Neoadjuvant HER2 inhibition induces ESR1 DNA methylation alterations resulting in clinically relevant ER expression changes in breast cancers
Gavin P. Dowling and Gordon R. Daly contributed equally to this work.
Abbreviations
-
- β
-
- beta value
-
- ASCO/CAP
-
- American Society of Clinical Oncology/College of American Pathologists
-
- Bx
-
- pre-treatment breast tumors
-
- CIs
-
- confidence intervals
-
- DFS
-
- disease-free survival
-
- EMT
-
- epithelial to mesenchymal transition
-
- ER
-
- estrogen receptor
-
- FFPE
-
- formalin-fixed paraffin-embedded
-
- FISH
-
- fluorescence in-situ hybridization
-
- HER2
-
- human epidermal growth receptor-2
-
- HR
-
- hazard ratios
-
- IHC
-
- immunohistochemistry
-
- m
-
- methylation intensity value
-
- Mx
-
- subsequent metastasis
-
- OS
-
- overall-survival
-
- pCR
-
- pathologic complete response
-
- PR
-
- progesterone receptor
-
- Rx
-
- metastatic surgical resection
-
- Sx
-
- post-treatment resection specimen
The expression of estrogen receptor (ER) and human epidermal growth factor receptor-2 (HER2) in breast cancer can change in response to treatment and pivotally influence tumor behavior and clinical management [1]. Receptor discordance has been observed at various distant metastatic site (bone, lung, liver, and brain), with a routine loss of ER and gains in HER2 reported [2]. This receptor discordance can influence tumor responsiveness to both HER2 inhibitor and endocrine therapies. Although the mechanisms underlying receptor expression changes are not fully understood, we recently reported gains in ESR1 promoter hypermethylation as a potential driver of ER loss during disease progression [3]. In this study, mechanisms underlying altered receptor expression and associated disease outcomes were examined following neoadjuvant trastuzumab treatment.
We investigated the impact of HER2 inhibition on ER expression. From a cohort of 2,917 patients, 527 tumors were HER2-positive. Of these, 161 patients received neoadjuvant trastuzumab with systemic chemotherapy (Supplementary Figure S1, Supplementary Table S1). In this cohort (n = 161), 89 patients (55.3%) achieved pathological complete response (pCR), while 72 patients (44.7%) had residual disease, and 18 patients developed metastases (Clinical Cohort, Figure 1A, Supplementary Table S1). A statistically significant higher proportion of patients with ER-negative tumors achieved pCR compared to their ER-positive counterparts (P = 0.016, Supplementary Table S1), consistent with previous studies [4]. In patients with residual disease, the most notable finding was the observed gain of ER protein expression and loss of HER2 in a number of patients (33% and 17%, respectively; Figure 1B). ER protein status remained unchanged in 38 patients (53%), with a loss of ER observed in 8 patients (11%) (Figure 1B).
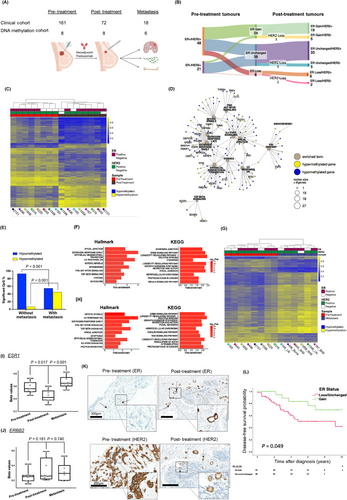
Epigenetic modifications are a potential mechanism underlying this observed receptor discordance. In this study, the role of DNA methylation in altered receptor expression was investigated (DNA methylation cohort, Figure 1A). Global DNA methylation was assessed in pre- and post-treatment samples (n = 16; Supplementary Table S2), of which 7 patient tumors were matched (biopsy and resection). Differentially methylated gene (DMG) analysis segregated patient tumors into pre- or post-treatment groups, illustrating the dominance of hypomethylation in post-treatment patient tumors (Figure 1C). Notably, pathway analysis identified Estrogen Response Early, Epithelial Mesenchymal Transition (EMT), ERRB, AMPK, and RAS signaling (P < 0.05) as statistically significant pathways (Supplementary Figure S2A-B). Network graph analysis revealed Estrogen Response Early, Estrogen Response Late and EMT as hub pathways (Figure 1D). Estrogen-responsive genes such as GREB1, FRK, IGFBP4 and IRS1 were predominantly hypomethylated, while oncogenic genes such as VEGFC, DEPTOR, TIAM1, and HDAC1 were hypermethylated (Figure 1D, Supplementary Table S3).
Analysis of pre- and post-neoadjuvant treated tumors from patients with no metastases (good outcomes) compared to those with metastases (poor outcomes) revealed significant divergent differential methylation profiles (P < 0.001; Figure 1E, Supplementary Table S4). In patients with subsequent metastases, reduced hypomethylation and enhanced hypermethylation were observed (Figure 1E). Pathway analysis of DMGs highlighted PI3K/AKT/mTOR (Hallmark) and ERRB2 (KEGG) as statistically significant pathways (P < 0.05; Figure 1F). Network graph analysis of hypermethylated Hallmark pathways (top 12) identified PI3K/AKT/mTOR signaling as a hub pathway with poor outcomes (Supplementary Figure S2C, Supplementary Table S4). Notably, differential hypermethylation was detected in genes such as SMAD2, AKT1, PAK4, EGFR, BRCA2, NF1 and PIK3R3 (Supplementary Figure S2C).
We also characterized DNA methylation profiles in pre-treatment biopsies and metastatic tumors. Global gains in hypermethylation were observed upon metastasis in both ER-positive and ER-negative matched tumors (Figure 1G). Pathway analysis revealed alterations in PI3K/AKT/MTOR (Hallmark), ERBB, focal adhesion, and Ras (KEGG) signaling pathways (P < 0.05; Figure 1H, Supplementary Table S5). Network analysis of DMGs identified PI3K/AKT/mTOR signaling as a hub pathway, with hypermethylation of SMAD2, MAPK8, PAK4, and hypomethylation of PRKCA, EGFR, and PIK3CD (Supplementary Figure S2D, Supplementary Table S6). Of note, global hypermethylation was more pronounced in metastatic tumors compared to post-treatment tumors (Supplementary Figure S3A), with differential methylation affecting key oncogenic pathways including EMT, PI3K/AKT/MTOR, and estrogen response (Supplementary Figure S3B).
Consistent with global hypomethylation observed post-neoadjuvant treatment (Figure 1C), ESR1 probe cg01715172 was identified as a statistically significant hypomethylated promoter CpG in ESR1 post-treatment (P = 0.0169), with subsequent significant gains in ESR1 promoter hypermethylation observed upon metastasis (P = 0.0014), (Figure 1I). These changes reflect altered CpG DNA methylation across ESR1 (Supplementary Figure S4). Conversely, ERBB2 promoter hypermethylation at cg15227682 was identified post-treatment, with promoter hypomethylation observed upon metastasis, though these differences did not reach statistical significance (Figure 1J). At a functional level, alterations in the promoter methylation status of ESR1 and ERBB2 were validated at the protein level, with relative gains in ER expression and a loss of HER2 expression observed post-neoadjuvant treatment (Figure 1K, Supplementary Table S7). Trastuzumab-induced ESR1 hypomethylation and related ER expression gains were further validated in HER2-positive cell line models, SKBR3 and T347 cells. No alterations were observed in HER2 non-amplified/trastuzumab-insensitive LY2 cells (Supplementary Figure S5).
The clinical relevance of protein expression changes as a consequence of dynamic methylation status of key breast cancer receptors ER and HER2 was determined. In patients with residual disease, gains in ER protein positivity following neoadjuvant therapy were associated with enhanced disease-free survival (DFS) (Gain HR = 0.37, 95% CI = 0.14-0.99, P < 0.05) (Figure 1L) and showed a trend toward an association with overall survival (OS) (Gain HR = 0.40, 95% CI = 0.15-1.10, P < 0.07, Supplementary Figure S6A). However, alterations in HER2 status post-neoadjuvant treatment did not have a statistically significant impact on either DFS or OS in this patient population (Supplementary Figure S6B-C).
We demonstrate dynamic alterations in receptor status in response to trastuzumab. Although the number of patient samples in this study is relatively limited, the data reported here support DNA methylation as a driver of expression changes, with global hypomethylation following neoadjuvant treatment and hypermethylation upon metastasis. While differential methylation of key signaling pathways is conserved between post-treatment primary surgery and metastasis, the direction of methylation shifts in core pathway genes, including ESR1, as well as known tumor suppressor genes, such as FRK, RARB, and SSBP2 [5], from hypomethylation post-neoadjuvant treatment to hypermethylation upon metastasis, ultimately driving an aggressive clinical phenotype.
The observed epigenetic changes in ESR1 and ERBB2 highlight the modifying nature of methylation in response to treatment and disease progression. Post-treatment hypomethylation of ESR1 may contribute to maintaining sensitivity to endocrine therapy, whereas hypermethylation during metastatic stages suggests a shift towards a more resistant phenotype.
Preclinical and clinical findings have demonstrated that, for HER2/ER-positive tumors, treatment with HER2-directed therapy in isolation increases ER expression through crosstalk between these receptors [6]. While this upregulation of ER and ER-related genes leads to a compensatory ‘escape’ pathway, it simultaneously creates an additional therapeutic target, with evidence that sustained anti-HER2 therapy sensitizes tumor cells to endocrine therapies [7]. Therefore, it has been suggested that dual blockade of HER2 and ER pathways may be necessary in HER2/ER-positive tumors to sustain an antitumor effect [7, 8]. Concomitant treatment with HER2-directed therapy and endocrine therapy has been reported to have greater antitumor activity than HER2-targeted therapy alone in multiple preclinical models [9]. Clinical trial data also suggest that the combination of endocrine therapy and anti-HER2 therapy is an effective therapeutic strategy in this cohort [10]. Therefore, repeat receptor analysis after treatment is crucial, as patients switching from ER-negative to ER-positive can benefit from the addition of endocrine therapy to their treatment regimen.
We report that an increase in ER expression post-neoadjuvant therapy is statistically significantly associated with better survival outcomes compared to tumors with decreased or unchanged ER expression. This suggests that the interplay between ER and HER2 is highly dynamic, with its influence on tumor behavior being more rapid in response to perturbations in cellular signaling than is currently understood. Global DNA methylation patterns shift in response to treatment and subsequent metastasis, altering the tumor phenotype. Consequential changes in core ER signaling not only influences outcomes but also provide opportunities for meaningful clinical intervention. Unlocking DNA methylation as a key process in breast cancer progression can provide important insights into the consequences of treatment and aid in the development of new therapeutic strategies.
AUTHOR CONTRIBUTIONS
Damir Varešlija and Leonie S. Young: study concept and design. Gavin P. Dowling, Gordon R. Daly, Damir Varešlija, Aisling Hegarty, Sinéad Cocchiglia, Sandra Hembrecht, Michael Flanagan, Mihaela Ola, Ramón Fallon, Fiona Bane, Vikrant Singh, Katherine M. Sheehan, Louise Watson, Jason McGrath, and Leonie S. Young: acquisition, analysis, or interpretation of data. Aisling Hegarty, Sinéad Cocchiglia, Patrick G. Morris, Bryan Hennessey, and Arnold D.K Hill: provision of administrative, technical, or material support. Gavin P. Dowling, Gordon R. Daly, Sinéad Cocchiglia, Damir Varešlija, and Leonie S. Young: drafting of the manuscript. All authors: critical revision of manuscript.
ACKNOWLEDGEMENTS
We thank all the patients who provided tumor tissue for our study and Lance Hudson and Joanna Faye who processed samples at Beaumont Hospital, Dublin. Funders had no role in the design of the study.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
FUNDING INFORMATION
We kindly acknowledge the funding support from Science Foundation Ireland Frontiers for the Future Award (19/FFP/6443), Science Foundation Ireland Strategic Partnership Program, Precision Oncology Ireland (18/SPP/3522) (L.S.Y.), Breast Cancer Now Fellowship Award with the generous support of Walk the Walk (2019AugSF1310) (D.V.), Science Foundation Ireland (20/FFP-P/8597) (D.V.), Breast Cancer Ireland program Grant (18239A01) (L.S.Y.).
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Written and informed consent was acquired prior to collection of patient tumor tissue under The Royal College of Surgeons Institutional Review Board approved protocol (CTI 09/07). All clinical material was collected as part of the prospective observational clinical trial NCT01840293 (https://clinicaltrials.gov).
Open Research
DATA AVAILABILITY STATEMENT
Raw DNA methylation data files can be found at the Gene Expression Omnibus (GEO) website under accession number GSE283272.




