related article was retracted (International BR, 2017). The bioinformatics analysis has shown KIAA0125 is downregulated in pancreatic cancer, but further investigations are required to elucidate the function (Yao et al., 2017). Therefore, the role of KIAA0125 in tumors may be cell-type dependent, although this possibility needs to be further investigated. Our results suggest that KIAA0125 may act as an oncosuppressor gene in CRC cells since overexpression of KIAA0125 significantly supressed cell growth and the potentials of invasion and migration whereas the reversal effects were showed in knockdown experiments. We chose HCT116 and SW480 cell lines because of the relative low expression that close to the results we found in tumor tissues. Also, the knockdown experiments indicated the low expressed KIAA0125 still obtained the function in cell proliferation, migration, and invasion. Thus, the relative low expression in CRC tissues and cells suggested loss of function and KIAA0125 may exist putative roles in balancing the cell microenvironment.
We also investigated the roles of KIAA0125 in EMT, the major phenotype happened in the tumorigenic process (Brabletz et al. 2018). Our results demonstrated that KIAA0125 suppressed the expression of EMT-related proteins and the regulation was through the canonical wnt/β-catenin pathway. In other studies, the main function of lncRNAs in the cancers is to serve as competing endogenous RNAs. We will focus on the KIAA0125-miRNA-mRNA networks in our further investigations.
Conclusions
In summary, our data demonstrated that KIAA0125 was downregulated in CRC tissues and cells. The ectopic expression of KIAA0125 inhibited the proliferation, migration, and invasion, whereas silencing KIAA0125 got the reversal effects. Overexpression of KIAA0125 inhibited EMT via Wnt/β-catenin signaling pathway. These findings suggested that KIAA0125 may be considered as a diagnosis biomarker in CRC.
Acknowledgments
This study was supported by Health Technology Innovation Project of Jilin Province (No.3D517ED43430).
Conflict of interest
The authors declare they have no conflict of interests.




