Comparative single-cell analysis of esophageal cancer subtypes reveals tumor microenvironment distinctions explaining varied immunotherapy responses
List of abbreviations
-
- AI
-
- artificial intelligence
-
- AKR 1
-
- aldo-keto reductase family 1
-
- AUROC
-
- area under the receiver operating characteristic curve
-
- CXCL
-
- C-X-C Motif Chemokine Ligand
-
- DC
-
- dendritic cell
-
- DEG
-
- differentially expressed gene
-
- EAC
-
- esophageal adenocarcinoma
-
- ESCC
-
- esophageal squamous cell carcinoma
-
- GAC
-
- gastric adenocarcinoma
-
- GCB
-
- germinal center B cell
-
- H&E
-
- hematoxylin and eosin
-
- HNSCC
-
- head and neck squamous cell carcinoma
-
- HSP
-
- heat-shock protein
-
- ICB
-
- immune checkpoint blockade
-
- MARCO
-
- macrophage receptor with collagenous structure
-
- MP
-
- metaprogram
-
- RNA-seq
-
- RNA sequencing
-
- TAM
-
- tumor-associated macrophage
-
- Tfh
-
- follicular helper T cell
-
- TLS
-
- tertiary lymphoid structure
-
- TME
-
- tumor microenvironment
Esophageal cancer comprises 2 anatomically shared but histologically different subtypes: esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). Previous bulk-level genomic and clinical studies have shown that ESCC shares molecular features with head and neck squamous cell carcinoma (HNSCC) [1] and is generally more responsive to immune checkpoint blockade (ICB) therapies than EAC [2], which is similar to gastric adenocarcinoma (GAC) [1]. Recent clinical trials have further demonstrated clinical benefits from various ICB therapies, including combination approaches, for ESCC [3].
To further expand the comparison at single-cell resolution of tumor microenvironment (TME), we conducted single-cell transcriptomic analysis on tumors from 35 patients representing 4 cancer types located near the esophagus: ESCC, EAC, HNSCC, and GAC (Supplementary Materials and Methods). By integrating newly generated single-cell datasets with published datasets (Supplementary Table S1) [4, 5], we analyzed more than 200,000 cells within TME (Supplementary Figure S1, Supplementary Figure S2A-C, Supplementary Table S2). This high-resolution approach allowed the dissection of cellular heterogeneity of malignant cells and various immune components within the TME (Figure 1A).
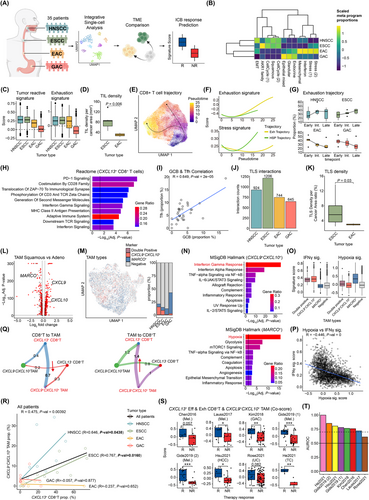
Comparative analysis of TME of esophageal cancer subtypes and nearby cancers. (A) Study scheme representing collection of 35 single-cell RNA-seq datasets, integration of these datasets for single-cell analysis, comparison of TME among cancer types, and prediction of ICB responses with gene signatures generated from comparative analysis. (B) Heatmap for proportions of “on” cells for each meta program calculated for each tumor type. Proportions are z-scaled column-wise. The rows and columns are clustered with hierarchical clustering. Separation of cancer types based on histological origins (Squamous, Glandular, Endocrine) as well as other cancer programs is observed. (C) Boxplots of tumor reactive signature scores (left) and exhaustion signature scores (right) from exhausted CD8+ T cells. The boxplots are visualized for each cancer type. Both squamous cell carcinomas (HNSCC, ESCC) show enrichment of those signatures while decreasing scores are observed for adenocarcinomas (EAC, GAC). (D) AI-guided quantification of H&E staining slide for TILs density per mm2 cancer area by Lunit SCOPE IO between ESCC and EAC. The significance of difference was calculated using the two-sided t-test. Slides from ESCC patients are significantly enriched with TIL compared to those from EAC patients. (E) UMAP of CD8+ T cells with pseudotimes and their trajectories into exhausted CD8+ T cells or HSP-high CD8+ T cells. The colors indicate pseudotimes for each cell. Two distinct trajectories for exhausted CD8+ T cells (left branch) and HSP-high CD8+ T cells (right branch) are observed. (F) Exhaustion signature scores and stress response signature scores along the exhaustion and HSP trajectories. These results confirm that each trajectory exhibits its expected characteristics. (G) Boxplots of proportions of cells for each cancer type in early, intermediate, and late stages of exhaustion trajectory for each patient. HNSCC and ESCC show increasing proportions of cells along the trajectory while EAC and GAC show the opposite trends. The results confirmed enrichment of exhausted yet tumor-reactive CD8+ T cell populations for squamous cell carcinomas. (H) Pathway analysis using Reactome (2022) database with DEGs from comparing CXCL13+ CD8+ T cells to CXCL13− CD8+ T cells. Colors of the bar indicate ratio of genes from the DEGs that are in gene list for each pathway term. Black vertical lines indicate an adjusted P-value threshold of 0.05. Pathways important for T cell activation and anti-tumoral T cell activities are enriched for CXCL13+ CD8+ T cells. (I) Correlation between proportions of GCB (out of B cells) and Tfh (out of CD4+ T cells) for each patient. “R” indicates Pearson correlation coefficient while “P-val” indicates P-values for significant of the correlation. For all patients, there is a significant positive correlation between proportions of GCB and Tfh, indicating close connections between these two TLS-forming populations. (J) Total cell-cell interaction counts calculated with CellChat for each tumor type among components of tertiary lymphoid structure; CD4+ Tfh cells, GCB cells, dendritic cells, and fibroblast. Self-interaction counts were excluded. ESCC exhibits the highest interaction counts while decreasing values are observed for HNSCC, EAC, and GAC. This result indicates possible enrichment of TLS through higher interactions among key populations. (K) AI-guided quantification of H&E staining slide for TLS density per cancer area by Lunit SCOPE IO between ESCC and EAC. The significance of difference was calculated using the two-sided t-test. Slides from ESCC patients are significantly enriched with TLS compared to those from EAC patients. (L) DEGs between TAMs from squamous cell carcinomas (ESCC and HNSCC) and adenocarcinomas (EAC and GAC). P-values for significance are calculated by Wilcoxon rank-sum test and corrected for multiple hypotheses with Bonferroni correction. Genes are considered DEGs if they have log2 fold change > 0.25 and adjusted P-values < 0.01. Genes colored red are DEGs from both comparisons of ESCC vs EAC and squamous vs adenocarcinomas. (M) UMAP plot of TAMs that are colored by expression of CXCL9/CXCL10 and MARCO. The boxplot indicates proportions of cells positive for both CXCL9/CXCL10 and MARCO (Double Positive), only CXCL9/CXCL10 or MARCO, or negative for both. EAC and GAC are highly enriched with MARCO+ TAMs while ESCC and HNSCC have 2∼4 times more CXCL9+CXCL10+ TAMs compared to MARCO+ TAMs. (N) Pathway enrichment analysis with MsigDB hallmark 2022 with DEGs from MARCO+ TAMs (Top) and CXCL9+CXCL10+ TAMs (Bottom). P-values for significance were calculated with Fisher's exact test and corrected for multiple hypotheses using Benjamini-Hochberg method. Colors of the bar indicate ratio of genes from the DEGs that are in gene list for each pathway term. Black vertical lines indicate an adjusted P-value threshold of 0.05. CXCL9+CXCL10+ TAMs are enriched for the pathway related to interferon-γ response while MARCO+ TAMs are enriched for the hypoxia-related pathway, indicating their possibly opposite roles in anti-tumor activities. (O) Boxplots of signature scores for each TAM classification with hypoxia, Tumor inflammation, macrophage-specific tumor inflammation, and interferon gamma signatures. The results confirm enrichment of interferon-γ response for CXCL9+CXCL10+ TAMs and hypoxia for MARCO+ TAMs from pathway analysis in Figure 1N. (P) Correlation between scores from hypoxia signature and interferon-γ signature for TAMs. TAMs not expressing MARCO, CXCL9, and CXCL10 are filtered. “R” indicates Pearson correlation coefficient while “P-val” indicates P-values for significant of the correlation. A significant negative correlation between interferon-γ signature and hypoxia signature is observed, indicating exclusiveness of TAMs enriched with these signatures. (Q) Circle plots of Intercellular communication probability generated by CellChat among CXCL13+ CD8+ T cells, CXCL13− CD8+ T cells, CXCL9+CXCL10+ TAMs, and CXCL9−CXCL10− TAMs. The arrows indicate interactions between CXCL13+ CD8+ T cells and CXCL9+CXCL10+ TAMs. We visualized only interactions from CD8+ T cells to TAMs (left) or TAMs to CD8+ T cells (right). The results indicate higher interactions between CXCL13+ CD8+ T cells and CXCL9+CXCL10+ TAMs. (R) Correlation between proportion of CXCL13+ CD8+ T cells and CXCL9+CXCL10+ TAMs. “R” indicates Pearson correlation coefficient while “P-val” indicates P-values for significant of the correlation. For all patients, there is a positive correlation between proportions of CXCL13+ CD8+ T cells and CXCL9+CXCL10+ TAMs. For each tumor type, there are higher positive correlations for ESCC and HNSCC while no correlations are observed for EAC and GAC. (S) Boxplots of GSVA scores calculated by averaging scores with gene signatures from CXCL13+ effector and exhausted CD8+ T cells and CXCL9+CXCL10+ TAMs (Co-score). The scores were calculated using bulk RNA-seq datasets from various cancer patients with response information for immunotherapy for each patient and grouped by ICB response (R, responder; NR, non-responder). For 6 out of 8 cohorts, there is positive enrichment of the Co-scores for immunotherapy responders. (T) AUROC scores for prediction of ICB responders with Co-score. These signatures can successfully predict immunotherapy responders with AUROC 0.7 for 7 out of 8 cohorts.
Differentially expressed gene (DEG) analysis of malignant cells revealed a clear separation based on epithelial cell origin (Supplementary Figure S2D). For detailed cancer cell states, we generated 14 malignant metaprograms (MPs) using non-negative matrix factorization (Supplementary Table S3). Histology-specific MPs, representing squamous or glandular differentiation, dominated the expression landscape, while additional MPs distinguished tumor types based on cell cycle dynamics, endocrine-like features, and activation of Aldo-keto reductase family 1 (AKR1) family genes, thereby providing insights into diverse and shared oncogenic processes (Figure 1B). Furthermore, we generated MPs from immune and stromal compartments and calculated correlations among them (Supplementary Figure S2E, Supplementary Table S3). We identified clusters of coordinated MPs, including an “immune activating” cluster characterized by interferon signaling and activation of adaptive immunity, predominantly enriched in HNSCC and ESCC. In contrast, heat-shock protein (HSP) MP, negatively correlated with the immune activating cluster, was more common in EAC and GAC, suggesting potentially immunosuppressive TMEs.
To better understand the immune compartments of the TME, we conducted in-depth analyses of each major immune cell type using subclustering approaches. We first focused on CD8+ T cells for their roles in anti-tumor immunity. We identified several key subtypes including naive/memory, effector, stress-response (HSP high), and exhausted populations (Supplementary Figure S3A-B, Supplementary Table S4). For exhausted populations, we evaluated whether each tumor type exhibited varying degrees of exhaustion and tumor reactivity. Indeed, both HNSCC and ESCC displayed higher tumor reactivity and exhaustion (Figure 1C, Supplementary Table S5). Higher levels of tumor infiltrating T cells for ESCC compared to EAC were validated with artificial intelligence (AI)-guided analysis of hematoxylin and eosin (H&E) slide (Figure 1D, Supplementary Figure S3C-D, Supplementary Table S6). Another key subset identified was a T cell population with high HSP expression, recently identified as a stress-responsive population and a poor indicator of ICB responses [6]. The trajectory analyses identified distinct exhaustion and HSP trajectories for these populations (Figure 1E-F, Supplementary Figure S3E-F). For exhaustion trajectory, there were increasingly more populations at the late stage for HNSCC and ESCC but opposite for EAC and GAC (Figure 1G), suggesting inadequate activation of tumor-reactive populations for adenocarcinomas.
To identify key genes contributing to the differences in CD8+ T cell populations, we performed DEG analysis comparing cells with high and low exhaustion and neoantigen-reactive scores (Supplementary Figure S3G, Supplementary Table S7). Among the top DEGs, we confirmed that C-X-C Motif Chemokine Ligand 13 (CXCL13) was highly enriched in effector and exhausted T cell populations, with GAC samples exhibiting very low percentages of CXCL13-expressing cells compared to all other samples (Supplementary Figure S3H). Furthermore, effector and exhausted CD8+ T cells lacking CXCL13 expression showed low signature scores for both exhaustion and neo-antigen reactivity, comparable to other non-effector CD8+ T cells (Supplementary Figure S3I). Pathway analysis revealed that CXCL13+ CD8+ T cells are associated with increased T cell infiltration and activation through interferon-γ signaling, costimulation by CD28 family, and antigen processing pathways (Figure 1H).
CD4+ T cells and B cells also play key roles in immunotherapy-related activities, particularly through the formation of tertiary lymphoid structure (TLS) [7]. Among their subtypes (Supplementary Figure S4A-D, Supplementary Tables S8-S9), we focused on CD4+ follicular helper T (Tfh) cells and germinal center B (GCB) cells. Our analysis revealed that ESCC, and to a lesser extent HNSCC, harbored higher proportions of Tfh cells with elevated CXCL13 expression (Supplementary Figure S4E), a chemokine known to promote early TLS maturation by attracting B cells [8]. We observed a positive correlation among these populations and an enrichment of GCBs and TLS signatures in ESCC (Figure 1I, Supplementary Figure S4F, Supplementary Table S5). Moreover, cell-cell interaction analyses confirmed that the crosstalk among TLS components including Tfh cells, GCBs, dendritic cells (DCs), and fibroblasts was stronger in HNSCC and ESCC than in EAC and GAC, supporting the notion that effective TLS formation contributes to superior immunotherapeutic responses (Figure 1J). These interactions were validated with a higher TLS density for ESCC quantified by AI-powered H&E slide analyzer (Figure 1K, Supplementary Figure S3D).
Among identified cell subtypes in the myeloid compartment (Supplementary Figure S4G-H, Supplementary Table S10), we focused on tumor-associated macrophages (TAMs) because they play roles in both immune activation and suppression. While canonical polarization markers failed to distinguish TAM states (Supplementary Figure S4I, Supplementary Table S5), DEG analysis revealed that Macrophage receptor with collagenous structure (MARCO) was enriched in EAC and GAC, while CXCL9 and CXLC10 were enriched in HNSCC and ESCC (Figure 1L, Supplementary Table S11). These markers appear mutually exclusive, with only 0.4% of TAMs co-expressing both (Figure 1M). Pathway analysis and gene signature scores further indicated that CXCL9+CXCL10+ TAMs were enriched with interferon-γ pathways, a positive indicator of ICB response, while MARCO+ TAMs were enriched in hypoxia pathways (Figure 1N-O). Recent studies linked hypoxia to resistance to ICB by interfering with other immune populations [9]. Negative correlations among the gene signature scores further validated exclusivity of these populations (Figure 1P).
Two cell types exhibited the most noticeable patterns: CD8+ T cells and TAMs. CXCL13+ CD8+ T cells and CXCL9+CXCL10+ TAMs displayed high levels of cellular interactions (Figure 1Q), consistent with a coordinated, interferon-γ-driven response that may underlie enhanced sensitivity to ICB therapies. In HNSCC and ESCC patients, these populations were significantly co-abundant (Figure 1R). By contrast, HSP-high CD8⁺ T cells and MARCO⁺ TAMs—associated with hypoxic and stress-related signaling—likely contribute to the formation of “cold” tumors that are less responsive to immunotherapy, as reflected by their higher interactions (Supplementary Figure S4J). Notably, GAC patients showed significant co-abundance of these 2 subtypes (Supplementary Figure S4K).
To validate the involvement of these subtypes in immunotherapy responses, we used bulk RNA sequencing (RNA-seq) datasets from 8 cohorts across diverse cancer types (Supplementary Table S12). With gene signatures extracted from these populations (Supplementary Table S13), we calculated co-enrichment scores (co-score) for CXCL13+CD8+ T cells and CXCL9+CXCL10+ TAMs. These co-scores showed significant differences between responders and non-responders in 6 cohorts (Figure 1S) and were highly predictive for responders (Area under the receiver operating characteristic curve [AUROC] > 0.7) in 7 cohorts (Figure 1T). We also calculated differences in 2 signature scores (diff-score) for CXCL9+CXCL10+ TAMs and MARCO+ TAMs. The diff-scores showed significant differences between responders and non-responders for 6 cohorts (Supplementary Figure S4L) and were predictive for responders in these cohorts (Supplementary Figure S4M). These results suggest that cellular signatures distinguishing esophageal cancer subtypes can predict ICB responses across diverse cancer types.
In summary, comprehensive single-cell analysis demonstrated that the distinct factors within the TME could explain the differential responses to ICB therapies between esophageal cancer subtypes. ESCC maintains a favorable TME characterized by interferon-γ-dominated, immune-activating populations while relative depletion of these favorable populations and higher prevalence of hypoxia-related populations in EAC could be linked to immune suppression. A recent single-cell study with neoadjuvant chemo-immunotherapy-treated ESCC patients similarly identified associations of interferon-γ-related genes to favorable treatment responses [10]. These contrasting TMEs not only provide insight into the mechanistic basis for the varied efficacy of immunotherapy but also suggest that tailoring treatment strategies to modulate specific immune subsets or to stratify patients based on the immune subsets could enhance clinical outcomes. This study lays the groundwork for developing predictive biomarkers and targeted interventions that consider unique immune landscapes associated with different tumor histology. However, due to the limited sample sizes and functional validations, future studies with larger cohorts and additional experimental validations are needed to further validate these results and to reduce possible confounders. Furthermore, future studies should explore additional factors influencing the TME, such as genetic mutations or environmental conditions, to provide further insight into tumor-immune interactions.
AUTHOR CONTRIBUTIONS
Seong Yong Park, Hye Ryun Kim, and Insuk Lee conceived the study. Seungbyn Baek performed single-cell transcriptome data analysis under the supervision of Insuk Lee. Junha Cha assisted bioinformatic analysis. Seong Yong Park and Hye Ryun Kim organized clinical samples and data collections. Min Hee Hong, Yoon Woo Koh, and Dahee Kim contributed to clinical sample collection. Gamin Kim contributed sample preparation. Wonrak Son, Chan-Young Ock, and Seungeun Lee performed AI-guided analysis of H&E slide. Martin Hemberg provided advice on single-cell data analysis. Insuk Lee and Hye Ryun Kim contributed to the financial and administrative support for this study. Seungbyn Baek, Seong Yong Park, Hye Ryun Kim, and Insuk Lee wrote the manuscript.
ACKNOWLEDGEMENTS
Not applicable.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflicts of interest.
FUNDING INFORMATION
This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation funded by the Ministry of Science and ICT (RS-2021-NR059674, RS-2022-CC125144, 2022M3A9F3016364, 2022R1A2C1092062, and RS-2025-00553825), Brain Korea 21 (BK21) FOUR program, and the Technology Innovation Program (20022947; funded by the Ministry of Trade Industry & Energy; MOTIE, Korea), Yonsei Fellow Program funded by Youn Jae Lee.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Institutional Review Board of Yonsei University Severance Hospital with IRB No 4-2016-0678. Written informed consent was obtained prior to enrollment and sample collection at Yonsei University Severance Hospital. The research conformed to the principles of the Helsinki Declaration.
Open Research
DATA AVAILABILITY STATEMENT
The single-cell RNA sequencing data generated in this study is deposited in the Gene Expression Omnibus database with accession number GSE273127 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc). The remaining data are available within the article or Supplementary Materials.