Nicotinamide N-methyltransferase negatively regulates metastasis-promoting property of cancer-associated fibroblasts in lung adenocarcinoma
Abstract
Background
Recurrence and metastasis remain significant challenges in lung adenocarcinoma (LUAD) after radical resection. The mechanisms behind the recurrence and metastasis of LUAD remain elusive, and deregulated cellular metabolism is suspected to play a significant role. This study explores the metabolic and epigenetic regulation mediated by nicotinamide N-methyl transferase (NNMT) in LUAD.
Methods
Untargeted metabolomic analyses were performed to detect metabolism irregularities. Single-cell RNA sequencing (RNA-seq) databases and multiplex immunofluorescence analysis were used to identify the location of NNMT within the tumor microenvironment. The biological functions of NNMT were investigated both in vitro and in vivo, with RNA-seq and chromatin immunoprecipitation-PCR providing insights into underlying mechanisms. Finally, single-cell RNA-seq data and immunohistochemistry of primary tumors were analyzed to validate the main findings.
Results
Untargeted metabolomic analyses revealed metabolic aberrations in amino acids, organic acids, lipids, and nicotinamide pathways, which are linked to metastasis of non-small cell lung cancer. NNMT is a key enzyme in nicotinamide metabolism, and we found the bulk tissue mRNA level of NNMT gene was inversely associated with LUAD metastasis. NNMT was proved to be predominantly expressed in cancer-associated fibroblasts (CAFs) within the stromal regions of LUAD, and a low stromal NNMT expression was identified as a predictor of poor disease-free survival following radical resection of LUAD. The isolation and primary culture of CAFs from LUAD enabled in vitro and in vivo experiments, which confirmed that NNMT negatively regulated the metastasis-promoting properties of CAFs in LUAD. Mechanistically, the downregulation of NNMT led to an increase in intracellular methyl groups by reducing the activity of the methionine cycle, resulting in heightened methylation at H3K4me3. This alteration triggered the upregulation of genes involved in extracellular matrix remodeling in CAFs, including those encoding collagens, integrins, laminins, and matrix metalloproteinases, thereby facilitating cancer cell invasion and metastasis. Reanalysis of single-cell RNA-seq data and immunohistochemistry assays of primary LUAD tissues substantiated NNMT's negative regulation of these genes in CAFs.
Conclusions
This study provides novel insights into the metabolic and epigenetic regulatory functions of NNMT in CAFs, expanding the current understanding of LUAD metastasis regulation and suggesting potential avenues for future research and therapeutic development.
Abbreviations
-
- AOX1
-
- Aldehyde oxidase 1
-
- CAF
-
- Cancer-associated fibroblast
-
- CCK-8
-
- Cell Counting Kit-8
-
- ChIP
-
- Chromatin immunoprecipitation
-
- COL3A1
-
- Collagen type III alpha 1 chain
-
- COL5A1
-
- Collagen type V alpha 1 chain
-
- DMEM
-
- Dulbecco's modified Eagle's medium
-
- ECM
-
- Extracellular matrix
-
- EGFR
-
- Epidermal Growth Factor Receptor
-
- FAP
-
- Fibroblast activation protein
-
- FBS
-
- Fetal bovine serum
-
- FFPE
-
- Formalin-fixed paraffin-embedded
-
- GO
-
- Gene ontology
-
- GSEA
-
- Gene Set Enrichment Analysis
-
- H3K4me3
-
- Histone H3 Lysine 4 Trimethylation
-
- HE
-
- Hematoxylin and Eosin
-
- IHC
-
- Immunohistochemistry
-
- ITGA
-
- Integrin alpha
-
- KEGG
-
- Kyoto Encyclopedia of Genes and Genome
-
- LC-MS
-
- Liquid chromatography-mass spectrometry
-
- LNM
-
- Lymph node metastasis
-
- log2FC
-
- log2FoldChange
-
- LUAD
-
- Lung adenocarcinoma
-
- LUSC
-
- Lung squamous cell carcinoma
-
- MMP11
-
- Matrix metalloproteinase 11
-
- MNA
-
- Methylnicotinamide
-
- NAD+
-
- Nicotinamide adenine dinucleotide
-
- NF
-
- Normal fibroblast
-
- NNMT
-
- Nicotinamide N-methyl transferase
-
- NSCLC
-
- Non-small cell lung cancer
-
- PBS
-
- Phosphate Buffered Saline
-
- QC
-
- Quality control
-
- RNA-seq
-
- RNA sequencing
-
- RT-qPCR
-
- Quantitative real-time PCR
-
- SAH
-
- S-adenosyl-L-homocysteine
-
- SAM
-
- S-adenosyl-methionine
-
- SET1/MLL
-
- Histone methyltransferases/mixed lineage leukemia
-
- siRNA
-
- Small interfering RNA
-
- SMA
-
- Smooth muscle actin (alpha)
-
- TP53
-
- Tumor Protein 53
-
- UHPLC
-
- Ultra-High Performance Liquid Chromatography
-
- WDR5
-
- WD repeat domain 5
1 BACKGROUND
Cancers are a major contributor to the global disease burden, which is projected to increase over the next two decades [1, 2]. In 2022, there were an estimated 20 million new cancer cases and 9.7 million cancer-related deaths worldwide [3]. Early diagnosis and treatment are critical for effective cancer management, with surgical resection being the primary curative approach for most solid tumors [4]. However, recurrence and metastasis of solid cancer remain significant challenges, frequently leading to treatment failure and increased mortality. Lung cancer is the most common cancer globally and the leading cause of cancer-related deaths [3]. Lung adenocarcinoma (LUAD) is the predominant subtype of non-small cell lung cancer (NSCLC) in East Asia [5, 6]. Although radical resections, such as anatomic lobectomy with systematic lymph node dissection, significantly improve patient prognosis [7], the complex nature of NSCLC metastasis challenges the conventional perspectives that the primary tumors only metastasize after reaching a certain size. Notably, many NSCLC cases exhibit lymph node or distant metastasis even when the primary tumor is small [8, 9]. Clinicians have also observed rapid recurrence in NSCLCs with small primary tumors following radical resection [10]. These cases underscore the complexity of recurrence and metastasis mechanisms in NSCLC.
Deregulation of cellular metabolism was recognized as a hallmark of cancer [11, 12], playing a critical role in various biological processes, particularly tumorigenesis and metastasis [13-15]. Altered metabolism in cancer cells, immune cells, and stromal cells within the tumor microenvironment can interactively influence numerous physiological and pathological processes, particularly metastasis [16, 17]. Metabolism reprogramming in cancer cells typically results from the deregulation of metabolic enzymes and transporters, which is induced by genome variations or environmental changes [13, 18, 19]. Abnormalities in metabolic substrates and products can directly affect biological processes [20, 21]. For instance, the reprogramming of aerobic glycolysis supports the extensive biosynthetic programs necessary for active cell proliferation [22]. However, the detailed mechanisms of deregulated cellular metabolism in NSCLC, particularly its link to invasion and metastasis, are not yet fully understood. Investigating these issues is essential for identifying novel approaches to cancer treatment.
Nicotinamide metabolism has garnered attention due to its connection with the salvage pathway of nicotinamide adenine dinucleotide (NAD+) and the methionine cycle, which can impact various biological and pathological processes [23, 24]. Nicotinamide N-methyl transferase (NNMT), a key metabolomic enzyme in nicotinamide metabolism, exhibits high heterogeneity in expression across different tumors, with elevated levels found in most gastrointestinal and urogenital cancers [23, 25]. In contrast, its expression in tumor tissues of LUAD and lung squamous cell carcinoma (LUSC) has been reported to be lower than in normal tissues [23, 25]. NNMT has also been reported to be associated with proliferation, metastasis, and anticancer resistance in several gastrointestinal and urogenital cancer cell types [26-28]. Growing evidence supports cancer-associated fibroblasts (CAFs) as the primary cellular sources of NNMT expression in solid tumors [21, 29]. However, to date, the specific location and function of NNMT in lung cancers have not been clearly defined.
This study aimed to reveal metabolomic irregularities associated with lung cancer recurrence and metastasis, with a particular focus on nicotinamide metabolism in LUAD. The specific location, biological function, and underlying mechanisms of NNMT in CAFs within LUAD were thoroughly elucidated. Investigating these factors may provide valuable insights into the metabolic regulation of LUAD metastasis.
2 MATERIALS AND METHODS
2.1 Patient samples
NSCLC patients included in this study underwent video-assisted thoracoscopic surgery, either lobectomy or segmentectomy, along with hilar and mediastinal lymph node dissection or sampling, as the primary treatment at Peking University People's Hospital (Beijing, P. R. China). Both tumor tissues and adjacent normal tissues were collected prospectively during surgery and were either stored at -80°C, in liquid nitrogen, or used for paraffin embedding. The inclusion criteria were as follows: 1) patients who were over 18 years of age; 2) those who had undergone pathologically confirmed radical resection; and 3) individuals who provided written informed consent for the utilization of their biological samples and medical records. Exclusion criteria included 1) patients with prior anticancer treatment; 2) a cancer history within five years; and 3) hepatorenal insufficiency, metabolic diseases, or rheumatic immune diseases. Tumor size, pathological stage, lymph node metastasis (LNM), and other pathological features were evaluated by pathologists after tumor resection using standard pathological tests. The American Joint Committee on Cancer's 8th edition of the tumor-node-metastasis (TNM) classification system was used for cancer staging or restaging. Follow-up after curative surgery included an outpatient visit every 3 months for the first 2 years, followed by visits at 6-month intervals thereafter. Recurrence was defined as any regional LNM, lung metastasis, or distant metastasis identified during the follow-up. In cohort studies, patients were generally matched by age, sex, smoking history, body mass index, surgical approach, and tumor size. Pathological stage and LNM at the time of treatment were not used for patient matching, as they are indicative of the primary tumors’ metastatic potential, given the assurance of comparable tumor size.
For the inclusion of NSCLC tumor tissues in the metabolomic and lipidomic analysis, beyond the previously mentioned criteria, only those tumors that were pathologically confirmed as stage T1 (with a diameter no greater than 3 cm) and had a postoperative follow-up period exceeding three years were included. The NSCLC tumor samples were segregated into recurrence and control groups based on the occurrence or non-occurrence of recurrence within three years, as depicted in Figure 1A. Ultimately, 21 tumor samples from the recurrence group and 23 tumor samples from the control group were selected for metabolomic and lipidomic analysis (Supplementary Table S1).
The use of clinical data and human tissue samples was approved by the Ethics Committee Board of Peking University People's Hospital (2021PHB155-001 and 2021PHB182-001). All patients were informed verbally and in writing about the research and provided their signed informed consent for the use of their biological samples and medical records.
2.2 Metabolite extraction for lipidomic analysis
The quality control (QC) sample was obtained by pooling together different samples. 100 µL of the liquid-liquid extraction solution (chloroform-methanol 2:1, v/v) was added to 20 mg of each tissue sample, including the QC sample. The samples were vortexed for 30 seconds, vibrated at 1,200 rpm for 8 min, and centrifuged at 12,000 rpm for 10 min. The lower organic phase of each sample containing hydrophobic metabolites was collected separately into a new test tube and evaporated under vacuum at room temperature. The residue was dissolved in a 25 µL dissolving solution (chloroform-methanol 1:1, v/v), vortexed for 30 seconds, and then diluted by adding 75 µL diluting solution (isopropanol-acetonitrile-H2O 2:1:1, v/v/v). The mixture was further vortexed for 30 seconds, centrifuged at 12,000 rpm for 15 min, and the supernatant was transferred into vials for liquid chromatography-mass spectrometry (LC-MS) analysis.
2.3 Metabolite extraction for metabolomic analysis
The QC sample was obtained by pooling different samples together. 200 µL of the pre-cooled methanol was added to 30 mg of each tissue sample, including the QC sample. The samples were vortexed for 30 seconds, vibrated at 1,200 rpm for 15 min, and centrifuged at 12,000 rpm for 15 min. The supernatant of each sample was collected into a new test tube and evaporated under vacuum at room temperature. The residue was dissolved in a 50 µL dissolving solution (acetonitrile-H2O 1:1, v/v). The mixture was then vortexed for 30 seconds, centrifuged at 12,000 rpm for 15 min, and the supernatant was transferred into vials for LC-MS analysis.
2.4 Reversed-phase liquid chromatography-mass spectrometry (RPLC-MS) analysis
The Ultimate 3000 Ultra-High Performance Liquid Chromatography (UHPLC) system (Thermo Fisher Scientific, Waltham, MA, USA) and CSH C18 column (Cat. No. 186005297, Waters, Milford, MA, USA) were used for untargeted lipidomic analysis. Column temperature was set to 50°C. Mobile phase A consisted of acetonitrile (Cat. No. A955-212, Fisher Scientific, Waltham, MA, USA) and water (60/40, v/v) with 10 mmol/L ammonium acetate (Cat. No. 238074, Sigma-Aldrich, St. Louis, MO, USA) and 0.1% formic acid (Cat. No. FX0450, Sigma-Aldrich, St. Louis, MO, USA). Mobile phase B comprised isopropanol (Cat. No. AC149320010, Thermo Fisher Scientific, Waltham, MA, USA) and acetonitrile (90/10, v/v) with 10 mmol/L ammonium acetate and 0.1% formic acid. The flow rate was set to 0.3 mL/min. The gradient of liquid phase was set as follows: 0 min - 40% B; 2 min - 43% B; 2.1 min - 50% B; 10 min - 60% B; 10.1 min - 75% B; 16 min - 99% B; 17 min - 99% B; 18 min - 40% B; and 19 min - 40% B. Q-Exactive (hybrid quadrupole-Orbitrap mass spectrometer) coupled with heated electrospray ionization source was used for mass analysis. Data-dependent acquisition mode was used. Each acquisition cycle consisted of one survey scan (MS1 scan) at 35,000 resolutions from 190 m/z to 1,200 m/z, followed by ten MS/MS scans in high-energy collisional dissociation mode at 17,500 resolutions. MS/MS parameters were set as follows: automatic gain control target of 5 × 106 (maximum injection time 80 ms) for MS1 scan and 1 × 105 (maximum injection time 70 ms) for MS/MS scan; fixed first mass at 50 m/z; dynamic exclusion of 8 seconds; stepped normalized collision energy set to 15, 30, and 45. The heated electrospray ionization source parameters were set as follows: spray voltage of 3.3 kV for positive ion mode and 3.0 kV for negative ion mode; ion source sheath gas at 40; aux gas at 10; capillary temperature at 320°C; probe heater temperature at 300°C; S-lens radiofrequency level at 55.5. QC samples were analyzed repeatedly during the batch of sample acquisition to evaluate the stability of the LC-MS instrument. All samples were acquired in the positive-negative switching ion mode.
2.5 Hydrophilic interaction liquid chromatography-mass spectrometry (HILIC-MS) analysis
The Ultimate 3000 UHPLC system (Thermo Fisher Scientific, Waltham, MA, USA) and Xbridge BEH Amide column (Cat. No. 186004455, Waters, Milford, MA, USA) were used for untargeted metabolomic analysis of hydrophilic metabolites. Column temperature was set to 30°C. Mobile phase A consisted of 5 mmol/L ammonium acetate in water and 5% acetonitrile, and acetonitrile was used as mobile phase B. The flow rate was set to 0.35 mL/min. The gradient of liquid phase was set as follows: 0 min - 95% B; 2 min - 95% B; 15 min - 50% B; 18 min - 50% B; 19 min - 95% B and 23 min - 95% B. Q-Exactive (hybrid quadrupole-Orbitrap mass spectrometer) coupled with heated electrospray ionization source was used for mass analysis. Data-dependent acquisition mode was used. Each acquisition cycle consists of one survey scan (MS1 scan) at 35,000 resolutions from 60 m/z to 800 m/z, followed by eight MS/MS scans in high-energy collisional dissociation mode at 17,500 resolutions. MS/MS parameters were set as follows: automatic gain control target of 5 × 106 (maximum injection time 100 ms) for MS1 scan and 2 × 105 (maximum injection time 64 ms) for MS/MS scan; dynamic exclusion of 8 seconds; stepped normalized collision energy set to 15, 25, and 35. The heated electrospray ionization source parameters were identical to that of RPLC-MS analysis. QC samples were analyzed repeatedly during the batch of sample acquisition to evaluate the stability of the LC-MS instrument. All samples were acquired in the positive-negative switching ion mode.
2.6 Analysis of metabolomic and lipidomic data
Both metabolomic and lipidomic analyses were performed using MetaboAnalyst 5.0 [30], following similar procedures. The matrix data of metabolites and peak intensities were uploaded to an online bioinformatics platform (https://www.metaboanalyst.ca/MetaboAnalyst/faces/home.xhtml). The data derived from QC samples, with a relative standard deviation threshold of 20%, were used for data filtering. Following sample normalization and auto-scaling, the processed data were used for bioinformatics analysis. The criteria of |log2FoldChange (log2FC)| > 0.3 and P-value < 0.05 were applied to identify differential metabolites. The Kyoto Encyclopedia of Genes and Genome (KEGG) pathway database was referred for pathway enrichment analysis based on differential metabolites identified [31]. Additionally, the Relational database of Metabolomics Pathways (RaMP), which integrates biological pathways from the KEGG, Reactome (https://reactome.org/), WikiPathways (https://www.wikipathways.org/), and the Human Metabolome Database (http://www.hmdb.ca/), was utilized for extensive enrichment analysis [32].
2.7 Quantification of 1-methylnicotinamide (MNA)
Intracellular 1-MNA levels were analyzed using a targeted metabolomic approach with LC-MS. Biological cell samples and standards (Cat. No. SML0704, Sigma-Aldrich, St. Louis, MO, USA) were treated with 1% zinc sulphate solution to precipitate protein, followed by shaking at 400 rpm for 30 min and centrifugation at 14,000 rpm for 15 min. The resulting supernatant was used for further analysis. UHPLC separation was performed using an Agilent 1290 Infinity II series UHPLC System (Agilent Technologies, Santa Clara, CA, USA), equipped with an Agilent ZORBAX Hilic Plus (Cat. No. 959961-901, Agilent Technologies, Santa Clara, CA, USA). Mobile phase A consisted of 0.1% formic acid in water, and mobile phase B was acetonitrile. The column temperature was maintained at 35°C, with the auto-sampler temperature set to 4°C and an injection volume of 1 µL. An Agilent 6460 triple quadrupole mass spectrometer (Agilent Technologies, Santa Clara, CA, USA), coupled with an Agilent Jet Stream electrospray ionization interface, was used for assay development. Multiple reaction monitoring parameters for the targeted analytes were optimized via flow injection analysis, by injecting the standard solutions of the individual analytes into the atmospheric pressure ionization source of the mass spectrometer. Four transitions were selected for 1-MNA and collision energies were optimized for maximum sensitivity. Agilent MassHunter Work Station Software (B.08.00, Agilent Technologies, Santa Clara, CA, USA) was used for multiple reaction monitoring data acquisition and processing, followed by data analysis using MultiQuant software (AB Sciex, Concord, Ontario, Canada). The areas under the extracted ion chromatograms for 1-MNA were calculated and normalized against internal standards.
2.8 DNA sequencing
Next-generation sequencing was utilized for DNA sequencing to explore the correlation between NNMT expression and genomic mutations in LUAD. A total of 89 LUAD tumor tissues were analyzed (Supplementary Table S2). Specifically, 18 tissues were examined using a 363-gene panel (Cat. No. HR363, Berry Oncology, Beijing, China), 35 tissues were assessed with a 457-gene panel (Cat. No. HR457, Berry Oncology, Beijing, China), and 36 tissues were screened with a 520-gene panel (Cat. No. RS520, Burning Rock Biotech, Guangzhou, China). The methodologies for genomic DNA extraction, library preparation, sequencing, QC, and variant calling have been thoroughly documented [5]. The total genomic mutation count was determined as the number of nonsynonymous mutations per sample, encompassing nonsynonymous single nucleotide variations, in-frame insertions and deletions, frameshift insertions and deletions, and stop-gain and splice site alterations. Particular attention was given to the commonly recognized driver genes in LUAD, such as those encoding Epidermal Growth Factor Receptor (EGFR) and Tumor Protein 53 (TP53).
2.9 RNA sequencing (RNA-seq)
Total RNA extraction from samples was performed using Trizol reagent (Cat. No. 15596026, Invitrogen, Carlsbad, CA, USA). RNA quality assessment was conducted using Agilent 2200 TapeStation System (Agilent Technologies, Santa Clara, CA, USA), and samples with an RNA integrity number > 7.0 were deemed suitable for cDNA library construction. The TruSeq Stranded mRNA Library Prep Kit (Illumina Inc., San Diego, CA, USA) was employed for the construction of cDNA libraries according to the manufacturer's instructions. The protocol encompassed several steps. Poly-A containing mRNA was purified from 1 µg total RNA using oligo (dT) magnetic beads and fragmented into 200-600 bp using divalent cations at 85°C for 6 min. The cleaved RNA fragments were used for first- and second-strand cDNA synthesis. dUTP mix was used for second-strand cDNA synthesis, allowing for the removal of the second-strand. The cDNA fragments were end-repaired, A-tailed, and ligated with indexed adapters. The ligated cDNA products were purified and treated with uracil DNA glycosylase to remove the second-strand cDNA. Purified first-strand cDNA was enriched by PCR to create the cDNA libraries. The libraries were quality-controlled using the Agilent 2200 TapeStation System (Agilent Technologies, Santa Clara, CA, USA) and sequenced on the DNBSEQ-T7 platform (BGI Genomics, Shenzhen, Guangdong, P. R. China) using a 150 bp paired-end run.
2.10 Analysis of RNA-seq data
The raw sequencing data were evaluated by FastQC (Babraham Bioinformatics, Cambridge, Cambridgeshire, United Kingdom) in terms of quality distribution of nucleotides, position-specific sequencing quality, GC content, the proportion of PCR duplication, and Kmer frequency. HTseq was used to call the gene counts and the Reads Per Kilobase Million was used to calculate gene expression [33]. The DESeq2 algorithm was used to identify the differentially expressed genes based on the criteria of |log2FC| > 1 and a false discovery rate < 0.05 [34]. Gene Ontology (GO) analysis [35] was conducted using the “clusterProfiler” R package [36] to clarify the biological significance of the differentially expressed genes. The GO annotations were sourced from the MSigDB Human collections (https://www.gsea-msigdb.org/gsea/msigdb/human/genesets.jsp?collection = C5). Fisher's exact test was utilized to identify significant GO terms with a P-value threshold of less than 0.05. GO terms exhibiting a P-value of less than 0.01, derived from both up- and down-regulated differentially expressed genes, were chosen to construct the GO-Tree. This tree summarizes the impacted biological functions observed in the experiment [37]. We performed pathway analysis with the KEGG database to identify significant pathways of differentially expressed genes [31], employing Fisher's exact test for pathway selection (P-value < 0.05). Gene-Act-Network analysis was performed using Cytoscape (https://cytoscape.org/), a widely recognized platform for visualizing complex networks. For this analysis, the KEGG database was utilized to construct a network that mapped the interactions among genes, proteins, and compounds as recorded in the database [38].
2.11 Multiplex immunofluorescence analysis
Multiplex immunofluorescence of 20 LUAD tumor tissues was conducted according to previously described methods (Supplementary Table S3) [5, 39]. The Opal 7-Color Manual IHC Kit (Cat. No. NEL811001KT, PerkinElmer, Waltham, MA, USA) was used for staining slides prepared from formalin-fixed paraffin-embedded (FFPE) LUAD tissues. The primary antibodies targeting fibroblast activation protein (FAP, Cat. No. ab53066, Abcam, Cambridge, MA, USA) and NNMT (Cat. No. ab119758, Abcam, Cambridge, MA, USA) were applied. PerkinElmer Vectra (Vectra 3.0.5; Akoya Biosciences, Marlborough, MA, USA) was employed for imaging. Hematoxylin and Eosin (HE) staining was used to distinguish tumor and stromal compartments, with evaluation by experienced pathologists. Fifteen high-resolution multiplex immunofluorescence images were randomly selected as training samples to develop an algorithm of tissue segmentation, cell segmentation, phenotyping, and positivity score using inForm Advanced Image Analysis software (inForm 2.5.0; Akoya Biosciences, Marlborough, MA, USA). The algorithm was subsequently applied in batch analysis to all images. Fifteen images of the tumor-stroma juncture were integrated to calculate the cell densities and fractions. Regarding the classification of staining intensities for FAP and NNMT, the positive threshold for fluorescence intensity was initially determined by two experienced pathologists. The cytoplasmic fluorescence intensity thresholds were set at 2.3 for FAP and 6.3 for NNMT. Staining intensities were then categorized as low (1-2-fold of the threshold), moderate (2-3-fold of the threshold), and high (over 3-fold of the threshold) based on their fluorescence intensity. The H-score, a semiquantitative scoring indicator, was calculated as 3 × percentage of high-intensity staining+2 × percentage of moderate-intensity staining+percentage of low-intensity staining [40, 41].
2.12 Immunohistochemistry (IHC) staining and analysis
Primary antibodies against NNMT (Cat. No. 33361T, CST, Danvers, MA, USA), collagen type III alpha 1 chain (COL3A1, Cat. No. 22734-1-AP, Proteintech, Rosemont, IL, USA), collagen type V alpha 1 chain (COL5A1, Cat. No. 86903T, CST, USA), and integrin alpha 5 (ITGA5, Cat. No. ab150361, Abcam, Cambridge, MA, USA) were utilized for IHC staining of 20 LUAD tumor tissues (Supplementary Table S4). Sections of FFPE tumor specimens were dewaxed at 60°C for 5 h and rehydrated. Endogenous peroxidase activity was blocked using 3% H2O2 for 20 min. Antigens retrieval was performed with a citrate buffer (10 mmol/L, pH 6.0) at 100°C for 15 min in a microwave oven. After washing with ddH2O, slides were blocked with 2% bovine serum albumin for 30 mins, and then incubated with primary antibodies overnight at 4°C. Following three washes with Phosphate Buffered Saline (PBS), slides were incubated with horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies (Proteintech, Rosemont, IL, USA) at 37°C for 30 min. Slides were then washed and incubated with diaminobenzidine for 5 min, and nuclei were counterstained with hematoxylin. Staining results were independently analyzed by two experienced pathologists using Image-Pro Plus software (version 6.0, Media Cybernetics, Rockville, MD, USA). Stromal and tumor compartments were distinguished based on HE staining. The average optical density of slides was calculated as the integrated option density divided by the distribution area [42] based on the comprehensive analysis of five randomly selected fields, and it was used for protein quantification.
2.13 Isolation and culturing of primary fibroblasts
Primary CAFs and normal fibroblasts (NFs) were obtained from surgically resected tumor tissues and adjacent normal tissues (≥ 2 cm away from the margin of tumors) of NSCLC, respectively. Samples were stored and transferred in Falcon tubes containing Dulbecco's modified Eagle's medium (DMEM, Cat. No. 11965-092, Gibco, Waltham, MA, USA) with a high concentration of antibiotics (300 U/mL Penicillin, 300 µg/mL Streptomycin, and 0.75 µg/mL Amphotericin B, Cat. No. 15240096, Gibco, Waltham, MA, USA). Tissues were thoroughly rinsed with PBS, minced into 1-3 mm3 fragments, and digested sequentially with trypsin (2.5 mg/mL, AQ5151, Aoqing Biotechnology, Beijing, P.R. China) and collagenase D (1 mg/mL, Cat. No. 11088858001, Roche Diagnostics GmbH, Basel, Switzerland). The digestion process was carried out in a 37°C constant temperature water bath with periodic mixing. The resulting mixture was filtered through 70 µm strainers to separate cells from undigested tissues. Cells were collected via centrifugation (1,000 rpm for 5 min), washed with PBS, resuspended, and plated in T25 culture flask containing DMEM supplemented with 10% fetal bovine serum (FBS) and high concentration antibiotics. Cultures were conducted at 37°C in a humidified incubator with 5% CO2, and the medium was replaced daily. If no signs of bacterial or fungal contamination were observed after two days, the concentration of antibiotics was reduced to normal levels (100 U/mL Penicillin and 100 µg/mL Streptomycin, Cat. No. 15140122, Gibco, Waltham, MA, USA). Typically, fibroblast cultures reach confluence within 10-15 days, after which the primary cultures are subcultured at a 1:2 ratio. For specific experiments, cells were treated with inhibitor of WD repeat domain 5 (WDR5, WDR5-0103, Cat. No. HY-19347, MedChemExpress, Monmouth Junction, NJ, USA), NNMT inhibitors, or a vehicle (DMSO) control for 72 h.
2.14 Lung cancer cell lines
The A549 and H1299 cells were purchased from the cell bank of the Chinese Academy of Sciences and cultured in DMEM supplemented with 10% FBS, 100 U/mL Penicillin, and 100 µg/mL Streptomycin. Cells are cultured at 37°C in a humidified incubator with 5% CO2. Regular testing ensured that all cell lines were free from mycoplasma contamination.
2.15 Construction and transfection of small interfering RNAs (siRNAs) and plasmids
siRNA targeted NNMT, ITGA5, and COL3A1, as well as negative control, were synthesized based on validated sequences (Supplementary Table S5) by Beijing Tsingke Biotech Co., Ltd. (Beijing, P. R. China). Plasmids were also constructed by Beijing Tsingke Biotech Co., Ltd., where pCDNA3.1(+) was assembled with (overexpression) or without (empty vector) the coding sequence of NNMT (Supplementary Table S6). The NheI-XhoI restriction site was selected, and the ampicillin-resistant gene was integrated into the pCDNA3.1(+) vector. Plasmids were amplified in Escherichia coli TOP10 strain cultured in LB medium (Cat. No. L1015, Solarbio, Beijing, P.R. China) supplemented with ampicillin (100 µg/mL, Cat. No. A8180-1, Solarbio, Beijing, P.R. China). Plasmid extraction was performed using the EndoFree Midi Plasmid Kit (Cat. No. 4992853, TIANGEN, Beijing, P.R. China).
siRNA transfection into CAFs was performed using Lipofectamine RNAiMAX Reagent (Cat. No. 13778150, Invitrogen, Waltham, MA, USA) according to the manufacturer's protocols, with a final siRNA working concentration of 12.5 nmol/L. Plasmids transfection into CAFs utilized X-tremeGENE HP DNA Transfection Reagent (Cat. No. 06366244001, Roche Diagnostics GmbH, Basel, Switzerland) according to the manufacturer's protocol, with a final plasmid DNA working concentration of 1 µg/mL. The efficacy of knockdown and overexpression was assessed through quantitative real-time PCR (RT-qPCR) and Western blotting 48 h post-transfection. Histone methylation tests and functional experiments with CAFs were typically conducted 72 h after transfection.
For sequential knockdown of NNMT and ITGA5 or COL3A1, NNMT-targeted siRNA was transfected first, followed by ITGA5- or COL3A1- targeted siRNA 6 h later. In experiments involving WDR5 inhibitors, WDR5-0103 was administered 6 h after NNMT knockdown.
2.16 Proliferation assays
Cell Counting Kit-8 (CCK-8, Cat. No. BS350A, Biosharp, Hefei, Anhui, China) was utilized to evaluate the proliferation of CAFs after transfection with siRNA or plasmid DNA, or after treatment with NNMT protein inhibitors. Specific processes are as follows: a total of 3,000 transfected CAFs in 100 µL medium was seeded and cultured in 96-well plates, with CCK-8 assays performed at 0 h, 24 h, 48 h, 72 h, and 96 h after cell seeding. CCK-8 reagent (10 µL) was added to each well and incubated at 37°C in a humidified incubator with 5% CO2 for 2 h. Absorbance at 450 nm was measured using a microplate reader.
Besides, we performed co-culture of CAFs and lung cancer cells as follows: the conditioned medium from CAFs was collected 48 h after transfection with siRNA or plasmid DNA, or after treatment with NNMT protein inhibitors, centrifuged (1,500 rpm, 5min) to remove cell debris, and used to resuspend A549 and H1299 cells. The resuspended cells were then seeded in 96-well plates (2,000 cells in 100 µL/well). We performed CCK-8 assay and EdU assay to assess the proliferation of A549 and H1299 cells. CCK-8 assays were conducted at 0 h, 24 h, 48 h, 72 h, and 96 h after co-culture with a 2-hour incubation of CCK-8 reagent. Before performing EdU assays, CAFs were co-cultured with A549 or H1299 cells at a 3:1 ratio. Tumor cells were initially added in 24-well plates, while CAFs were subsequently added to the upper compartment of polycarbonate membrane inserts with 0.4 µm pores (Cat. No. 3412, Corning Incorporated, Corning, NY, USA). After 48 h of co-culture, cancer cell proliferation was assessed using EdU-594 (Cat. No. C0078L, Beyotime, Shanghai, China). Briefly, the polycarbonate membrane insert was removed, and cancer cells were incubated with EdU buffer (10 µmol/L) at 37°C for 2 h, followed by fixation with 4% paraformaldehyde for 20 min and permeabilization with 0.3% Triton X-100 for 20 min, and subjected to click additive solution for 30 min. Nuclear staining was performed with Hoechst (Cat. No. C0078S, Beyotime, Shanghai, China), and cells were visualized using fluorescence microscopy.
2.17 Transwell assay
Transwell assays were employed to evaluate the migration of cancer cells in indirect co-culture with CAFs. CAFs were combined with A549 or H1299 cells at a 3:1 ratio, with CAFs (6 × 105 in 600 µL medium) initially added to 24-well plates, followed by the addition of cancer cells (2 × 105 in 100 µL medium) to the upper compartment of polycarbonate membrane inserts with 8.0 µm pores (Cat. No. 3422, Corning Incorporated, Corning, NY, USA). CAFs were cultured in DMEM supplemented with 10% FBS, while cancer cells were cultured in serum-free DMEM. After incubating for 24 h (A549) or 16 h (H1299) at 37°C in a humidified incubator with 5% CO2, cells attached to the upper surface of the inserts were removed using a cotton swab. The remaining cells were fixed with 4% paraformaldehyde for 20 min, stained with 0.5% crystal violet for 10 min, and counted using a microscope in five predetermined fields for each membrane.
Transwell assays were also utilized to assess the invasion of cancer cells in indirect co-culture with CAFs. However, before co-culture, the inserts were covered with Matrigel (Cat. No. 356234, Corning Incorporated, Corning, NY, USA). Matrigel was diluted with serum-free DMEM at a ratio of 1:8, and 100 µL of the diluted Matrigel was evenly applied to the upper surface of the polycarbonate membrane insert. After incubating for 2 h at 37°C in a humidified incubator with 5% CO2, cancer cells were seeded onto the Matrigel-coated membrane. The incubation time for A549 and H1299 cells with CAFs was 48 h and 32 h, respectively.
2.18 Immunofluorescence
CAFs, NFs, and cancer cells were cultured on coverslips for 24 h, followed by fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and blocked with 3% bovine serum albumin. After washing with PBS, cells were incubated overnight with primary antibodies at 4°C. Primary antibodies against FAP (Cat. No. sc-65398, Santa Cruz, Dallas, TX, USA), alpha-smooth muscle actin (SMA, Cat. No. 67735-1-Ig, Proteintech, Rosemont, IL, USA), alcohol dehydrogenase 1B (Cat. No. 66939-1-Ig, Proteintech, Rosemont, IL, USA), myosin heavy chain 11 (Cat. No. sc-6956, Santa Cruz, Dallas, TX, USA), pan-cytokeratin (Cat. No. ab7753, Abcam, Cambridge, MA, USA), CD31 (Cat. No. ab76533, Abcam, Cambridge, MA, USA), and NNMT (Cat. No. ab119758, Abcam, Cambridge, MA, USA) were used. Following incubation with CoraLite488 or CoraLite594-conjugated secondary antibodies (Proteintech, Rosemont, IL, USA), cells were stained with Hoechst (Cat. No. C0078S, Beyotime, Shanghai, China) for nuclear staining and visualized via fluorescence microscopy.
2.19 Western blotting
The Cell Lysis Buffer for Western blotting or Immunoprecipitation (Cat. No. P0013J, Beyotime, Shanghai, China) was employed for total cellular protein extraction, followed by quantification using the bicinchoninic acid protein assay kit (Cat. No. KGPBCA, KEYGEN, Nanjing, Jiangsu, P. R. China). The extracted proteins were resolved on precast sodium dodecyl sulphate-polyacrylamide gel (Tris-Glycine, 4%-20%) and transferred onto a hydrophobic PVDF transfer membrane (Cat. No. IPVH00010, Millipore, Billerica, MA, USA). Blots were blocked with 5% non-fat milk for 1 h and then incubated overnight at 4°C with primary antibodies. Primary antibodies against FAP (Cat. No. sc-65398, Santa Cruz, Santa Cruz, Dallas, USA), SMA (Cat. No. 14395-1-AP, Proteintech, Rosemont, IL, USA), NNMT (Cat. No. ab119758, Abcam, Cambridge, MA, USA), β-actin (Cat. No. 66009-1-Ig, Proteintech, Rosemont, IL, USA), and β-tubulin (Cat. No. TDY043, TDYbio, Wuxi, Jiangsu, P. R. China) were used. Subsequently, blots were probed using horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies (Proteintech, Rosemont, IL, USA), followed by blot development using an enhanced chemiluminescence reagent and visualization via ChemiScope 6100 (ClinX, Shanghai, P. R. China).
For histone immunoblots, a histone extraction kit (Cat. No. ab113476, Abcam, Cambridge, MA, USA) was used following the manufacturer's protocols. Briefly, cells were harvested, pelleted by centrifugation, and then resuspended and treated with pre-lysis buffer and lysis buffer to dissolve membrane and nuclei. Histones were extracted, mixed with balanced-DTT buffer, quantified, and subjected to immunoblotting. The employed primary antibodies used were anti-histone H3 (Cat. No. 39064, Active Motif, Carlsbad, CA, USA) and anti-histone H3 Lysine 4 Trimethylation (H3K4me3, Cat. No. 61979, Active Motif, Carlsbad, CA, USA). Immunoblots for H3 and H3K4me3 were performed separately due to their identical molecular weight. Equal amounts of proteins were employed for immunoblots across all samples.
2.20 RT-qPCR
RNA was extracted from cells using Trizol reagent (Cat. No. 15596026, Invitrogen, Carlsbad, CA, USA), followed by reverse transcription using the PrimeScript RT reagent Kit (Cat. No. RR037A, TaKaRa, Shiga Prefecture, Otsu, Japan) to prepare cDNA. A total of 1,000 ng of RNA was added to a 20-µL reaction system of reverse transcription. RT-qPCR was conducted using SYBR Green qPCR Master Mix (Universal) (Cat. No. HY-K0501A, MedChemExpress, Marlborough, MA, USA). Primers for RT-qPCR of selected genes were derived from published literatures and databases (Supplementary Table S7). A total of 0.5 µL of cDNA from the reverse transcription was added to the 10 µL PCR system with a primer concentration of 0.2 µmol/L. Relative gene expression was evaluated using the 2−ΔΔCt method.
2.21 Chromatin immunoprecipitation (ChIP)-PCR
ChIP assays were conducted using IgG (Cat. No. B900620, Proteintech, Rosemont, IL, USA) and H3K4me3 (Cat. No. 61979, Active Motif, Carlsbad, CA, USA) antibodies with the ChIP Assay Kit (Cat. No. P2078, Beyotime, Wuxi, Jiangsu, China) following the manufacturer's guidelines. In summary, the DNA and proteins in CAF cells were cross-linked with a 1% formaldehyde solution for 10 min at room temperature, followed by quenching and lysis using a glycine solution. The cell lysate was then sonicated to shear the genomic DNA. Agarose gel electrophoresis was used to evaluate the size of the genomic DNA fragments, which helped to adjust the sonication conditions. The final sonication parameters were set to 200 W power, with 20 seconds pulses and 40 seconds intervals, repeated three times, resulting in DNA being predominantly fragmented into sizes ranging from 200 to 800 base pairs. The lysate was then immunoprecipitated with anti-H3K4me3 or anti-IgG antibodies. Immunoprecipitated DNA was then washed, eluted, purified, and analyzed by RT-qPCR. ChIP primers targeting the promoters of selected genes were designed and validated (Supplementary Table S8).
2.22 Enzyme-linked immunosorbent assay (ELISA)
The levels of S-adenosyl-methionine (SAM) and S-adenosyl-L-homocysteine (SAH) in CAFs were assessed using the SAM and SAH ELISA Combo Kit (Cat. No. MET-5151-C, Cell Biolabs, San Diego, CA, USA) following the manufacturer's instructions. Cells were harvested, centrifuged at 3,000 rpm for 10 min at 4°C, sonicated on ice in 1 mL cold PBS, and then centrifuged at 12,000 rpm for 15 min at 4°C. Samples and standards were added to the appropriate conjugated coated plate with a volume of 50 µL. Subsequently, 50 µL of diluted anti-SAM or anti-SAH antibodies were added, and the mixture was incubated at room temperature for 1 h on an orbital shaker. After washing microwell strips, 100 µL of diluted horseradish peroxidase-conjugate secondary antibody was added to each well, followed by incubation at room temperature for 1 h on an orbital shaker. Following another wash, 100 µL of substrate solution was added to each well and incubated at room temperature for 15 min on an orbital shaker. The reaction was stopped by adding a stop solution, and the absorbance of each well at 450 nm was immediately measured using a microplate reader. Standard curves for SAM and SAH were constructed based on optical density and standard concentration, and the concentrations of SAM and SAH in samples were calculated accordingly.
2.23 Cell spheroid assays
Cell spheroid assays were conducted to evaluate the proliferation and invasion of cancer cells directedly co-cultured with CAFs [43, 44]. CAFs were mixed with A549 or H1299 cells at a 2:1 ratio and suspended in DMEM supplemented with 10% FBS and 2.5% Matrigel (Cat. No. 356234, Corning Incorporated, Corning, NY, USA). A total of 100 µL of the mixture containing 8,000 CAFs and 4,000 cancer cells was added to each well of a Nunclon Sphere 96 U Bottom Plate (Cat. No. 174925, Thermo Fisher Scientific, Waltham, MA, USA). After incubating for 24 h at 37°C in a humidified incubator with 5% CO2, cell spheroids were formed. The cell spheroids were gently washed with serum-free DMEM before subsequent assays. For the proliferation assay, spheroids were cultured in 150 µL DMEM supplemented with 10% FBS for 8 to 10 days, and the maximum cross-sectional area was measured as an indicator of proliferation. For invasion assessment, spheroids were encapsulated with 50 µL of Matrigel diluted with serum-free DMEM at a ratio of 1:1 and incubated for 30 min at 37°C to allow for gelation. Subsequently, 100 µL of DMEM with 20% FBS was added, and spheroids were cultured at 37°C in a humidified incubator with 5% CO2 for 4 to 6 days. The maximum expansion area of the spheroids was recorded as an indicator of invasion capacity [45].
2.24 Xenograft models
All animal experiments were approved by the Animal Care Committee of Peking University People's Hospital (2022PHE076), and the experiments adhered to regulations on animal care set by the State Food and Drug Administration of China. In the xenograft model, 6-week-old female nude mice received orthotopic injection in the left lung with 100 µL a mixture of PBS and Matrigel (Cat. No. 356234, Corning Incorporated, Corning, NY, USA) in a 1:1 ratio containing a mixture of 5 × 105 A549 cells and 10 × 105 CAFs. The procedure involved inhalation anesthesia with sevoflurane, right lateral position, a surgical incision 1 cm above the lower edge of the rib arch on the left axillary front line, blunt dissection of subcutaneous tissue and pectoralis, and the injection of cell suspension into the left lung through intercostal muscles using insulin needles. Following these steps, the layered structure of the pectoralis and skin was restored, and anesthesia was discontinued. Termination criteria for the experiments included weight loss exceeding 20% of baseline weight, respiratory or movement difficulties, or reaching the observation endpoint. Finally, euthanasia was performed using CO2 at six weeks for NNMT-knockdown experiments and at four weeks for NNMT-overexpression experiments. Dissection of thoracic and abdominal cavities and the head revealed orthotopic tumors and metastasis, which were assessed through visual recognition and HE staining. Primary tumor volume was calculated using the formula: volume = 0.5 × length × width2. Lung metastases were identified as tumors distinct from the orthotopic lesion, with intrathoracic implantation defined as metastasis located on the parietal pleura and pericardium. The organs detected for distant metastasis included the liver, spleen, kidneys, intestines, mesentery, and brain.
2.25 Utilization of public databases
The public 22Q4 database from the Cancer Cell Line Encyclopedia (https://www.broadinstitute.org/ccle) was queried to evaluate NNMT mRNA expression levels across lung cancer cell lines and fibroblast lines. The Cistrome Data Browser (http://cistrome.org/db/#/) and the UCSC Genome Browser (https://genome.ucsc.edu/) were utilized for searching ChIP-sequencing data, examining the enrichment of H3K4me3 at gene promoters, and providing initial evidence for epigenetic regulation. Publicly accessible single-cell RNA-seq data were analyzed to investigate the expression of NNMT and AOX1 in various cells within the tumor microenvironment and to explore the regulatory effects of NNMT on downstream genes in CAF cells. These single-cell RNA-seq datasets included GSE140819 [46], GSE126111 [47], and E-MTAB-6149 & E-MTAB-6653 [48]. CAFs and other cell types have already been annotated by the data publisher using established markers. The single-cell RNA-seq data underwent initial QC, followed by filtering and normalization using R software (version 4.2.0, R Core Team, Vienna, Austria). Uniform Manifold Approximation and Projection analysis was conducted to discern clusters of various cell types. Simple linear regression was employed to explore the correlation between the normalized expression levels of NNMT and ECM-related genes within CAFs. The median expression level of NNMT served as the threshold to categorize CAFs into high or low expression groups. Gene Set Enrichment Analysis (GSEA) was then performed to investigate the gene sets from GO and KEGG databases that are regulated by NNMT expression in CAFs. The sort order of all genes was established using the Signal2Noise method, with all other parameter settings set to their default values. GSEA analysis was performed using the “clusterProfiler” R package [36] with the OmicShare Analysis Platform (https://www.omicshare.com/).
2.26 Virtual screening of small molecule inhibitors for NNMT protein
The virtual screening was conducted using the D001 database from Topscience (Shanghai, P. R. China), which contains approximately 1.5 million molecules. This process was carried out through a workflow application of Glide within Maestro (Schrödinger 2018, Schrödinger LLC, New York, NY, USA). The program Epik39 was utilized to predict their protonation states at pH 7.2 ± 2.0. The three-dimensional conformations of the molecules were prepared using the LigPrep module in Maestro. The structure of NNMT (PDB ID: 6PVE) was employed to create the receptor grid for docking simulations. The center of the grid was set based on the position of LL319 (OZP) within the structure. We sequentially applied the Glide high-throughput virtual screening, Glide standard precision, and Glide extra precision models for the screening process. At each stage, the top 10% of compounds were retained. Ultimately, candidate molecules were selected manually based on their scores and by analyzing the predicted binding modes. The potential small molecule inhibitors were then synthesized by ChemDiv Inc. (San Diego, CA, USA).
2.27 Statistics and reproducibility
Statistical analyses were performed using IBM SPSS Statistics for Windows (version 22.0, IBM Corp., Armonk, NY, USA), the R software (version 4.2.0, R Core Team, Vienna, Austria), and GraphPad Prism (version 9.0, GraphPad Software, San Diego, CA, USA). Continuous variables were reported as mean ± standard deviation and compared using either Student's t-test or ANOVA. Alternatively, they were reported as median with a 10th to 90th percentile range and compared using the Mann-Whitney U test. Categorical variables were presented as numbers (percentages) and compared using Pearson's chi-square test, Fisher's exact test, or the Mann-Whitney U test. The correlation between two continuous variables was assessed with Pearson correlation analysis. Survival analysis was conducted using the Kaplan-Meier method with log-rank statistics. Disease-free survival was determined from the time of surgery to the first recurrence of the index cancer or to death from any cause. Overall survival was calculated from the time of surgery to the time of death from any cause. The X-tile software was utilized for survival-based cut-point optimization [49]. Unless otherwise indicated, experiments were independently replicated at least three times, with each replicate consisting of three technical replicates. P-values less than 0.05 were considered as statistical significance.
3 RESULTS
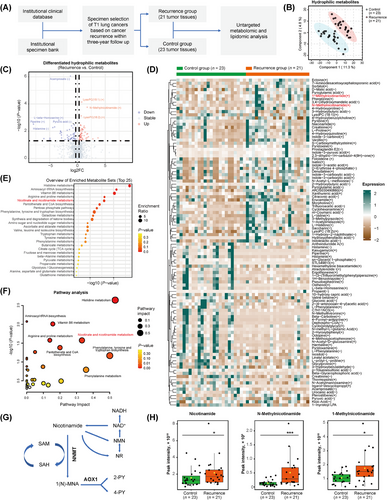
3.1 Metabolic abnormalities associated with NSCLC recurrence
A total of 44 NSCLC tumor tissues were included in the metabolomic and lipidomic analyses, comprising 21 from the recurrence group and 23 from the control group (Figure 1A). The characteristics of the sample cohort are detailed in Supplementary Table S1. The majority of the cases were LUAD, accounting for 93.2% of the total. Patients in the recurrence group showed significantly shorter disease-free survival and overall survival than those in the control group (Supplementary Figure S1A). Despite comparable tumor size, the recurrence group showed increased LNM and more advanced cancer stages at the time of surgery, suggesting higher metastatic potential. The untargeted metabolomic analysis identified 763 hydrophilic metabolites and 1,510 lipids (Figure 1B, Supplementary Figure S1B). Further exploration revealed 103 hydrophilic metabolites and 174 lipids with significantly differential levels between the two groups (|log2FC| > 0.3 and P < 0.05) (Supplementary Table S9-S10, Figure 1C, Supplementary Figure S1C). The differentiated hydrophilic metabolites were mainly amino acids and organic acids involved in multiple metabolic pathways (Figure 1D-F). Deregulated lipid metabolism was primarily related to glycosphingolipid, glycerophospholipid, and sphingolipid (Supplementary Figure S1D-F). The metabolism of nicotinate and nicotinamide was highlighted due to its link to the NAD+ salvage cycle and methionine cycle (Figure 1E-G) [23, 24]. Notably, tumors in the recurrence group exhibited increased levels of nicotinamide and its product 1(N)-MNA (Figure 1H). These findings implied significant metabolic abnormalities were linked to the recurrence and metastasis of NSCLC.
3.2 Downregulation of NNMT and AOX1 was linked to LUAD metastasis
NNMT catalyzes the conversion of nicotinamide to MNA, while AOX1 is responsible for the decomposition of MNA (Figure 1G) [21]. In a cohort of 68 LUAD and 35 LUSC cases (Supplementary Table S11), the mRNA levels of NNMT in LUAD were comparable to those in paired normal tissues, and the mRNA level of AOX1 in LUAD was lower than in paired normal tissues. In contrast, NNMT and AOX1 mRNA levels in LUSC were significantly lower than in paired normal tissues (Figure 2A). Downregulation of NNMT and AOX1 mRNA levels were associated with poor disease-free survival, specifically in LUAD, not in LUSC (Figure 2B, Supplementary Figure S2A). The occurrence of LNM at the time of treatment is a strong predictor for lung cancer recurrence and metastasis [8]. Low mRNA levels of NNMT and AOX1 were significantly associated with the occurrence of LNM in LUAD, but not in LUSC (Supplementary Table S11, Figure 2C, Supplementary Figure S2B). Moreover, mRNA levels of NNMT and AOX1 in LUAD showed synchronous changes (Figure 2D). Collectively, the downregulation of NNMT and AOX1 may be a reason for the increased abundance of nicotinamide and MNA in tumor tissues and is associated with the recurrence and metastasis of LUAD.
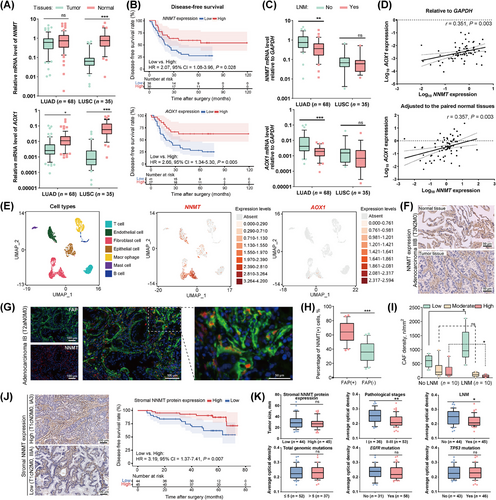
3.3 NNMT expression in CAFs and its decline during LUAD metastasis
NNMT serves as an upstream metabolic enzyme connecting NAD+ and methionine metabolism (Figure 1G). Its expression in NSCLC cell lines is remarkably heterogeneous, with consistently high expression in fibroblasts (Supplementary Figure S2C-D). We reanalyzed the NSCLC single-cell RNA-seq data of GSM4186957 [46] and found that both NNMT and AOX1 were primarily expressed in CAFs (Figure 2E). IHC staining showed significant NNMT protein expression in the stroma of both tumor and paired normal tissues of LUAD (Figure 2F). Multiplex immunofluorescence analysis of 20 LUAD tumor tissues indicated that both NNMT protein and FAP protein, a classical marker of CAFs, were primarily expressed in the stroma with noticeable co-expression (Supplementary Table S3, Figure 2G, Supplementary Figure S2E-F). Over 60% of NNMT-expressing cells were identified as FAP-positive CAFs in the stroma (Figure 2H). We then investigated the association between NNMT expression and the occurrence of LNM at the time of treatment in LUADs. The stroma of LUADs with LNM had increased densities of CAFs with low NNMT expression and decreased densities of CAFs with high NNMT expression, compared to LUADs without LNM (Figure 2I, Supplementary Figure S2G).
We further explored the association between stromal NNMT expression and LUAD characteristics in a cohort of 89 patients (Supplementary Table S2). Stromal NNMT expression was assessed using IHC and categorized based on the median value of average optical density. Low stromal NNMT expression correlated to poor disease-free survival after radical resection of LUAD (Figure 2J). Reduced stromal NNMT expression was linked to an increased risk of LNM and more advanced cancer stage at the time of treatment, despite comparable tumor size (Figure 2K). However, stromal NNMT expression was not related to the total number of genomic mutations or mutations of driver genes like EGFR and TP53. These findings differed from a previous report that NNMT upregulation in cancer cells favored EGFR-TKI resistance [50]. Our findings indicated different roles of NNMT protein in CAFs in cancer cells. After adjusting for pathological stage, LNM, and other confounding factors (Supplementary Table S2), low stromal NNMT expression remained a predictor of poor disease-free survival (multivariable Cox proportional hazards regression models, hazard ratio = 3.41, 95% confidence interval = 1.33-8.77, P = 0.011). These findings suggested that the reduced NNMT expression in LUAD may be attributed to its downregulation in CAFs, which could significantly influence the recurrence and metastasis of LUAD.
3.4 Primary culture of CAFs from LUAD and NNMT expression validation
To explore the biological role of NNMT gene in CAFs within lung cancers, we prospectively collected and isolated 15 NSCLC tumor tissues alongside paired ten adjacent normal tissues for establishing primary cultures of both CAFs and corresponding NFs (Supplementary Table S12, Figure 3A). The primary culture cells from LUAD comprised 11 CAFs and 8 pairs of matched NFs. Unless otherwise indicated, CAFs derived from LUAD were employed for subsequent experiments and analyses.
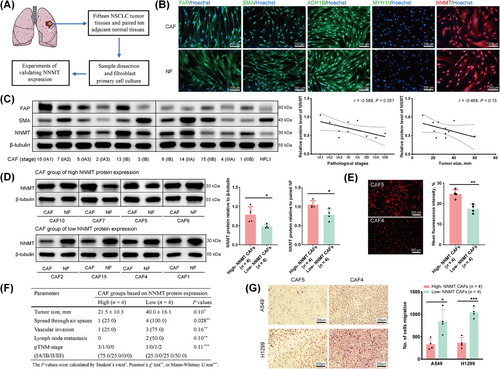
Primary cultured CAFs and NFs exhibited similar morphological features and proliferation processes, however, CAFs had remarkably shorter culture cycles than NFs (Supplementary Figure S3A). CAFs were then confirmed for the absence of cancer cell or immune cell contamination (Supplementary Figure S3B-C). Classic CAF markers, particularly FAP and SMA [51, 52], were expressed in both CAFs and NFs (Figure 3B-C, Supplementary Figure S3D). However, no significant differences in mRNA levels of FAP, SMA, NNMT, AOX1, and other markers were detected between CAFs and NFs (Supplementary Figure S3D). The expression of classic markers and representative functional genes in CAFs remained stable over eight continuous passages (Supplementary Figure S3E). Therefore, CAFs within eight passage culturing were used for subsequent experiments. Previous research has identified the majority of CAFs in tumors as myofibroblasts characterized by the expression of FAP and SMA [51, 53]. Adjacent tissues also showed low percentages of CAF subpopulations, and CAFs have been proven to evolve from NFs [48, 50]. Collectively, these pieces of evidence indicate that our primary cultured CAFs and NFs are derived from the same myofibroblast populations [51-53].
The mRNA level of NNMT was positively correlated with that of AOX1 (Supplementary Figure S3F) in CAFs isolated from LUADs, corresponding to the synchronous downregulation of NNMT and AOX1 in LUAD tissues. The mRNA level of NNMT was positively correlated with, but not highly consistent with, the protein level of NNMT in CAFs (Supplementary Figure S3G), suggesting the existence of post-transcriptional regulation of NNMT expression. Within the established primary cultures of CAFs from LUADs, NNMT protein expression showed a trend of downregulation with advanced pathological stage and increased tumor size (Figure 3C). After adjusting for NNMT protein expression in the paired NFs, NNMT protein expressions in CAFs were categorized into high (CAF 5, 6, 7, 10) or low (CAF 1, 2, 4, 15) groups based on the median value (Figure 3D-E). The primary tumors in the low NNMT group showed trends toward larger tumor sizes, more advanced cancer stages, and higher frequencies of early metastasis events, including spread through air space, vascular invasion, and LNM [54-56] (Figure 3F). CAFs with low NNMT protein expression exhibited an enhanced ability to promote the migration of A549 and H1299 cells (Figure 3G). These findings suggest the potential involvement of NNMT in regulating the metastasis-promoting properties of CAFs.
3.5 NNMT downregulation enhances the metastasis-promoting property of CAFs
Experiments were conducted to knock down NNMT in CAFs with high NNMT protein expression and to overexpress NNMT in CAFs with low NNMT protein expression (Figure 4A). Neither NNMT knockdown nor overexpression had a significant impact on CAF self-proliferation (Supplementary Figure S4A) or the property of CAFs in promoting the proliferation of cancer cells under indirect co-culture (Supplementary Figure S4B-D). However, NNMT-knockdown significantly enhanced CAFs’ ability to promote migration and invasion of cancer cells during indirect coculture (Figure 4B, Supplementary Figure S4E), whereas NNMT overexpression reduced this effect (Figure 4C, Supplementary Figure S4F). Cell spheroid assays combining CAFs and cancer cells were then conducted. In these direct coculture models, NNMT-knockdown in CAFs significantly enhanced the invasion of cell spheroids (Figure 4D-E), while NNMT overexpression in CAFs significantly inhibited the invasion of cell spheroids (Figure 4F). Consistently, neither NNMT knockdown nor overexpression of CAFs significantly influenced the proliferation of cell spheroids (Supplementary Figure S4G-H).
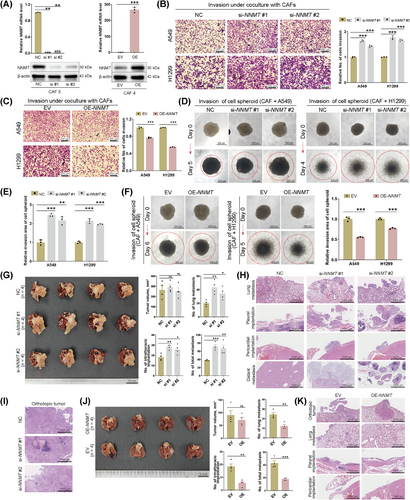
In vivo experiments involved injecting a mixture of NNMT-knockdown or NNMT-overexpression CAFs and A549 cells into the left lung of nude mice. Models of co-injection of NNMT-knockdown CAFs and A549 cells showed increased metastases, including lung metastasis, intrathoracic implantation, and total metastasis (Figure 4G-I). Distant metastases to the liver and mesentery were specifically observed in the NNMT-knockdown groups. Conversely, models of co-injection of NNMT-overexpression CAFs and A549 cells showed decreased lung metastasis, intrathoracic implantation, and total metastasis (Figure 4J-K). Orthotopic tumor size was not significantly influenced by NNMT knockdown or overexpression in co-injected CAFs. Overall, both in vitro and in vivo experiments supported NNMT negatively regulates CAFs in promoting lung cancer metastasis.
3.6 NNMT downregulation in CAFs could induce oncogenic extracellular matrix (ECM) remodeling
By connecting the salvage pathway of NAD+ and SAM metabolism, NNMT has the potential to impact CAF function in multiple ways, particularly by regulating intracellular methyl sink and epigenetic profiles [23, 24]. (Figure 5A). RNA-seq was initially performed on four CAF cell lines following NNMT-knockdown, yielding a total of twelve samples, which led to the identification of 14,408 mRNAs and 4,701 long non-coding RNAs (Supplementary Figure S5A-B). Applying the criteria of |log2(FC)| > 1 and false discovery rate < 0.05, 493 up-regulated and 492 down-regulated genes were identified in two NNMT-knockdown groups compared to the negative control group (Supplementary Table S13, Figure 5B). After NNMT-knockdown, GO analysis, including biological process, molecular function, and cellular component, highlighted activated functions related to ECM remodeling (Figure 5C, Supplementary Figure S5C), and pathway analysis highlighted the activated pathways of protein digestion and absorption and ECM receptor interaction (Figure 5D). Further analyses of the GO-Tree and Gene-Act-Network highlighted the remodeling of ECM organization and function caused by NNMT-knockdown in CAFs (Supplementary Figure S5D-E). Genes enriched in ECM-related function classifications or pathways, mainly producing collagens, integrins, laminins, and matrix metalloproteinase, were confirmed to be upregulated in NNMT-knockdown CAFs and down-regulated in NNMT-overexpression CAFs (Figure 5E-F). CAFs with low NNMT protein expression showed trends of increased expression of these genes compared to CAFs with high NNMT protein expression (Supplementary Figure S5F). Notably, these genes have been widely reported to be associated with cancer invasion and metastasis [57-60]. Collectively, these results indicated that ECM remodeling may be involved in NNMT's negative regulation of the metastasis-promoting property of CAFs in LUAD.
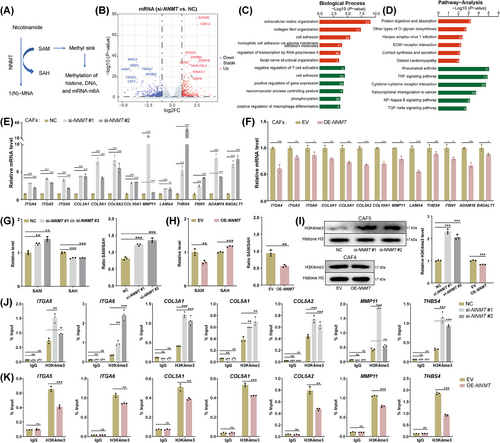
3.7 H3K4me3 mediates the regulation of NNMT on ECM-related genes in CAFs
SAM is the principal biological methyl donor in cells [61]. The transfer of a methyl group from SAM to nicotinamide by NNMT protein could influence intracellular active methyl sink and the methylation of DNA, RNA, and histones (Figure 5A). DNA hypermethylation generally represses gene transcription [62], while RNA methylation affects post-transcriptional expression [63]. Histone lysine methylations can confer either active or repressive transcription, depending on their positions and methylation states [64, 65]. Our exploration of the Cistrome Data Browser and the UCSC Genome Browser revealed a broad presence of H3K4me3 at the promoters of the mentioned ECM-related genes (Supplementary Figure S5G). H3K4me3, a hallmark of actively transcribing and poised genes, could mediate the negative regulation of NNMT on the expression of ECM-related genes in CAFs [64, 65].
NNMT-knockdown led to an increased level of SAM and a decreased level of SAH, along with an increased SAM/SAH ratio (Figure 5G). Additionally, the intracellular level of 1-MNA significantly decreased after NNMT-knockdown (Supplementary Figure S5H). Conversely, NNMT-overexpression resulted in reverse changes in SAM, SAH, the SAM/SAH ratio, and 1-MNA levels (Figure 5H, Supplementary Figure S5H). Interestingly, CAFs with low NNMT protein expression exhibited higher intracellular levels of 1-MNA compared to those with high NNMT protein expression (Supplementary Figure S5I). This observation aligns with the increased organizational level of 1-MNA and decreased NNMT expression observed in LUADs, which are at a higher risk of recurrence and metastasis. The reduced expression of AOX1 likely plays a significant role in this phenomenon (Supplementary Figure S3F). A significantly increased overall level of H3K4me3 caused by NNMT knockdown and a moderately decreased level caused by NNMT overexpression was confirmed (Figure 5I). Enrichment of H3K4me3 at the promoters of representative ECM-related genes increased in NNMT-knockdown CAFs (Figure 5J) and decreased in NNMT-overexpression CAFs (Figure 5K). In addition, CAFs with low NNMT protein expression exhibited a higher overall level of H3K4me3 than those with high NNMT protein expression (Supplementary Figure S5J). The enrichment of H3K4me3 at the promoters of ECM-related genes in CAFs with low NNMT protein expression was higher than that in CAFs with high NNMT protein expression (Supplementary Figure S5K). Collectively, H3K4me3 could mediate the negative regulation of NNMT on ECM-related genes in CAFs.
3.8 H3K4me3 as the central program in NNMT-regulated metastasis-promoting properties of CAFs
The histone methyltransferases/mixed lineage leukemia (SET1/MLL) family is responsible for the methylation of H3K4me3 [64]. WDR5-0103, a cell-permeant inhibitor of SET1/MLL histone methyltransferases, effectively antagonizes their catalytic activity by competing for binding sites on WDR5, a conserved core subunit of SET1/MLL family members [66]. At a concentration of 10 µmol/L, WDR5-0103 significantly reduced the overall H3K4me3 level in both naïve and NNMT-knockdown CAFs (Figure 6A). The subsequent treatment of NNMT-knockdown CAFs with WDR5-0103 markedly diminished their ability to promote the migration and invasion of cancer cells (Figure 6B, Supplementary Figure S6A) and the invasion of cell spheroids (Figure 6C). Concurrent with WDR5-0103 administration, the expression of ECM-related genes in NNMT-knockdown CAFs was significantly down-regulated (Figure 6D, Supplementary Figure S6B). A reduction in H3K4me3 enrichment at the promoters of ECM-related genes was identified in NNMT-knockdown CAFs treated with WDR5-0103 (Figure 6E, Supplementary Figure S6B).
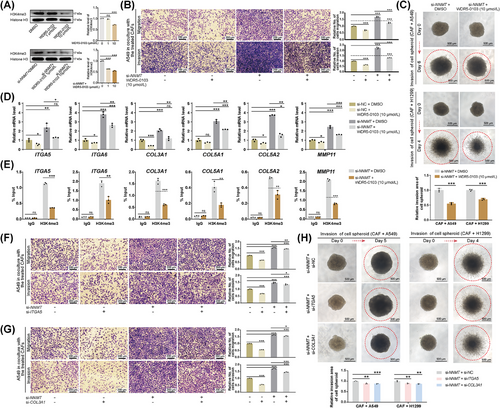
ITGA5 and COL3A1, recognized as canonical genes associated with ECM remodeling, are acknowledged for their roles in cancer metastasis [57, 67], and thus were subjected to further investigation. Sequential knockdown of NNMT, followed by ITGA5 or COL3A1, was conducted and confirmed (Supplementary Figure S6C-D). Sequential knockdown of ITGA5 or COL3A1 after NNMT-knockdown only modestly diminished the metastasis-promoting property of CAFs in both indirect and direct coculture models (Figure 6F-H, Supplementary Figure S6E-F). The efficiency of reversing NNMT-knockdown-induced metastasis via WDR5-0103 treatment was significantly higher than that achieved by subsequent knockdown of ITGA5 and COL3A1. (Supplementary Figure S6G-H). Collectively, H3K4me3 emerges as the central regulatory program in the negative regulation of NNMT for the metastasis-promoting properties of CAFs, characterized by widespread regulation of ECM-related genes.
3.9 Inhibitor of NNMT enhances metastasis-promoting properties of CAFs
Eight candidate small molecule inhibitors targeting the NNMT protein were ultimately selected and synthesized (Supplementary Table S14). NNMTi-4 (E856-0169) was selected for further investigations due to its superior performance in increasing the overall H3K4me3 level in CAFs; the working concentration utilized was 10 µmol/L (Figure 7A). NNMTi-4 showed no significant cytotoxicity and did not affect the self-proliferation of CAFs (Supplementary Figure S7A-B), nor did it impact the CAF's ability to promote the proliferation of cancer cells (Supplementary Figure S7C). An increased SAM level, a decreased SAH level, an increased SAM/SAH ratio, and a decreased 1-MNA level were observed in CAFs treated with NNMTi-4, indicating effective inhibition (Figure 7B, Supplementary Figure S7D). Treatment with NNMTi-4 significantly enhanced the ability of CAFs to promote the migration and invasion of lung cancer cells (Figure 7C) and the invasion of cell spheroids (Figure 7D). NNMTi-4 treatment was further confirmed to increase mRNA levels of ECM-related genes in CAFs (Figure 7E). Increased enrichment of H3K4me3 at the promoters of ECM-related genes was detected in CAFs following NNMTi-4 treatment (Supplementary Figure S7E). These results confirm the negative regulation of NNMT on the metastasis-promoting properties of CAFs through H3K4me3-mediated epigenetic programs.
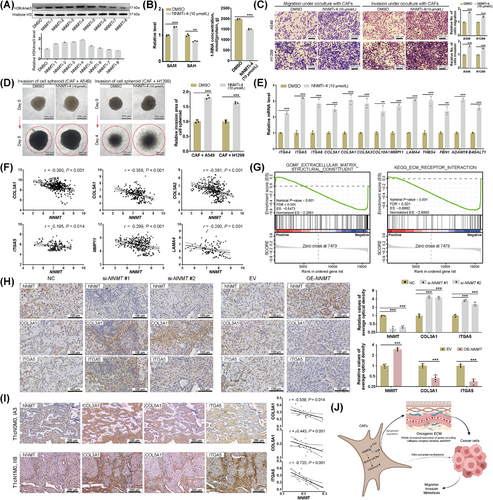
3.10 Organizational validation of main findings
We initially reanalyzed publicly available single-cell RNA-seq datasets of NSCLCs, specifically GSE140819 [46], GSE126111 [47], and E-MTAB-6149 & E-MTAB-6653 [48], to identify fibroblast subclusters. Within these subclusters, we observed a negative correlation between NNMT expression and the expression of ECM-related genes (Supplementary Figure S7F). The single-cell RNA-seq data of CAFs from NSCLCs, sourced from GSE140819 [46] and E-MTAB-6149 & E-MTAB-6653 [48], were integrated, filtered, and normalized (Supplementary Table S15). Analysis of annotated CAFs demonstrated a negative correlation between the expression of NNMT and representative ECM-related genes (Figure 7F, Supplementary Figure S7G). GSEA, incorporating both GO and KEGG gene sets, was subsequently performed on CAFs derived from the single-cell RNA-seq data of E-MTAB-6149 and E-MTAB-6653 (Supplementary Table S16) [48]. CAFs with low NNMT expression showed significant activation of gene sets related to remodeling of ECM organization and function (Supplementary Tables S17-S18), particularly the “Molecular function: Extracellular matrix structure constituent” and the “KEGG: ECM receptor interaction” (Figure 7G). Following IHC staining of primary tumor tissues derived from orthotopic co-injection of A549 cells and CAFs in the left lung of nude mice, as reported in chapter 3.5, NNMT-knockdown was associated with an increased expression of COL3A1 and ITGA5 in stroma regions, while NNMT overexpression was associated with decreased expression of COL3A1 and ITGA5 (Figure 7H). IHC staining was then conducted in a cohort consisting of 20 LUADs (Supplementary Table S4), and the expression of NNMT protein in the stromal compartments was found to be negatively associated with that of representative ECM-related genes encoding COL3A1, COL5A1, and ITGA5 (Figure 7I). These results have partially validated the negative regulation of NNMT on the expression of ECM-related genes in CAFs at the organizational level.
These findings collectively support the roles of NNMT in the metabolic epigenetic regulation of CAFs in LUAD, influencing their metastasis-promoting properties (Figure 7J). The downregulation of NNMT leads to heightened methylation at H3K4me3. This occurs through the redirection of the methyl group flux away from the methionine cycle, which in turn results in broad transcriptional activation of ECM-related genes in CAFs. The activation of these genes can produce an oncogenic ECM that ultimately facilitates the invasion and metastasis of cancer cells.
4 DISCUSSION
This study utilized metabolomic analysis to reveal multiple metabolomic abnormalities associated with the recurrence and metastasis of NSCLC following curative resection, with a particular focus on nicotinamide metabolism. The downregulations of NNMT and AOX1 in CAFs were speculated to be key factors contributing to deregulated nicotinamide metabolism. NNMT in CAFs was shown to negatively influence the invasion and metastasis of LUAD through downstream genes related to ECM remodeling, with H3K4me3 acting as a mediator. Comprehensive organizational validations were conducted thereafter. These findings provide insights into the epigenetic regulation initiated by metabolism in CAFs, which is related to LUAD metastasis.
Metabolites are closely linked to the biological programs of cancers [13, 14]. This underscores the advantages of metabolomics in detecting phycological and pathological irregularities, although it also leads to analytical complexities due to the comprehensive influences from multiple processes. Typically, derangements in glucose, amino acid, and lipid metabolism are observed during tumor development [22, 68, 69]. Abnormalities in glucose and amino acid metabolism have been identified in NSCLC and are linked to metastasis [70, 71]. Deregulation of fatty acid and cholesterol metabolism has been reported to be associated with the incidence and prognosis of NSCLC [72]. The metabolism of cancer tissues is significantly affected by multiple cells, components, and biological processes within the local environment [17, 73]. Single-cell RNA-seq of early-stage NSCLC has demonstrated the downregulation of glycerolipid and glycerophospholipid metabolism in different cells within tumor environment [74]. In contrast, the current study, utilizing tissue metabolomic analysis, revealed differentiated lipid metabolism associated with NSCLC metastasis. Specifically, there is an upregulation of fatty acids, along with certain glycerolipids and glycerophospholipids, and a downregulation of sphingolipids. Deregulation of multiple amino acids, organic acids, and the nicotinate/nicotinamide pathway is also evident. These findings suggest a significant reprogramming of metabolism in NSCLC, which may enhance the risk of cancer recurrence and metastasis.
Deregulation of nicotinate and nicotinamide has been observed in various cancers, drawing attention due to their connection with the salvage pathway of NAD+ and SAM metabolism [23]. SAM is the principal methyl donor for transmethylation reactions that are fundamental to epigenetic processes [61]. NNMT, a central metabolic enzyme in nicotinamide metabolism, is reported to influence epigenetic programs by regulating SAM metabolism [75, 76]. The enzyme also impacts NAD+-dependent redox reactions and signaling pathways, exerting multiple effects on cancer development [23, 77]. Most studies have explored the roles of NNMT in cancer cells, associating its upregulation with tumor progression and unfavorable prognosis [25, 75]. In A549 cells, NNMT was reported to promote proliferation and migration through pro-inflammatory effects mediated by the STAT3/IL1β/PGE2 axis [77]. Researchers have largely confirmed NNMT's impact on histone methylation, particularly H3K9me3 and H3K27me3, which alleviate silenced chromatin states [78-81]. However, CAFs have been notably understudied in the context of NNMT research. Previous studies in ovarian and oral cancers [21, 24, 29], together with our current investigations in LUAD, consistently identify stromal CAFs as the primary sites of NNMT expression. Eckert et al.’s study [24] on high-grade serous ovarian cancers demonstrated that upregulation of NNMT in CAFs can reduce levels of H3K27me3 histone methylation, leading to the activation of pro-tumorigenic cytokines and several ECM-related genes, which in turn enhances the proliferation and metastasis of cancer cells. Similarly, Zhao et al.’s research [29] on oral squamous cell carcinoma revealed that elevated NNMT expression can activate the expression of lysyl oxidase and the focal adhesion kinase signaling pathway by reducing H3K27me3 levels, which can promote the interaction between CAFs and tumor cells during cancer development.
The current study presents different findings in LUAD. The expression of NNMT in LUADs is not higher than in normal tissues, which is distinct from most gastrointestinal and urogenital cancers [25]. The downregulation of NNMT in CAFs is shown to facilitate an oncogenic ECM, with H3K4me3 serving as a mediator. Notably, NNMT consistently regulates the methylation of histones with poor specificity [24, 82]. Changes in H3K4me3, H3K9me3, and H3K27me3 levels, induced by NNMT, may occur simultaneously but regulate the transcription of distinct genes differently [64, 65]. H3K4me3 is a well-documented hallmark of actively transcribing and poised gene promoters. Oncogenic ECM-related genes, particularly integrins, and collagens, are commonly regulated by H3K4 methylation rather than H3K9 or H3K27, as indicated by public ChIP-seq databases (Supplementary Figure S5G). NNMT-knockdown in CAFs has also been observed to repress multiple processes, such as the TGF-β, MAPK, and NF-kappa B signaling pathways, potentially associated with hypermethylation of H3K9 and H3K27 (Figure 5D, Supplementary Figure S5E). Collectively, the roles of NNMT in CAFs of LUAD differ from those in other cancers, possibly due to the heterogeneity of organization and differences in the epigenetic regulation of histones. On the other hand, direct interventions targeting NNMT in CAFs in LUAD may be less applicable, as overexpression may not be a suitable strategy. Instead, more attention should be focused on the mechanisms leading to the downregulation of NNMT during cancer development and metastasis. These issues require further investigation.
CAFs are known to play a pivotal role in shaping the ECM in the tumor microenvironment, influencing cancer proliferation, growth, invasion, metastasis, angiogenesis, and immunity [83]. However, the specific subgroups of CAFs in lung cancers remain inadequately characterized [83]. Hanley et al. [53] identified and characterized three major fibroblast subpopulations in NSCLC: adventitial, alveolar and myofibroblasts, with myofibroblasts being particularly prominent in cancer tissues and adventitial and alveolar fibroblasts in adjacent tissues. NNMT is commonly expressed in adventitial, alveolar, and myofibroblasts, with a similar positive fraction [53]. Grout et al. [51] categorized NSCLC CAFs into four subgroups using classic biomarkers of FAP, SMA, alcohol dehydrogenase 1B, and myosin heavy chain 11, revealing distinct functions of these subpopulations and demonstrating that CAFs characterized by the expression of FAP and SMA as myofibroblasts. Notably, adjacent tissues also showed low percentages of myofibroblasts, and it has been proven that myofibroblasts can evolve from adventitial and alveolar fibroblasts [51, 53]. Our experiments showed that primarily cultured CAFs and NFs from NSCLC exhibited similar morphological characteristics and comparably significant expression of FAP and SMA, while NFs had a longer culture period than the paired CAFs (Figure 3B-C, Supplementary Figure S3A-D). Collectively, both CAFs and NFs used in this study are hypothesized to be derived from tissue myofibroblasts. However, discrepancies between CAFs in primary culture and in situ must be considered, due to issues such as decreased diversity of fibroblast subgroups, lack of tissue context, and genomic abnormal activation during primary culture [47].
Myofibroblasts, marked by the expression of FAP and SMA, have been revealed to play an important role in ECM remodeling, particularly in the formation and biosynthesis of collagens and integrin-mediated interactions [51, 53]. These CAFs exhibit unique properties in producing matrix molecules of collagen III and V and MMP11 [51]. These previously confirmed findings align closely with our current results. The ECM-related genes found to be negatively regulated by the NNMT-H3K4me3 axis in our investigations are primarily integrins (such as ITGA5 and ITGA6), collagens (such as COL3A1 and COL5A1), and MMP11. Integrins act as key cell adhesion receptors for ECM components, and integrin-dependent processes are implicated in nearly every aspect of cancer metastasis [57]. Collagens III and V, mainly produced by CAFs in the tumor microenvironment, are fibrillary collagens embedded in the interstitial matrix [58], directly interacting with cancer cell receptors and supporting cancer cell migration and invasion [67]. MMP11, predominantly produced and secreted by CAFs, plays a crucial role in ECM degradation, promoting cancer cell invasion and metastasis [59]. The negative association between the expression of NNMT and that of collagens and integrins was further validated through reanalysis of single-cell RNA-seq data, in vivo experiments, and LUAD cohorts. These insights reinforce the credibility of NNMT's regulatory role in CAFs concerning cancer metastasis through ECM remodeling, although the specific details of remodeling and the underlying mechanisms warrant further investigation.
Several limitations should be taken into account in this study. Firstly, the reported oncogenic role of NNMT in lung cancer cells [77] differs from its function in CAFs, adding complexity and casting doubt on the reliability of overall NNMT mRNA levels for prognosis based on bulk tissue assessments. Assessment of stromal NNMT expression is thus recommended. Differences between primary cultured CAFs and their in situ counterparts require attention, even though our findings are based on the predominant myofibroblasts. The non-specific regulation of NNMT, which broadly affects the methylation of histones, may lead to heterogeneity in NNMT-modulated phenotypes across various cells and cancers. These uncertainties prompt questions regarding the targeting of histone methylation by NNMT and its viability as a therapeutic target. The influence of NNMT in CAFs on the remodeling of ECM has only been hinted at by preliminary investigations of relevant genes and warrants further direct validations. Importantly, the mechanisms underlying the regulation of NNMT expression in CAFs remain unclear, representing a significant knowledge gap that is crucial for the development of novel treatments. The small number of patients and samples in survival analyses and correlation analyses also raises concerns about the stability of the findings. Additionally, the significance of multiple metabolic deregulations in NSCLC, especially amino acids, organic acids, and lipids, deserves further research.
5 CONCLUSIONS
This study underscores metabolic irregularities in amino acids, organic acids, lipids, and nicotinamide, which are linked to the recurrence and metastasis of NSCLC. NNMT is predominantly expressed in CAFs within the stromal compartments of LUAD. NNMT in CAFs negatively regulates the invasion and metastasis of LUAD through the epigenetic regulation of ECM-related genes mediated by H3K4me3. These findings highlight the interplay between metabolic and epigenetic regulatory roles of NNMT in CAFs concerning LUAD metastasis, suggesting potential avenues for research and therapeutic development.
AUTHOR CONTRIBUTIONS
Peiyu Wang, Yun Li, Fan Yang, Jun Wang, Kezhong Chen, and Mantang Qiu contributed to the study design, data analysis, and data interpretation. Peiyu Wang, Guangxi Wang, Haoran Li, Yuyao Yuan, Haiming Chen, Shaodong Wang, Zewen Sun, and Fanjie Meng contributed to experiments, data collection, and data analysis. All authors participated in manuscript writing and approved the final version. Mantang Qiu, Kezhong Chen, and Jun Wang verified the underlying data. All authors had full access to the study data and assumed responsibility for submitting it for publication. The work reported in the paper has been conducted by the authors unless explicitly specified in the text.
ACKNOWLEDGMENTS
The study received support from the National Natural Science Foundation of China (grant number 92059203 and 82173386), National Key R&D Program of China (grant number 2023YFF0723500), the Research Unit of Intelligence Diagnosis and Treatment in Early Non-small Cell Lung Cancer, Chinese Academy of Medical Sciences (grant number 2021RU002), Medical and Health Science and Technology Innovation project, grant number (2021-I2M-002), Beijing Nova Program (grant number 20230484314), and the Innovation Fund for Excellent PhD student of Peking University Health Science Center (grant number 2022-58).
CONFLICT OF INTEREST STATEMENT
The authors affirm that there are no potential conflicts of interest associated with this study.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The use of clinical data and human tissue samples was approved by the Ethics Committee Board of Peking University People's Hospital (2021PHB155-001 and 2021PHB182-001). All patients were informed verbally and in writing about the research and provided their signed informed consent for the use of their biological samples and medical records. Approval for all animal experiments was obtained from the Animal Care Committee of Peking University People's Hospital (2022PHE076), and the experiments adhered to regulations on animal care set by the State Food and Drug Administration of China.
Open Research
DATA AVAILABILITY STATEMENT
The data supporting the findings of this study are available from the corresponding author upon reasonable request. The raw RNA-seq data have been submitted to the National Center for Biotechnology Information under the accession number PRJNA1182950 (SRA accession: SUB14851662). The raw data from the metabolomic and lipidomic analyses have been deposited in MetaboLights under the accession number MTBLS11595.




