Noncovalent Spin-Labeling of DNA and RNA Triplexes
Abstract
Studying nucleic acids often requires labeling. Many labeling approaches require covalent bonds between the nucleic acid and the label, which complicates experimental procedures. Noncovalent labeling avoids the need for highly specific reagents and reaction conditions, and the effort of purifying bioconjugates. Among the least invasive techniques for studying biomacromolecules are NMR and EPR. Here, we report noncovalent labeling of DNA and RNA triplexes with spin labels that are nucleobase derivatives. Spectroscopic signals indicating strong binding were detected in EPR experiments in the cold, and filtration assays showed micromolar dissociation constants for complexes between a guanine-derived label and triplex motifs containing a single-nucleotide gap in the oligopurine strand. The advantages and challenges of noncovalent labeling via this approach that complements techniques relying on covalent links are discussed.
Graphical Abstract
Introduction
Investigating the function of biomacromolecules usually requires a specific kind of monitoring. Most biomacromolecules, including nucleic acids, do not produce strong spectroscopic signals that distinguishes them from background signals and, therefore, monitoring them often requires labeling.1 Traditionally, radioactive labeling has been used to detect proteins or nucleic acids,2 but this method is hazardous, costly, and requires special training and laboratory settings. Alternatively, one or several chromophores can be covalently linked to the biomolecules of interest to obtain a UV/VIS signal that reports on the location, folding or binding state of a biomacromolecule.3, 4 Besides the need for selective labeling chemistry and post-synthetic purification, this changes the covalent structure of the analyte. Moreover, conjugation of labels to biopolymers usually calls for a considerable synthetic effort and skill. A complementary approach is noncovalent labeling. Here, binding motifs are engineered into the biomacromolecule of interest and small fluorogenic or otherwise detectable small molecules are added as ligands.
Aptamers are one class of nucleic acid binding motifs that have been used to bind fluorophores.5 A range of aptamers are available for this purpose.6, 7 Optimized aptamers that bind brightly fluorescent dyes allow for intracellular tracking of RNA via super-resolution, even for bacterial cells.8 Another class of binding motifs are based on rational design, for example by using canonical structures, such as duplexes and triplexes that contain nucleotide or nucleobase gaps as binding sites. This class of binders includes DNA duplexes with abasic sites that can bind larger aromatic ligands9 and modified DNA triplexes that act as sensors10, 11 or receptors12 for nucleobases. Triplexes with a single-nucleotide gap in the oligopurine segment have been shown to bind purine nucleotides with low micromolar affinity.13 The gap may be unbridged, as in four-strand triplexes, or it may be bridged by a linker. Further, the individual strands of the triplex may be covalently linked to produce intramolecular folding motifs. The high affinity of such rationally designed DNA or RNA triplex motifs for purine nucleotides has been used to capture and release cofactors14 and to modulate the level of a second messenger in vivo.15 Reinforcing the gap with linkers can improve binding,16 and the binding capacity can be optimized by introducing several binding sites per triplex.17 Rationally designed triplex binding motifs can be turned into functional materials operating in macroscopic devices by precipitating them with protamines.18 Thus far, however, the binding properties of DNA and RNA triplexes have not been used for spin labeling.
Nitroxide spin-labels are good reporter groups for electron paramagnetic resonance (EPR) spectroscopy, which is a useful spectroscopic technique for the study of the structure and dynamics of biomolecules.19-23 There are examples of noncovalent spin-labeling of nucleic acids, such as the malachite green aptamer24 and designed abasic sites in nucleic acid duplexes.25-29 Of several purine-nitroxide derivatives that were recently prepared for noncovalent labeling (Figure 1, 1–5 and Ǵ), Ǵ (G-spin) was found to have high affinity to abasic sites in RNA opposite C, with an equilibrium dissociation constant (Kd) of 1.46 ⋅ 10−7 m at 20 °C.26 Here, we report the evaluation of purine-nitroxides as spin labels for rationally designed purine-binding triple helices containing nucleotide gaps (I–III, Figure 1). The binding of the purine-derived spin labels to triplex-DNA was investigated by continuous wave (CW)-EPR spectroscopy and equilibrium filtration. The spin label Ǵ showed nearly full binding to triplexes II and III in the presence of Mg2+ ions at low temperatures, making it a viable spin-label for distance measurements with pulsed dipolar EPR spectroscopy, and equilibrium filtration assays with a DNA and an RNA triplex confirmed tight binding for Ǵ.
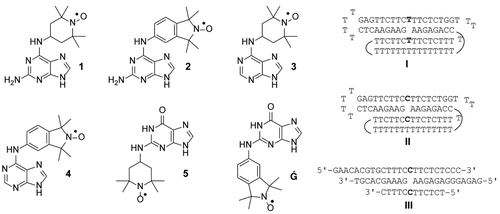
Purine-derived nitroxide spin labels (left) and sequences of purine-binding DNA triplex motifs (right). The curved lines in triplex motifs I and II indicate the connectivity of the T15 loops.
Results and Discussion
EPR Measurements
Three different DNA triplex motifs were evaluated as binders for the purine spin-labels shown in Figure 1 (1–5 and Ǵ). Triplex I was designed to bind adenine by introducing a dA gap opposite T while triplexes II and III were designed to be binders for guanine, with a dG nucleotide gap opposite dC. Triplex II differs from III, in that it contains two strands that form a triplex through intramolecular folding, rather than using four individual strands as in III. High concentrations of monovalent salt, such as NaCl, in addition to inclusion of divalent metal ions (e. g., Mg2+) are known to stabilize triplex helices and, therefore, we evaluated binding of the spin labels in a Mg2+-containing buffer.13
Figure 2 shows results from the evaluation of nitroxide binding to the DNA triplexes by EPR spectroscopy. Spin-label binding to the triplexes can be readily detected because of the large change in rotational correlation time of the nitroxide upon binding, which results in considerable broadening of the EPR spectrum. When partial binding takes place, the spectrum contains two components, the so-called ‘fast one’ corresponding to the unbound label and the slow component representing the bound label. The degree of binding can subsequently be determined by double integration of the spectra.29 In practice, the normalized spectra of the fully bound label and the unbound label are manually added together to get a best fit of the experimental spectrum.30, 31
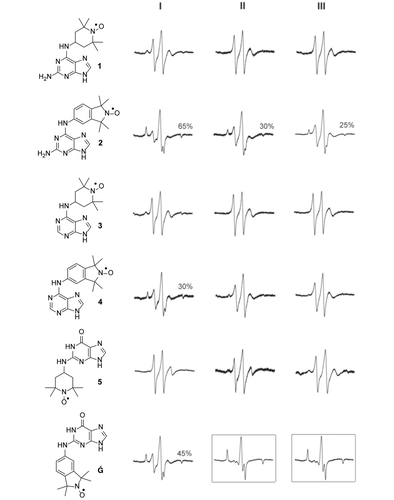
EPR spectra of spin labels Ǵ and 1–5 in the presence of DNA motifs I, II or III. EPR spectra inside the black boxes show nearly full binding of Ǵ to DNA triplexes II and III. Each spin label (1–5, Ǵ; 200 μm) was incubated with DNA motifs I–III (400 μm) in Breaker's buffer (20 mm Tris⋅HCl, 450 mm NaCl, 100 mm KCl, 10 mm MgCl2, 1 mm MnCl2, 5 mm CaCl2, pH 6.7) containing 30 % ethylene glycol and 2 % DMSO and the EPR spectra were recorded at −30 °C.
The TEMPO-derived nitroxide spin labels (1, 3 and 5) showed very limited binding to the DNA motifs, whereas isoindoline-derived nitroxides 2, 4 and Ǵ showed extensive or full binding (Figure 2). A similar trend was observed for binding of these spin labels to abasic sites in duplex DNA and RNA, which is probably due to the increased steric requirements of the saturated nitroxide-containing ring of TEMPO.26 Nitroxide 4, an isoindoline derivative of A, binds better to triplex I. This was to be expected because triplex I was designed to bind A. In contrast, the 2-aminoadenine derivative 2 showed extensive binding to all three triplexes (50–70 %). This may be due to the fact that although 2 is a derivative of A, it bears structural similarity to guanine in that it also has an amino group in position 2. The best binder was found to be Ǵ, which bound 45 % to triplex I and almost fully (>98 %) to triplexes II and III (Figure 2, boxed spectra).
Equilibrium Filtration Measurements
The spin label with the most favorable binding properties, as determined in the EPR study, was Ǵ. To quantify binding, we sought experiments that provide a dissociation constant (Kd) for the complex of this spin label and triplex motifs. We have recently described equilibrium filtration assays for this purpose that involve centrifugal filters with a molecular weight cut-off of 3 kDa.13, 14 The filters retain the triplexes and let the solution with unbound ligand pass, the ligand content of which can then be quantified by absorption spectroscopy. In the present case, a co-solvent had to be identified that dissolves the spin label, does not interfere with the membrane filtration, and does not lead to a significant denaturation of the triplexes. We identified ethylene glycol as such a solvent. Spin label Ǵ dissolves in ethylene glycol upon heating to 100 °C, whereas up to 15 % of ethylene glycol as co-solvent is tolerated by the polyether sulfone membranes used in the filtration assays. Figure 3 shows UV-melting curves of DNA triplex motif II and RNA triplex motif IV in the presence and the absence of this co-solvent. Lithium phosphate buffer was used to avoid any issues with G quartets. In the presence of 15 % ethylene glycol, the melting point of the triplex-to-duplex transition for II was found to be 29 °C, and that of the duplex-to-single strands was 68 °C, confirming that the motif is folded at room temperature. For IV, the addition of the diol solvent slightly increased the triplex stability, so that the shoulder detectable for the triplex-to-duplex transition was shifted to above 30 °C, where it became indistinguishable from the duplex-to-single strands transition, with a resulting Tm of 56 °C. Again, this confirmed the stability of the motif in the presence of the alcohol.
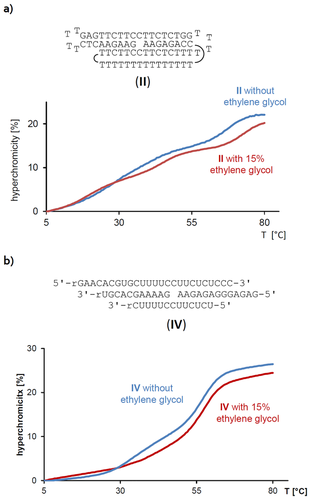
UV-melting curves of a) triplex DNA motif II, or b) RNA motif IV. Conditions: 1 μm strands in lithium phosphate buffer (10 mm, 150 mm NaCl, pH 6.0), with or without 15 % ethylene glycol as co-solvent.
The N2-labeled guanine Ǵ has an absorption maximum at 274 nm in the filtration buffer, which can be used to quantify it. At 15 % ethylene glycol, the recovery of the spin label was 98 % in control filtration assays, indicating that the guanine derivative does not aggregate or stick to the filter membrane under these conditions. Aliquots of a 3 mm stock solution of Ǵ were combined with solutions of motif II, and spin filtration gave the absorption spectra shown in Figure 4. From the decrease in concentration of the label in the presence of the DNA motif, a dissociation constant (Kd) of 2 μm was calculated for the complex between label and folded DNA.
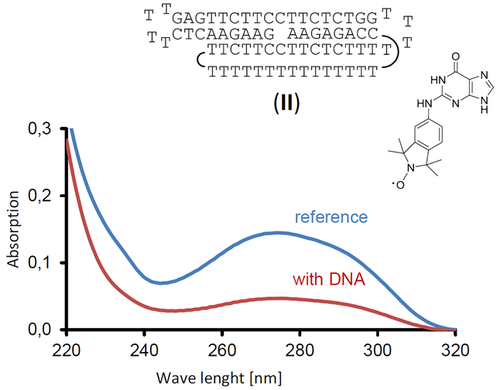
Absorption spectra of a solution of spin label Ǵ, either incubated with DNA motif II or by itself, after being subjected to filtration with a molecular weight cut-off of 3 kDa. Conditions: 9 μm Ǵ with or without 9 μm DNA motif II, 10 mm lithium phosphate buffer, pH 6, 150 mm NaCl, with 15 % ethylene glycol as co-solvent, after filtration at 4 °C.
To test the scope of the method, we then performed exploratory assays with the same guanine-based label Ǵ and an intermolecular, four-strand RNA triplex motif that was modelled after motif III. Figure 5 shows the UV-spectra of the solutions of the neat label and the label incubated with the triplex RNA (IV). Again, significant binding of the label was detected, even though some leaching of RNA from this more labile motif, with its shorter strands, was measured in the shorter wavelength range of the spectrum from the solution with the triplex. Several assays were run to confirm binding, which gave apparent Kd values in the range of 21–49 μm. Because residual oligoribonucleotides in the solution result in a higher apparent absorption than the true value, these are the upper limits of the dissociation constant of the complex between Ǵ and IV. As such, they confirm that Ǵ is suitable for noncovalent labeling of triplex binding motifs at NMR concentrations (100 μm or higher).
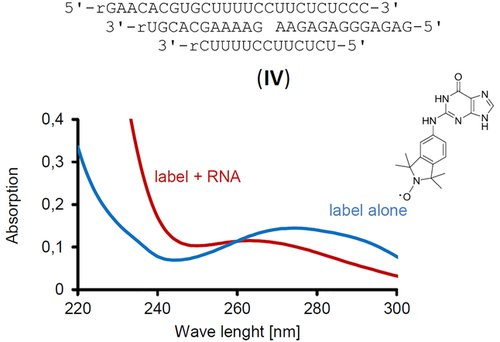
Absorption spectra of solutions of spin label Ǵ with or without intermolecular RNA motif IV, after filtration with a molecular weight cut-off of 3 kDa. Conditions: 9 μm Ǵ with or without 9 μm IV, 10 mm lithium phosphate buffer, pH 6, 150 mm NaCl, with 15 % ethylene glycol, with filtration at 4 °C.
Conclusions
The purine spin label based on guanine (Ǵ) binds well to triplex motifs with a single-nucleotide gap in the central oligopurine strand. Most probably, it engages in Watson–Crick and Hoogsteen base pairing with the complementary cytosine bases in the triplex. Binding can be detected by EPR and by equilibrium filtration. The latter technique gave dissociation constants in the lower micromolar range. Among the advantages of the method are the ease of performing the labeling and the straightforward design that is suitable for DNA and RNA motifs. One of the experimental challenges is the solubility of the label, which was overcome in the current study by using a known cryoprotectant32 as co-solvent. The noncovalent labeling technique is suitable for EPR and PELDOR/DEER measurements of distances as well as for paramagnetic relaxation enhancement (PRE) experiments using NMR spectroscopy, and complements other methods for monitoring and structure elucidation.
Experimental Section
General
Ethylene glycol, phosphoric acid, LiOH and distilled water were purchased from Sigma-Aldrich (Munich, Germany). Spin labels were prepared as previously described.26, 27 Amicon Ultra centrifugal filters with a molecular weight cut-off (MWCO) of 3 kDa were from Merck/Millipore (Darmstadt, Germany). Filtration assays were performed using Roti-Spin Mini-3 spin filters, 3 kDa MWCO from Carl Roth (Karlsruhe, Germany). The UV/VIS spectra and UV-melting curves were measured on a Lambda 25 spectrophotometer (PerkinElmer, Überlingen, Germany). Oligonucleotides were purchased from Biomers (Ulm, Germany) in HPLC-purified form and were used without modification.
EPR Experiments
Solutions for CW-EPR experiments were prepared by mixing each spin label (1–5, Ǵ; 200 μm) with the DNA motifs I–III (400 μm) in Breaker's buffer (20 mm Tris⋅HCl, 450 mm NaCl, 100 mm KCl, 10 mm MgCl2, 1 mm MnCl2, 5 mm CaCl2, pH 6.7) containing 30 % ethylene glycol and 2 % DMSO and placed in a 50 μL quartz capillary (BLAUBRAND intraMARK). The EPR spectra were recorded using 100–200 scans with a MiniScope MS200 (Magnettech Germany) spectrometer (100 kHz modulation frequency, 1.0 G modulation amplitude and 2.0 mW microwave power). Magnettech temperature controller M01 (±0.5 °C) was used to regulate the temperature at −30 °C.
Stock Solutions for Kd Measurements
A stock solution of buffer containing lithium phosphate (100 mm) and NaCl (1.5 m) was prepared as described below. Phosphoric acid (85 % in water, 0.68 mL) and NaCl (8.8 g) were dissolved in distilled water (90 mL); the pH was adjusted to a value of 6, using a stock solution of LiOH (5 m), and the solution was brought to a volume of 100 mL. The spin-label Ǵ (1.0 mg, 2.95 μmol) was dissolved in ethylene glycol (1 mL) at 100 °C under constant vortexing to give a stock solution (3 mm) that was allowed to cool and used for subsequent assays.
Assembly of Triplex Motifs for Kd Measurements
Equimolar mixtures of all oligonucleotides required for a given motif were prepared from aqueous stock solutions. The solution of DNA strands (10 nmol each) were treated with a tenth of the buffer stock solution (end concentration: 10 mm lithium phosphate buffer, 150 mm NaCl, pH 6). The solution was heated to 90 °C in a thermocycler and cooled to 4 °C in 2 h, using a linear temperature gradient. The triplex motifs were then freed from remaining free strands by filtration with Amicon ultracentrifugal filters with a MWCO of 3 kDa (0.5 mL capacity). The centrifugation was performed at 14 000 g for 30 min at 4 °C. The spin filters were washed three times with buffer (500 μL, 10 mm lithium phosphate buffer, 150 mm NaCl, pH 6). The triplex motifs were isolated by reverse spinning of the filter at 14 000 g for 5 min at 4 °C.
Filtration Assays
An aliquot of the solution of the triplex motif (5 nmol, 9 μm solution) was treated with buffer (400 μL, 10 mm lithium phosphate buffer, 150 mm NaCl, pH 6) and ethylene glycol (82.5 μL). An aliquot of the stock solution of Ǵ that gave an equimolar amount (5 nmol, 9 μm solution) was added, and the resulting solution was adjusted to a volume of 550 μL with buffer (10 mm lithium phosphate buffer, 150 mm NaCl, pH 6). For the reference assay that provided the background reading, Ǵ (5 nmol, 9 μm solution) was dissolved in the same buffer with ethylene glycol to give a total volume of 550 μL. The solutions were incubated for 16 h at 4 °C and then transferred to Roti-Spin Mini-3 spin-filters and centrifuged at 14 000 g and 4 °C for 30 min. The filtrate was immediately transferred to a quartz cuvette with a 10 mm pathlength and the UV/VIS spectrum was measured.
UV-Melting Curves
Solutions containing the stated concentration of the triplex motif in buffer (1.5 mL, 10 mm lithium phosphate buffer, 150 mm NaCl, pH 6) were transferred to a quartz cuvette with 10 mm pathlength. For measurements regarding the stability of the motifs toward the co-solvent, up to 15 % ethylene glycol was added. The temperature profile for the melting curve was applied with the help of the Peltier elements of the six-cell holder of the spectrophotometer. Melting points are extrema of the first derivative of the melting curve.
Author Contribution Statement
N.K., M.W., S.T.S. and C.R. planned the experiments. N.K. synthesized the spin labels. N.K. and T.H. collected and analyzed the EPR data. M.W. conducted the equilibrium binding assays. N.K., M.W., T.H., S.T.S. and C.R. contributed to writing of the manuscript.
Acknowledgements
The authors thank Anna-Lena J. Segler for technical assistance. This work was supported by the Icelandic Research Fund, grant No. 141062-051 to S.T.S. and DFG, grant No. RI 1063/16-1 to C.R. T.H. thanks the Deutsche Forschungsgemeinschaft (DFG) for a postdoctoral fellowship (414196920).