Vascular smooth muscle cell plasticity in the tumor microenvironment
Abbreviations
-
- BST2
-
- Bone marrow stromal antigen 2
-
- PIRA2
-
- Paired Immunoglobulin-like Receptor A2
-
- scRNA-seq
-
- Single cell RNA sequencing
-
- SMC
-
- Smooth muscle cell
-
- TME
-
- Tumor microenvironment
-
- UMAP
-
- Uniform manifold approximation and projection
Smooth muscle cell (SMC) plasticity plays a prominent role in the pathogenesis of multiple diseases. This phenomenon is characterized by the loss of canonical SMC marker gene expression (such as Acta2 and Myh11), increased proliferation and migration, and the upregulation of genes typically associated with other cell types, such as macrophages [1-3]. This process is best described in atherosclerosis, where phenotype switching, clonal expansion, and the aberrant expression of inflammatory and matrix proteins contribute to lesion progression and plaque instability [1-4]. However, this phenomenon has not been studied in the context of tumorigenesis. Here, we investigated whether SMC diversity and plasticity play a role in the tumor microenvironment (TME) using well-established SMC-lineage tracing mouse models, single cell RNA sequencing (scRNA-seq), and in silico ligand-receptor predictions. Detailed study methods are described in the supplementary materials and methods section. The goal of this work was to determine if vascular SMC plasticity should be prioritized as a translational target in oncology.
Two-colored Myh11 lineage tracing mice have native cells that express tdTomato at baseline. Following tamoxifen administration, any cell expressing MYH11 will lose tdTomato and instead express eGFP (Supplementary Figure S1A-B). Syngeneic colon cancers (MC38) implanted subcutaneously into the flanks of these two-colored mice showed a marked and progressive investment of SMCs into the tumor over an 11-day period (Figure 1A-B, Supplementary Figure S1C). High-resolution fluorescent microscopy revealed the loss of the canonical SMC marker ACTA2 in the eGFP+ lineage traced cells, indicating that they may have been misidentified using traditional histological approaches (Figure 1C). eGFP+ cells were noted far from discernible vasculature within the TME (Figure 1D-E), suggesting their migration away from endothelial networks into the tumor interstitium. Experiments using a separate Rainbow lineage tracer revealed that the expansion of these cells did not occur in a clonal fashion (Supplementary Figure S1D-E) [5].
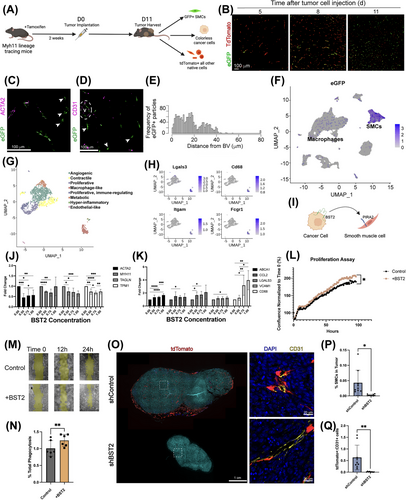
To more precisely define the diversity of these cells, scRNA-seq was performed. Unbiased clustering and uniform manifold approximation and projection (UMAP) analysis of the tumor data showed the representation of all anticipated cell types, identified by their gene expression profiles (Supplementary Figure S1F). As expected, eGFP-expressing cells were concentrated in the SMC cluster but were also surprisingly prevalent within the larger macrophage cluster (Figure 1F), representing 10% of eGFP+ cells in total. To define the diversity of SMC-derived cells in the TME, all cells expressing an eGFP transcript ≥ 1 were subset and reanalyzed, identifying eight distinct groups of tumor-associated lineage-traced SMCs (Figure 1G). We then used Monocle3 pseudotemporal analysis to map the trajectory of transitioning SMC (Supplementary Figure S1G). The trajectory starts with high contractile gene expression, which diminishes as the SMC adopt a more proliferative and non-traditional phenotype, consistent with our immunofluorescent staining. The cluster furthest from the original contractile cell state appears to take on a ‘macrophage-like’ phenotype, upregulating genes related to antigen presentation and immune response relative to other SMC-derived cells (Figure 1H, Supplementary Figure S1H). Studies using a Dre-Cre reporter, which maps sequential upregulation of macrophage-related genes in SMC-derived cells [4], suggested that this phenomenon was not merely the result of cell fusion (Supplementary Figure S1I-J). These results demonstrate an unexpected level of SMC plasticity in the TME, including the ability to transition toward a macrophage-like cell (macSMC).
To understand the mechanism underlying the plasticity from a contractile to a macSMC state, we utilized CellChat to predict ligand-receptor interactions. A comparison of predicted ligand-receptor pairs between the tumor and typical tumor-associated macrophages (TAMs) versus the tumor and macSMCs revealed exactly one unique cell-cell interaction (Figure 1I, Supplementary Figure S2A-C): the cell surface ligand bone marrow stromal antigen 2 (BST2) on tumor cells binding to the cell surface receptor paired immunoglobulin-like receptor A2 (PIRA2) on macSMCs. BST2 is a type II transmembrane protein that is upregulated in several cancer subtypes and thought to partly promote tumorigenesis through its propensity for vascular invasion [6, 7]. PIRA receptor complexes have been shown to activate macrophage pathways via phosphorylation of an immunoreceptor tyrosine-based activation motif and ERK signaling [8].
To test these in silico predictions, primary murine aortic SMCs were cultured with increasing concentrations of recombinant BST2. Quantitative reverse transcription polymerase chain reaction (RT-qPCR) showed decreased expression of contractile genes and increased expression of macrophage-associated genes, consistent with the in vivo results (Figure 1J-K). SMCs incubated with recombinant BST2 also exhibited increased proliferation and chemokinesis in migration assays (Figure 1L-M). When apoptotic MC38 cells were cocultured with SMCs, the presence of BST2 induced a modest but significant increase in the phagocytic ability of SMCs, as assessed by FACS phagocytosis assay (Figure 1N). BST2 also increased ERK phosphorylation in SMCs, aligning with prior literature on downstream PIRA signaling [8] (Supplementary Figure S2D-E). Loss-of-function studies using siRNA targeting PIRA2 diminished or abolished the effects of BST2 in several assays, implicating PIRA2 as the interacting receptor (Supplementary Figure S2F-J). Together, these data suggest that signaling between BST2 and PIRA2 contributes to the loss of canonical contractile markers in SMCs while promoting behaviors typical of TAM, such as proliferation, migration, and capacity for tumor cell phagocytosis.
To explore the translational relevance of this phenomenon, MC38 cells were transfected with either a BST2-targeting shRNA or a non-targeting shRNA (shControl) (Supplementary Figure S3A), then implanted into the flanks of single-colored Myh11 lineage-tracing mice. Consistent with previous studies [6], BST2-deficient tumors were smaller, with reduced volume and weight at the time of harvest (Figure 1O, Supplementary Figure S3B). SMC content in the shBST2 tumors was significantly reduced, both overall and as a percentage of the total quantified cell number (Figure 1P). When using CD31+ endothelial cell content to adjust for total tumor vascularization, there were still significantly fewer SMC-derived cells in the BST2-deficient tumors (Figure 1Q). By lineage-traced scRNA-seq, the SMC-derived cells which persisted in the TME were less likely to adopt a macrophage-like profile and expressed fewer genes typically associated with immunosuppressive TAMs (Supplementary Figure S3C-F). These findings suggest that tumor cell expression of BST2 may contribute to pathological SMC investment in tumors, even after adjusting for differences in tumor size and vessel ingrowth.
Limitations of this work include the possibility that these macSMCs reflect cellular fusion within the TME or macrophage phagocytosis. However, because we used multiple lineage-specific reporter systems and conservative doublet thresholds for SMC identification, we consider this explanation less likely. Definitive exclusion of this possibility would require chimeric or parabiotic models. Additionally, our studies used only male mice, as the lineage tracer is located on the Y chromosome.
In summary, these studies provide novel insights into the dynamic nature and remarkable plasticity of vascular SMCs during tumorigenesis. We found that, in addition to contractile and proliferative states, a small subset of SMCs can adopt a phenotype with cancer-promoting, macrophage-like features, possibly in response to direct cell-cell interactions with the tumor interface. This phenomenon, which would not have been observed without the use of indelible lineage tracing systems, may represent a novel determinant of tumor progression. These results also demonstrate that SMCs are potential constituents of the TME and highlight their ability to function beyond their classic role in vascular homeostasis, similar to what has been observed in cardiovascular conditions such as atherosclerosis [9]. Future studies will determine whether targeting SMC plasticity can have a therapeutic impact on the vascular compartment signature within the TME [10], beyond what is currently achievable with anti-VEGF therapies. If successful, targeting cancer neovascularization may not only reduce nutrient delivery to the developing tumor, but also alter its immunomodulatory landscape, thus providing an additional translational strategy for oncologists.
AUTHOR CONTRIBUTIONS
Caitling F. Bell, Richard A. Baylis, and Nicholas J. Leeper designed research. Caitlin F. Bell, Richard A. Baylis, Nicolas G. Lopez, Wei Feng Ma, Hua Gao, Fudi Wang, Sharika Bamezai, Changhao Fu, Yoko Kojima, Shaunak S. Adkar, Lingfeng Luo, and Nicholas J. Leeper, performed research. Caitlin F. Bell, Richard A. Baylis, Nicolas G. Lopez, Wei Feng Ma, Clint L. Miller, and Nicholas J. Leeper contributed new reagents/analytic tools. Caitlin F. Bell, Richard A. Baylis, Nicolas G. Leeper, Wei Feng Ma, Hua Gao, Fudi Wang, and Nicholas J. Leeper analyzed data. Caitlin F. Bell, Richard A. Baylis, and Nicholas J. Leeper wrote the paper. All authors read and approved the final manuscript.
ACKNOWLEDGEMENTS
No further acknowledgements.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
FUNDING INFORMATION
This study was supported by the Damon Runyon Cancer Research Foundation (PST 33-21 to CFB), the National Institutes of Health (R35 HL144475 to N.J.L.), the American Heart Association (EIA34770065 to N.J.L.), and the Fondation Leducq (‘PlaqOmics’ 18CVD02 to N.J.L. and C.L.M.).
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
All animal studies were approved by the Stanford University Administrative Panel on Laboratory Animal Care (Protocol 27279) and conform to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 1996).
Open Research
DATA AVAILABILITY STATEMENT
The raw and processed single-cell RNA sequencing datasets are made available on the Gene Expression Omnibus (GEO) database, accession code GSE262942. The data that support the findings of this study are available from the corresponding author upon reasonable request.




