Local TSH/TSHR signaling promotes CD8+ T cell exhaustion and immune evasion in colorectal carcinoma
Sisi Zeng, Huiling Hu, Zhiyang Li, and Qi Hu contributed equally to this work.
Abstract
Background
Dysfunction of CD8+ T cells in the tumor microenvironment (TME) contributes to tumor immune escape and immunotherapy tolerance. The effects of hormones such as leptin, steroid hormones, and glucocorticoids on T cell function have been reported previously. However, the mechanism underlying thyroid-stimulating hormone (TSH)/thyroid-stimulating hormone receptor (TSHR) signaling in CD8+ T cell exhaustion and tumor immune evasion remain poorly understood. This study was aimed at investigating the effects of TSH/TSHR signaling on the function of CD8+ T cells and immune evasion in colorectal cancer (CRC).
Methods
TSHR expression levels in CD8+ T cells were assessed with immunofluorescence and flow cytometry. Functional investigations involved manipulation of TSHR expression in cellular and mouse models to study its role in CD8+ T cells. Mechanistic insights were mainly gained through RNA-sequencing, Western blotting, chromatin immunoprecipitation and luciferase activity assay. Immunofluorescence, flow cytometry and Western blotting were used to investigate the source of TSH and TSHR in CRC tissues.
Results
TSHR was highly expressed in cancer cells and CD8+ T cells in CRC tissues. TSH/TSHR signaling was identified as the intrinsic pathway promoting CD8+ T cell exhaustion. Conditional deletion of TSHR in CD8+ tumor-infiltrating lymphocytes (TILs) improved effector differentiation and suppressed the expression of immune checkpoint receptors such as programmed cell death 1 (PD-1) and hepatitis A virus cellular receptor 2 (HAVCR2 or TIM3) through the protein kinase A (PKA)/cAMP-response element binding protein (CREB) signaling pathway. CRC cells secreted TSHR via exosomes to increase the TSHR level in CD8+ T cells, resulting in immunosuppression in the TME. Myeloid-derived suppressor cells (MDSCs) was the main source of TSH within the TME. Low expression of TSHR in CRC was a predictor of immunotherapy response.
Conclusions
The present findings highlighted the role of endogenous TSH/TSHR signaling in CD8+ T cell exhaustion and immune evasion in CRC. TSHR may be suitable as a predictive and therapeutic biomarker in CRC immunotherapy.
Abbreviations
-
- AKT
-
- protein kinase B
-
- ANOVA
-
- one-way analysis of variance
-
- BCA
-
- bicinchoninic acid
-
- CD62L
-
- lymphocyte adhesion molecule 62L
-
- CFSE
-
- carboxyfluorescein diacetate succinimidyl ester
-
- ChIP
-
- chromatin immunoprecipitation
-
- ChIP-qPCR
-
- chromatin immunoprecipitation-quantitative polymerase chain reaction
-
- CM
-
- conditioned medium
-
- CRC
-
- colorectal cancer
-
- CREB
-
- CAMP -response element binding protein
-
- CTDR
-
- cellTracker deep red
-
- CTLA4
-
- cytotoxic T lymphocyte-associated antigen-4
-
- CTLs
-
- cytotoxic T lymphocytes
-
- DAB
-
- 3,3’-diaminobenzidine tetrahydrochloride
-
- DAPI
-
- 4',6-diamidino-2-phenylindole
-
- dNTP
-
- deoxy-ribonucleoside triphosphate
-
- EDTA
-
- ethylenediaminetetraacetic acid
-
- ERK
-
- extracellular regulated protein kinases
-
- ERK
-
- extracellular regulated protein kinases
-
- FACS
-
- fluorescence activated cell sorting
-
- GAPDH
-
- glyceraldehyde-3-phosphate dehydrogenase
-
- GFP
-
- green fluorescent protein
-
- GM-CSF
-
- granulocyte-macrophage colony-stimulating factor
-
- GO
-
- Gene Ontology
-
- HAVCR2 or TIM3
-
- hepatitis A virus cellular receptor 2
-
- HE
-
- hematoxylin-eosinstaining
-
- HRP
-
- horseradish peroxidase
-
- hTSH
-
- human thyroid stimulating hormone; thyroid stimulating hormone
-
- ICB
-
- immune checkpoint blockade
-
- IFNγ
-
- Interferon γ
-
- IHC
-
- immunohistochemistry
-
- IL6
-
- interleukin 6
-
- Iso
-
- isotype control
-
- KEGG
-
- kyoto encyclopedia of genes and genomes
-
- LAG3
-
- Lymphocyte Activation Gene-3
-
- LN
-
- lymph node
-
- MDSCs
-
- myeloid-derived suppressor cells
-
- MFI
-
- mean fluorescence intensity
-
- mTSH
-
- murine thyroid stimulating hormone
-
- p-AKT
-
- phospho-protein kinase B
-
- PBMCs
-
- peripheral blood mononuclear cell
-
- PBS
-
- phosphate buffered saline
-
- p-CERB
-
- Phospho CAMP -response element binding protein
-
- PCR
-
- polymerase chain reaction
-
- PD-1
-
- programmed cell death 1
-
- PDCD1
-
- Programmed cell death protein 1
-
- p-ERK
-
- phospho extracellular regulated protein kinases
-
- PKA
-
- protein kinase A
-
- PKC
-
- protein kinase C
-
- PKH67
-
- paul karl horan 67
-
- PVDF
-
- polyvinylidene fluoride
-
- RNA-seq
-
- RNA sequencing
-
- RT-PCR
-
- reverse transcription-polymerase chain reaction
-
- SDS–PAGE
-
- SDS polyacrylamide gels
-
- shCtrl
-
- short hairpin RNA of Control
-
- shTSHR
-
- short hairpin RNA of TSHR
-
- SPL
-
- spleen
-
- TCR
-
- T cell receptor
-
- TIGIT
-
- T cell immunoreceptor with Ig and ITIM domains
-
- TILs
-
- tumor-infiltrating lymphocytes
-
- TIM3
-
- T cell immunoglobulin domain and mucin domain-3
-
- TIMER
-
- Tumor Immune Estimation Resource
-
- TME
-
- tumor microenvironment
-
- TNFα
-
- tumor necrosis factor
-
- TSHR o/e
-
- TSHR-overexpress
-
- TSHR
-
- thyroid stimulating hormone receptor
-
- Tshr-KD
-
- Tshr-knockdown
-
- Tshr-KO
-
- Tshr-knockout
-
- Tshr-WT
-
- Tshr-wildtype
-
- UTR
-
- 3' Untranslated Regions
-
- WCL
-
- whole-cell proteins
1 BACKGROUND
Immune checkpoint blockade (ICB) therapy in colorectal cancer (CRC) is indicated in patients with gene mismatch repair or a highly unstable microsatellite type. However, these patients account for only approximately 5% of the population with advanced CRC, and programmed cell death 1 (PD-1)/programmed cell death 1 ligand 1 (PD-L1) blockade therapy is only effective in 30%–50% of cases [1]. In addition, some patients experience disease progression even after the tumor is controlled [1]. ICB therapy achieves clearance of tumor cells by restoring the function of depleted CD8+ T cells. Thus, it is important to explore the molecular mechanism of CD8+ T cell exhaustion in the tumor microenvironment (TME) to identify novel immunotherapeutic targets and improve the effect of tumor immunotherapy.
Loss of effector CD8+ T cell function in the TME was caused mainly by long-term high-intensity tumor antigen stimulation and the effect of various immunosuppressive factors [2]. CD8+ T cell exhaustion not only decreased the tumor clearance capacity, but also led to an immunosuppressive state in the TME [3, 4]. Tumor cells and immunosuppressive cells in the TME secreted many active substances (exosomes [5], microRNAs [6], hormones [7]) that directly affected the function of CD8+ T cells, thus promoting tumor immune escape. The effects of hormones (leptin, lipocalin, insulin, steroid hormones, and glucocorticoids) on T cell function had been reported previously [7]. For example, glucocorticoid signaling played a role in the antitumor response of CD8+ T cells [8]. However, the effects of TSH/TSHR signaling pathway on CD8+ T cell function have not been reported.
TSH is synthesized primarily in the distal pituitary and thyroid nodules, and several types of extrapituitary cells are sources of TSH [9, 10]. Once released by the cells, TSH binds to TSHR in the cell membrane, activating TSHR and associated signaling cascades. TSHR mRNA and protein were expressed in other human and animal extrathyroidal tissues, including neural tissues, immune cells, ocular muscle, bone, adipocytes, erythrocytes, ovaries, and so on. The expression and functional roles of TSHR had been reported in a variety of nonthyroidal cancer tissues, including melanoma [11], glioma [12], lung [13], breast [14], ovarian [15], and hepatocellular carcinomas [16]; however, the expression of TSHR in CRC has not been reported. TSH could be synthesized in the subvillous crypt region and localized epithelial regions of the mouse small intestine, and TSH staining was increased in the epithelial cells of virus-infected regions during acute rotavirus infection, indicating that TSH might be involved in intestinal immune regulation [9, 17-19].
Here, we evaluated the TSHR expression in CD8+ T cells in CRC tissues. Subsequently, the roles of TSHR in CD8+ T cell were explored through in vitro and in vivo assays. We further investigated the mechanism underlying TSHR-induced CD8+ T cell exhaustion and CRC immune invasion. Finally, we investigated the reasons for the high expression of TSHR in CD8+ T cells and the source of TSH in the TME.
2 METHODS AND MATERIALS
2.1 Cells
CRC cell lines (MC38, CMT93, CT26, SW480, RKO, and LoVo), a human normal intestinal epithelial cell line (NCM460), T cell lines (Jurkat T, EL4 and 293T), and a macrophage cell line (THP-1), were originally obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). These cell lines were confirmed by short tandem repeat profiling in August 2019. MC38 and 293T cells were maintained in Dulbecco's modified eagle's medium high glucose (DMEM; Gibco, Grand Island, NY, USA), and other cells were maintained in roswell park memorial institute-1640 (RPMI-1640; Gibco) medium supplemented with 10% fetal bovine serum (FBS; ExCell Bio, Suzhou, Jiangsu, China) and 1% penicillin‒streptomycin (Biosharp, Hefei, Anhui, China). Cells were cultured at 37°C in a humidified atmosphere containing 95% air and 5% CO2.
Mouse CD8+ T cells or CD4+ T cells were isolated from the spleen and lymph nodes of C57BL/6 mice (Guangdong Medical Laboratory Animal Center, Guangzhou, Guangdong, China) with an EasySep Mouse Naïve CD8+ T Cell Isolation Kit (Stemcell Technologies, Cambridge, MA, USA) or EasySep™ Mouse CD4+ T Cell Isolation Kit (Stemcell Technologies).
For generation of effector T lymphocytes, 1 × 106 CD8+ T cells were seeded per well of a 24 well plate and stimulated for 48 h with 2 µg/mL anti-CD3 (Clone 145-2C11, BD Biosciences, San Jose, CA, USA) and anti-CD28 (Clone 37.51, BD Biosciences). Then, Interleukin-2 (IL-2; Peprotech, Cranbury, NJ, USA) was used to induce differentiation for 3 days in the presence of 0.5 mIU/mL mouse TSH (mTSH; R&D Systems, Minneapolis, MN, USA).
For generation of exhausted cells, CD8+ T cells were isolated and stimulated with 2 µg/mL anti-CD3 and anti-CD28, After 48 h, cells were treated with IL-2 for another 72 h. On day 5, CD8+ T cells were stimulated with 2 µg/mL anti-CD3 and anti-CD28 in the presence of 0.5 mIU/mL mTSH. High expression of exhaustion markers (PD-1 and TIM3) were considered to indicate exhausted CD8+ T cells.
2.2 Patients and clinical specimens
Paraffin-embedded CRC and para-tumor tissues were obtained from the tissue bank of the Department of Pathology, Nanfang Hospital, Southern Medical University (Guangzhou, Guangdong, China) that had been clinically diagnosed as colorectal adenocarcinoma between January 2012 and June 2017. Samples were collected CRC cases after surgery and TNM stage was based on the 8th Edition Colorectal Cancer Stage Classification released by American Joint Committee on Cancer (AJCC). Those without concurrent other malignancies and neoadjuvant chemoradiation therapy were included, whereas those with multiple primary CRC or hereditary CRC were excluded. Written informed consent was obtained from all patients.
Peripheral blood mononuclear cell (PBMCs) were obtained from healthy subjects in the Health Management Center of Nanfang Hospital, and lymphocytes were isolated by Ficoll (Thermo Fisher Scientific, Waltham, MA, USA) through density gradient centrifugation (400 × g, 20°C, 40 min), The study was approved by the Clinical Research Ethics Committee of Southern Medical University under authorization approval No: NFEC-2024-127.
CRC immunotherapy included intravenous injection of anti-PD-1 antibodies, including Nivolumab (360 mg, Bristol-Myers Squibb, Princeton, NJ, USA), Sintilimab (200 mg, Innovent, Suzhou, Jiangsu, China), Camrelizumab (200 mg, Suncadia Biopharmaceuticals, Suzhou, Jiangsu, China), Toripalimab (240 mg, Junshi Biosciences, Shanghai, China) and Pembrolizumab (200 mg, Merck, Darmstadt, Hessen, Germany) before and after surgery. Immunotherapy was performed every 3 weeks for 6 months or longer, and a computed tomography (CT) examination was performed after three treatments. The effect of immunotherapy was evaluated by measuring the size of the primary lesion or metastasis. Partial response (PR) means tumor volume reduction >30%; stable disease (SD) means tumor volume reduction <20% or increase <20%; PR and SD mean the tumor responds to immunotherapy; progressive disease (PD) means tumor lesion increase >20%, and tumors are not sensitive to immunotherapy.
2.3 Animal models
Wild-type C57BL/6 and BALB/c nude mice were purchased from the Guangdong Medical Laboratory Animal Center, TSHRfl/fl mice (06467) was originally purchased from Cyagen Biosciences (Guangzhou, Guangdong, China), CD4Cre mice (022071, JAX, Bar, ME, USA), and OT-1 T cell receptor (TCR) transgenic mice (003831, JAX) were originally purchased from the Jackson Laboratory (Bar, ME, USA). TSHRfl/flCD4Cre mice were obtained by crossing CD4Cre mice with TSHRfl/fl mice, The offspring were tail-clipped to validate the genotypes by PCR. Because CD4+ and CD8+ T cells are originated from precursor T cells that undergo positive selection and negative selection during development, TSHRfl/flCD4Cre mice had knockout of TSHR in both CD4+ and CD8+ T cells. All mice used in this study were maintained under specific pathogen-free conditions. For all animal experiments, mice were randomly allocated to specific groups (six mice per group) matched for age and sex. Animal experiments were approved by the Ethics Committee on the Use and Care of Animals of Southern Medical University (No. 00301532).
MC38 or CMT93 cells were washed with phosphate buffered saline (PBS; Corning, NY, USA) and filtered through a 70 µm strainer. Before tumor cell inoculation, age- and sex-matched mice (5-6 weeks) were shaved, and then 1 × 106 MC38 or CMT93 cells were subcutaneously injected into the dorsal part of the mice. Tumor size was measured every 2 or 3 days and calculated as length × width2/2.
Mice were euthanized by injected overdose barbiturates in accordance with the American Veterinary Medical Association (AVMA) Guidelines when the tumor size was >1500 mm3 for ethical considerations.
2.4 Immunohistochemistry(IHC)
Paraffin-embedded tissue blocks were cut into 2.5 µm sections and transferred to glass slides. Sections were immersed in 3% hydrogen peroxide to block endogenous peroxidase activity and incubated with primary antibodies overnight at 4°C, TSHR (1:200, GTX33561, GeneTex, Irvine, CA, USA) and TSH beta (1:200, 66750-1-Ig, Proteintech, Rosemont, NJ, USA) antibodies were used for IHC staining, followed by goat anti-IgG horseradish peroxidase (HRP; ZSGB-Bio, Beijing, China) antibody for 1 h at room temperature. The expression of TSHR and TSH beta was visualized with 3,3-diaminobenzidine tetrahydrochloride (DAB; ZSGB-Bio) and counterstaining with hematoxylin.
The degree of IHC staining was reviewed and scored independently by two pathologists. The intensity of staining was graded according to the following criteria: 0 (no staining), 1 (weak staining = light yellow), 2 (moderate staining = yellow brown), and 3 (strong staining = brown). The proportion of tumor cells or stromal cells was scored as follows: 1 (<25% positive tumor/stromal cells), 2 (25%–50% positive tumor/stromal cells), 3 (51%–75% positive tumor/stromal cells), and 4 (>75% positive tumor/stromal cells). The IHC score was calculated as the staining intensity score × the proportion of positive tumor or stromal cells. The cutoff for high or low expression of the indicated molecule was determined on the basis of median score. Optimal cutoff values were identified, and a staining index ≥8 was used to define tumors with high TSHR expression, whereas an index ≤7 was used to define tumors with low TSHR expression.
2.5 Immunofluorescence
Human tissue samples and mouse tumor tissues were embedded in paraffin and sectioned longitudinally at 2 µm. Tissue sections were dewaxed and rehydrated, and antigens were retrieved with ethylenediaminetetraacetic acid (EDTA; Sigma, St Louis, MO, USA) in a pressure cooker. Sections were incubated in 3% H2O2 for 30 min to block endogenous peroxidase activity. The slides were blocked with goat serum and then incubated with primary antibodies overnight at 4°C, followed by goat anti-IgG HRP antibody. Opal Fluorophore Working Solution (Akoya Biosciences, Marlborough, MA, USA) was pipetted onto each slide, and the slides were incubated at room temperature for 30 min. The slides were placed in a microwave for 4 min at 100% power, blocked with goat serum (ZSGB-Bio), and then incubated with the following antibodies: goat anti-IgG HRP antibody and Opal Fluorophore Working Solution. 4',6-diamidino-2-phenylindole (DAPI; Thermo Fisher Scientific) Working Solution was applied for 5 min at room temperature in a humidity chamber, and coverslips were applied with mounting medium.
Quantification of TSHR or TSH in CD8+ T or myeloid-derived suppressor cells (MDSCs) was performed using ZEN software (Zeiss, Oberkochen, Baden-Württemberg, Germany). Five 200× magnification images were obtained for each section, and after circling the target cells, the average fluorescence value was calculated as fluorescence intensity/area.
Antibodies in the following were used: TSHR (1:200, GTX33561, Genetex), TSH (1:200, 66750-1-Ig, Proteintech), CD8 (1:500, 66868-1-Ig, Proteintech), CD4 (1:500, ab133616, Abcam, Cambridge, Cambs, UK), CD11b (1:200, 48893, Cell Signaling Technology, Boston, MA, USA), CD33 (1:200, ab199432, Abcam), CD19 (1:200, ab134114, Abcam), CD68 (1:200, ab955, Abcam), CD11c (1:200, ab52632, Abcam), CD66b (1:200, 305102, Biolegend, San Diego, CA, USA), CD14 (1:200, ab183322, Abcam).
2.6 Flow cytometry
Cells were stained with live/dead cell staining (Fixable Aqua Dead Cell Stain Kit, Life Tech, Waltham, MA, USA), and anti-mouse CD16/32 antibody (Biolegend) was used to block nonspecific binding with Fc receptors before all surface staining. For cell surface staining, cells were washed with staining buffer (1% FBS in PBS) and incubated with the indicated antibodies on ice for 30 min. Cells were washed two additional times with staining buffer, fixed in 1% paraformaldehyde in PBS, and analyzed with a BD LSR-Fortessa (BD Biosciences). For intracellular cytokine staining, cells were stimulated with phorbol 12-myristate13-acetate (PMA; 50 ng/mL, Sigma) and ionomycin (500 ng/mL, Sigma) for 6 h in the presence of Golgi-plug (BD Bioscience) at 37°C for the last 4 h. Cells were then subjected to live/dead cell staining (Thermo Fisher Scientific), fixed/permeabilized with a Cytofifix/Cytoperm kit (BD Bioscience), and stained with antibodies against the indicated cytokines.
To determine the expression of TSHR, cells were stained with the respective primary antibodies followed by allophycocyanin (APC)-conjugated anti-rabbit IgG (Life Tech).
The following fluorescent dye-labeled antibodies were used in this study: APC anti-human PD-1 (1:200, MIH4, BD Biosciences), PE anti-human TIM3 (1:200, F38-2E2, Biolegend), APC anti-human tumor necrosis factor α (TNF α; 1:200, Mab11, Biolegend), PE anti-mouse TIM3 (1:200, B8.2C12, Biolegend), APC-anti-mouse PD-1 (1:200, RMP1-30, Biolegend), PerCP/Cyanine5.5 anti-mouse CD45 (1:200, 30-F11, Biolegend), BV510 anti-mouse CD45 (1:200, S18009F, Biolegend), FITC anti-mouse CD8a (1:200, 53-6.7, Biolegend), PE-Cy7 anti-mouse interferon γ (IFN γ; 1:200, XMG1.2, Biolegend), PE anti-mouse/human CD44 Antibody (1:200, IM7, Biolegend), PerCP anti-mouse CD62L (1:200, MEL-14, Biolegend). APC-Cy7 anti-mouse F4/80 (1:200, QA17A29, Biolegend), PE anti-mouse Ly6G (1:200, S19018G, Biolegend), PE-Cy7 anti-mouse Gr1 (1:200, RB6-8C5, Biolegend), FITC anti-mouse CD11b (1:200, M1/70, Biolegend), BV421 anti-mouse CD19 (1:200, 6D5, Biolegend), BV605 anti-mouse CD3 (1:200, 17A2, Biolegend), PerCP/Cyanine5.5 anti-mouse Ly6C (1:200, HK1.4, Biolegend), BV421 anti-mouse CD11c (1:200, N418, Biolegend), PE anti-mouse MHC II (1:200, M5/114.5.2, Biolegend).
2.7 Western blotting
Tissue and cell lysates were prepared in RIPA buffer (KeyGEN BioTECH). The homogenate was centrifuged at 4°C for 15min at 13,000 × g, and the supernatant was collected and quantified with the Bradford Protein Assays (KeyGEN BioTECH). The lysates were separated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and transferred onto polyvinylidene fluoride membranes (PVDF; Millipore, Billerica, MA, USA), The membranes were blocked with 5% skimmed milk for 1 h and then incubated with primary antibodies overnight at 4°C.
Antibodies in the following were used: TSHR (1:1,000, A6781, Abclonal, Wuhan, Hubei, China), PD-1 (1:1,000, 86163, Cell Signaling Technology), cytotoxic T lymphocyte-associated antigen-4 (CTLA4; 1:1,000, sc-376016, Santa Cruz, Dallas, TX, USA), beta-Actin (1:10,000, 66009-1-Ig, Proteintech), TSH (1:500, sc-365801, Santa Cruz), cAMP-response element binding protein (CREB; 1:1,000, 9197, Cell Signaling Technology), phosphorylated-CREB (p-CREB; 1:500, ab220798, Abcam), protein kinase B (AKT; 1:500, ab38449, Abcam), phosphorylated-protein kinase B (p-AKT; 1:1,000, 3192,Abcam), extracellular regulated protein kinases (ERK; 1:1,000, 4370, Cell Signaling Technology), phosphorylated-extracellular regulated protein kinases (p-ERK; 1:1,000, 3192, Cell Signaling Technology), and CD63 (1:1,000, 25682-2-AP, Proteintech).
2.8 RT-PCR
Total RNA was extracted from cells using the TRIzol reagent (TaKaRa, Otsu, Shiga, Japan) according to the manufacturer's instructions. Reverse transcription was performed using PrimeScript™ RT Master Mix for general genes (TaKaRa). Real-time PCR was performed under conditions of 95°C for 3 min, and 45 cycles of 95°C for 5 s and 60°C for 30 s, with SYBR Green PCR Master Mix (TaKaRa) on an Applied Biosystems 7500 Fast Real-Time RCR System (Biosystems, Foster City, CA, USA). Relative mRNA levels were calculated with the 2–ΔΔCt method. The primer sequences were listed in Supplementary Table S1.
2.9 Transfection and infection
293T cells were seeded and grown overnight to approximately 70% confluence at the time of transfection. Short hairpin RNA of TSHR vectors (shTSHR; Umine-Bio, Guangzhou, Guangdong, China), TSHR-overexpressing plasmids (TSHR-oe; Umine-Bio), GFP-Ctrl (GFP-Ctrl plasmids contains a CD28 response element that locates GFP to the cell membrane), and GFP-TSHR plasmids (Genecopoeia, Beijing, China) together with the lentiviral packaging vectors PAX2 and pMD2.G were transfected into 293T cells using Lipofectamine 3000 according to standard procedures.
Viral particles were collected and filtered after transfection for 48h. For infection, lentiviral supernatant was added to cultured cells with 10 µg/mL Polybrene (Sigma-Aldrich). After incubation for 24 h, infected cells were selected for 3 to 5 days with puromycin (Beyotime, Shanghai, China).
For plasmid transient transfection, 293T cells were transfected with CREB-overexpressing plasmids (Umine-Bio) using reduced-serum medium (Opti-MEM, Gibco) and lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) for 6 h.
2.10 Tumor-infiltrating lymphocyte (TIL) isolation and analysis
Tumor tissues were dissected, cut into pieces and digested in RPMI 1640 medium containing collagenase VI (210 U/mL, Thermo Fisher Scientific), DNase I (100 U/mL, Thermo Fisher Scientific) and hyaluronidase (0.5 mg/mL, Thermo Fisher Scientific) for 30 min at 37°C. The dissociated cells were passed through a 70 µm strainer. Then, the supernatant was transferred to a new tube and centrifuged at 1,000 × g for 10 min at 4°C. The cells were resuspended for density gradient centrifugation with 40% Percoll and 70% Percoll. The interphase of the gradient was harvested and centrifuged at 1,000 × g for 5 min at 4°C. The isolated TILs were then used in subsequent experiments. To measure the cytokine production of isolated TILs, the cells were stimulated with 50 ng/mL PMA, 1 µmol/L ionomycin, and 5 µg/mL GolgiPlug for 4 h at 37°C.
2.11 Measurement of the cytotoxicity of cytotoxic T lymphocytes (CTLs)
OT-1 mouse splenocytes were harvested and homogenized using sterile techniques. Red blood cells were then lysed with red blood cell lysis buffer (Biosharp) for 5 min at room temperature. The splenocytes were pelleted, resuspended at 1 × 106/mL in RPMI-1640 medium supplemented with 10% FBS, 1% penicillin-streptomycin and 2-mercaptoethanol and treated with 10 nmol/L Ovalbumin257-264 peptide (OVA257-264; Thermo Fisher Scientific) and 10 ng/mL IL-2 for 3 days. Then the cells were cultured in fresh medium containing 10 ng/mL IL-2 for 2 more days to do the subsequent experiment.
To measure the cytotoxicity of CTLs, EL4 cells were pulsed with 10 nmol/L OVA257-264 for 30 min at 37°C. Then, the antigen-pulsed EL4 cells were washed with PBS and labeled with 1 µmol/L CellTracker Deep Red (CTDR; Thermo Fisher Scientific) in serum-free medium for 15 min at 37°C in the dark. Meanwhile, EL4 cells were labeled with 0.5 µmol/L carboxyfluorescein diacetate succinimidyl ester (CFSE; Selleck, Shanghai, China) in PBS for 10 min at room temperature in the dark. After washing EL4 cells with PBS three times, CTDR-labeled and CFSE-labeled EL4 cells were mixed at a ratio of 1:1 in RPMI 1640 supplemented with 2% FBS. CTLs were added to the plate at the indicated ratio. After 4 h, the cytotoxic efficiency was measured by quantifying the value of one minus the ratio of the CTDR-labeled/CFSE-labeled EL4 cells.
2.12 Human CD8+ T cell culture
CD8+ T cells were isolated from human PBMC, and stimulated with 2 µg/mL anti-CD3 and anti-CD28 in a 24-well plate. On day 5, CD8+ T cells were stimulated with 2 µg/mL anti-CD3 and anti-CD28 (Peprotech) in the presence of 0.5 mIU/mL human TSH (hTSH; R&D Systems). The expression of PD-1 and TIM3, as well as IFNγ or TNFα was detected by flow cytometry (Becton Dickinson, Bergen County, NJ, USA).
2.13 In vitro inhibition of PKA with H89 and blockade protein kinase C (PKC) with GO6983
Mouse CD8+ T cells or Jurkat T cells were pretreated with vehicle control, the PKA inhibitor H89 (30 nmol/L, HY-15979, MedChemExpress, Monmouth Junction, NJ, USA), or the PKC inhibitor GO6983 (100 nmol/L, HY-13689, MedChemExpress) for 1 h, then treated with TSH as indicated for 24 h. The cell lysates were subjected to Western blotting to detect the amounts of PD-1 and CTLA4.
2.14 Chromatin Immunoprecipitation-Quantitative Polymerase Chain Reaction (ChIP-qPCR)
ChIP assays were performed using the SimpleChIP Plus Sonication Chromatin IP Kit (Cell Signaling Technology) following the manufacturer's guidelines. Briefly, Jurkat T and CD8+ T cells were fixed with formaldehyde to crosslink DNA and protein and sonicated to yield 150-bp to 900-bp fragments. The protein-DNA complexes were precipitated using the normal rabbit IgG antibody and polyclonal antibodies. For each immunoprecipitation, 10 µg of antibody were added to the lysate and incubated overnight at 4°C with rotation. Then, 30 µL of protein G magnetic beads were added and incubated at 4°C for 2 h with rotation. Precipitin G beads were precipitated and washed sequentially with low-salt and high-salt wash buffer. The protein-DNA complex was reversed at 65°C overnight followed by DNA purification. Enrichment of the DNA sequences was detected using qPCR. The data were normalized and analyzed using the percent input method as follows: Percent Input = 2%×2(C [T] 2%Input Sample−C [T] IP Sample). C [T] = Threshold cycle of PCR.
2.15 Luciferase activity assay
The 3′ untranslated regions (UTR) segments of the PD-1 and TIM3 genes were amplified by polymerase chain reaction (PCR) and inserted into the vector. Co-transfections of PD-1 and TIM3 3′ UTR plasmids with CREB-overexpressing plasmids into cells was performed with Lipofectamine 2000. Luciferase activity was measured 48 h after transfection with a Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA).
2.16 Orthotopic tumor formation
In the orthotopic primary tumor model, a midline incision was made in the abdomen, and the cecum was gently exteriorized onto gauze impregnated with saline. MC38 cells were injected into the cecal wall of mice with a 30-gauge needle (Braun, Melsungen, Germany) in 5 µL volume (2 × 104 cells). The cecum was returned to the abdominal cavity, and the incision was closed.
2.17 In vivo depletion of CD8+ or CD4+ T cells or MDSCs
For CD8+ T cell depletion, Tshr-WT and Tshr-KO mice were administered 200 µg of monoclonal anti-mouse CD8 antibody (BioXCell, West Lebanon, NH, USA) or normal rat IgG (BioXCell) through the intraperitoneal route 7 days after tumor inoculation and three times per week. For CD4+ T cell depletion, Tshr-WT and Tshr-KO mice were administered 200 µg anti-mouse CD4 antibody (BioXCell) or normal rat IgG through the intraperitoneal route 7 days after tumor inoculation and two times per week. For MDSC depletion, mice were administered 200 µg anti-mouse ly6G antibody (BioXCell) or normal rat IgG through the intraperitoneal route on days -3, 0, 3, 6, 9, 12 after tumor implantation.
2.18 Anti-PD-1 treatment
MC38/shTSHR cells were inoculated into the flank of C57BL/6 mice. On days 7, 10, 13 and 16 after tumor inoculation, the mice were treated with anti-PD-1 (200 µg per mouse) or isotype control (BioXcell) intraperitoneal Injection (i.p). Seventeen days after tumor inoculation, the effect of anti-PD-1 on tumor growth was analyzed.
2.19 Exosome isolation, characterization, and treatment
Exosomes were purified from CRC-derived conditioned medium by ultracentrifugation. CRC cells were cultured in 1640 medium supplemented with 10% FBS. The fetal bovine serum was depleted of exosomes by ultracentrifugation at 110,000 × g overnight at 4°C prior to use. Conditioned medium was collected after 48 h and centrifuged at 500 × g for 10 min at 4°C, followed by 16,800 × g for 30 min at 4°C. The supernatants were passed through a 0.45 µm filter (Millipore) and ultracentrifuged at 110,000 × g for 70 min at 4°C. The exosome pellets were washed with PBS, followed by a second ultracentrifugation at 110,000 × g for 70 min at 4°C and then resuspended in PBS. The amount of exosomes were measured with a Bicinchoninic Acid protein assay kit (BCA; KeyGEN BioTECH, Nanjing, Jiangsu, China). For transmission electron microscopy, exosomes were fixed with 2% paraformaldehyde and placed on 200-mesh Formvar-coated grids. Then, the cells were stained using 2% phosphotungstic acid for 2 min and observed on a transmission electron microscope (H-7500, Hitachi, Tokyo, Japan). For exosome labeling, exosomes were fluorescently labeled with Paul Karl Horan 67 (PKH67; Sigma). Labeled exosomes were washed in 10 mL of PBS, collected by ultracentrifugation, and resuspended in PBS. For cell treatment, 2 µg of exosomes were incubated with 2 × 105 recipient cells for 24 h.
To capture the process of cellular internalization of CD8+ T cells, we co-incubated GFP-TSHR-containing exosomes with CD8+ T cells for 1, 3, 6, 12 or 24 h. Flow cytometry was performed to observe the endocytosis of GFP-TSHR-containing exosomes by CD8+ T cells. To investigate the pathway of uptake of GFP-TSHR-containing exosomes by CD8+ T cells, 10 µg/mL of the clathrin-mediated endocytosis inhibitor chlorpromazine (CPZ; Macklin, Shanghai, China), 500 µmol/L of the caveolin-mediated endocytosis inhibitor methyl-β-cyclodextrin (MβCD; Macklin) and 1 mmol/L of the macropinocytosis inhibitor amiloride (Macklin) were added to exhausted CD8+ T cells. After 2 h, GFP-TSHR-containing exosomes were added, and the cells were cultured for 4 h. Next, the cells were harvested and the GFP was detected by flow cytometry.
2.20 Blockade exosome biogenesis or release
For in vivo experiments, GW4869 (MedChemExpress), an inhibitor of exosome biogenesis/release, was injected (25 µg/g) intraperitoneally on days 8, 10, 12, 14, 16 after tumor inoculation. For in vitro experiments, GW4869 was added to the cell culture medium at a final concentration of 20 nmol/L for 48 h before exosome collection.
2.21 Preparation of human peripheral neutrophils
To investigate whether GFP-TSHR-containing exosomes affected other cell types such as neutrophils, we isolated neutrophils from peripheral anticoagulated blood samples through density gradient centrifugation. Red blood cells were removed with lysis buffer. Total neutrophils were suspended in complete RPMI-1640 medium supplemented with 10% FBS and 1% penicillin–streptomycin, and seeded on 24-well plates.
2.22 In vitro stimulation of THP-1 cells with exosomes
THP-1 cells were cultured in RPMI-1640 medium supplemented with 10% FBS and 1% penicillin-streptomycin. The cells were differentiated into M0 macrophages by incubating with 50 ng/mL PMA for 48 h. For M2 polarization, M0 macrophages were stimulated with 20 ng/mL IL-4 (Peprotech) for 48 h with GFP-Ctrl or GFP-TSHR-containing exosomes (10 µg/mL). After stimulation, the cells were collected and the RNA expression levels of CD206, ARG1 and TGF β were measured.
2.23 Enzyme-linked Immunosorbent Assay (ELISA)
Neutrophils promote tumor progression by forming neutrophil extracellular traps (NETs). Myeloperoxidase (MPO) is a major component of NETs. To investigate whether GFP-TSHR-containing exosomes affected neutrophil function, neutrophils were cultured in complete 1640 medium supplemented with 10% FBS, 1% penicillin–streptomycin and stimulated with vehicle or exosomes (10 µg/mL) for 12 h. After stimulation, the supernatant was collected, and MPO levels were measured with a commercial ELISA kit (SEA601Mu, Cloud-Clone, Wuhan, Hubei, China).The protein expression of TSH in serum and tumor tissue was measured with a mouse ELISA kit (CEA463Mu, Cloud-Clone) according to the manufacturer's instructions.
2.24 Monocyte-derived MDSC culture
Bone marrow cells were isolated from mouse femurs and were suspended in complete DMEM supplemented with 10% FBS and 20 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF; Peprotech). On day 4, non-adherent cells were discarded and adherent cells were further cultured for 3 days with fresh medium supplemented with 20 ng/mL GM-CSF. On day 7, adherent cells confirmed to be Gr-1+CD11b+ were considered to be mature MDSCs.
2.25 Co-culture of CD8+ T cells and MDSCs
For MDSC-T cell co-culture, MDSCs differentiated under the above conditions were seeded together with CD8+ T cells from spleen of Tshr-WT or Tshr-KO mice in the presence of IL-2, After 48 h, the cells were collected by centrifugation (300 × g, 4°C, 5 min), stained and immunophenotyped by flow cytometry.
2.26 Bioinformatics analysis
Correlations between TSHR expression and T cell exhaustion signature genes (HAVCR2, PDCD1(Programmed cell death 1), and CTLA4) were calculated with Spearman correlations on The Tumor Immune Estimation Resource (TIMER) website (http://timer.cistrome.org). The TIMER 2.0 website used gene expression data downloaded from the corresponding GDC portal (https://gdc.cancer.gov/about data/publications/panimmune; n = 10,496, 33 cancer types).
Gene Ontology (GO) enrichment analysis was performed to determine functional annotations of differentially expressed genes (DEGs), including biological processes, cellular components and molecular functions, using the clusterProfiler R package (v4.0.0). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was also further performed to identify the enriched pathways among these DEGs. The threshold of significance was defined as P value ≤ 0.05 for both GO and KEGG analyses.
2.27 RNA-sequencing (RNA-seq) and data analysis
Total RNA was extracted using a TRIzol reagent kit according to the manufacturer's protocol. RNA quality was assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and confirmed using RNase-free agarose gel electrophoresis. After total RNA was extracted, eukaryotic mRNA was enriched using oligo (dT) beads, whereas prokaryotic mRNA was enriched by removing rRNA with a Ribo-ZeroTM Magnetic Kit (Epicenter, Madison, WI, USA). Then, the enriched mRNA was fragmented into short fragments using fragmentation buffer and reverse transcribed into cDNA using random primers. Second-strand cDNA was synthesized using DNA polymerase I, RNase H, deoxy-ribonucleoside triphosphate (dNTP), and buffer. The cDNA fragments were purified with a QiaQuick PCR extraction kit (Qiagen, Dusseldorf, Germany), end repaired, treated with A base, and ligated to Illumina sequencing adapters. The ligation products were size selected by agarose gel electrophoresis, PCR amplified, and sequenced using Illumina NovaSeq6000 by Gene Denovo Biotechnology Co (Guangzhou, Guangdong, China).
2.28 Prediction of CREB target gene
We obtained the CREB target gene from the website: http://natural.salk.edu/CREB/, CREB regulates cellular gene expression by binding to a conserved CRE that occurs either as a palindrome (TGACGTCA) or half site (CGTCATGACG). A comprehensive scan of the human genome revealed 10,447 full CREs and 740,390 half CREs.
2.29 Statistical analysis
Statistical analyses were performed using GraphPad Prism (v9) software. Data are presented by descriptive statistics such as mean ± standard error of mean (SEM). Data distribution was assumed to be normal with the one sample Kolmogorov-Smirnov test, but this was not formally tested. Data collection and analysis were not performed in a blinded manner to the conditions of the experiments. For continuous endpoints, such as flow cytometry data or qPCR data, a Student's t-test or paired t-test was used to compare two independent or matched conditions/groups, and one-way analysis of variance (ANOVA) models were used to compare three or more independent conditions/groups. P < 0.05 was considered statistically significant.
3 RESULTS
3.1 TSHR expression was elevated in CD8+ T cells in CRC
To investigate the expression pattern of TSHR protein in CRC, we performed IHC in clinical samples of human CRC tissues and paired adjacent normal tissues. Tumor cells and tumor stromal cells showed high TSHR expression, whereas epithelial cells and stromal cells in normal intestinal epithelia did not (Figure 1A-B). We also found higher TSHR expression in CRC tissues compared with paired adjacent normal tissues by using TCGA-CRC dataset GSE21510 (Supplementary Figure S1A). The results of clinicopathological analysis showed that high TSHR expression was related to advanced TNM stage, lymph node metastasis, and distant metastasis (Supplementary Table S2), suggesting that TSHR might influence CRC progression. Logistic regression analysis showed that the high TSHR expression was associated with microsatellite stability (MSS) and high TNM stage (Supplementary Table S3).

The TIMER 2.0 database showed that TSHR expression was positively correlated with the expression of T cell exhaustion signature genes, including HAVCR2 (TIM3), PDCD1 (PD-1), and CTLA4 (Figure 1C). Because these immunosuppressive molecules were mainly expressed on exhausted T cells in tumor tissues [20], we examined the expression of TSHR in CD4+ and CD8+ T cells in CRC tissues. Interestingly, TSHR protein was highly expressed in CD8+ but not in CD4+ T cells (Figure 1D-E). Moreover, macrophages and neutrophils showed low expression of TSHR, whereas monocytes, B cells, dendritic cells (DCs), fibroblasts did not express TSHR in CRC tissues (Supplementary Figure S1B). Although the TSHR protein expression was moderate in CD8+ T cells from normal intestinal tissue, it was lower than that in CD8+ TILs (Figure 1E-F). Meanwhile, we analyzed TSHR protein expression in CD8+ T cells from normal lymph nodes, which mainly contain naïve and effector CD8+ T cells. TSHR protein expression was almost undetectable in CD8+ T cells from normal lymph nodes compared with that in CD8+ TILs (Figure 1E-F).
In agreement with the expression pattern in human CD8+ TILs, TSHR protein expression was higher in CD8+ TILs from mouse CRC subcutaneous tumors than in CD4+ T cells from mouse CRC subcutaneous tumors and in CD8+ T cells from mouse spleen and lymph nodes (Figure 1G-H). As reported previously, PD-1−TIM3−CD8+ T cells exhibit full effector function, PD-1+TIM3−CD8+ T cells exhibit partial dysfunction, and PD-1+TIM3+CD8+ T cells exhibit severe dysfunction [21]. Among CD8+ TILs, PD-1+TIM3+CD8+ TIL subsets showed high expression of the TSHR protein compared with the PD-1−TIM3− and PD-1+TIM3−CD8+ TIL subsets (Figure 1I). Collectively, these results indicated that increased TSHR expression was associated with a dysfunctional phenotype in exhausted CD8+ T cells.
3.2 TSH/TSHR signaling promoted functional exhaustion of CD8+ T cells
To further explore the potential roles of TSHR in CD8+ T cells, we generated Tshr conditional knockout mice (Tshr-KO mice) and confirmed the efficient deletion of TSHR in CD8+ T cells and CD4+ T cells (Supplementary Figure S2A-B). T cells in the Tshr-KO mice developed normally in the thymus (Supplementary Figure S2C) and displayed similar activation states in the spleen and lymph nodes (Supplementary Figure S2D-I). In vitro cultured CD8+ T cells exhibited effector phenotypes after acute TCR activation and developed exhaustion after TCR reactivation [8]. We observed low expression of checkpoint receptors and high production of proinflammatory cytokines in effector CD8+ T cells, which were not affected by Tshr knockout (Supplementary Figure S3A-B). Nonetheless, loss of TSHR decreased the expression of the immunosuppressive molecules PD-1 and TIM3 and facilitated the production of the cytokines TNFα and IFNγ in exhausted CD8+ T cells (Figure 2A-B), but not in exhausted CD4+ T cells (Supplementary Figure S3C-D). Loss of TSHR also did not affect T cell proliferation (Supplementary Figure S3E), suggesting that TSHR was a regulator of CD8+ T cell exhaustion and dysfunction. Conversely, overexpression of TSHR in murine CD8+ T cells increased both the mRNA and protein levels of the checkpoint receptors PD-1 and CTLA4 (Supplementary Figure S3F-G).
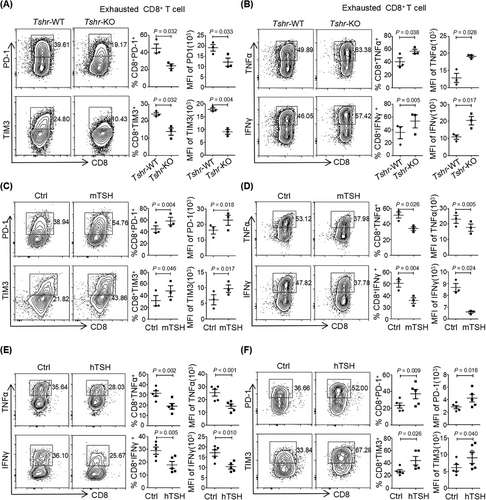
In agreement with the observations in Tshr-KO CD8+ T cells, mTSH or hTSH treatment increased the protein levels of the checkpoint receptors PD-1 and CTLA4 in CD8+ T cells or Jurkat T cells (Supplementary Figure S3H-I). mTSH treatment significantly promoted the expression of PD-1 and TIM3 and inhibited the production of the proinflammatory cytokines TNFα and IFNγ in exhausted CD8+ T cells (Figure 2C-D), but not in effector CD8+ T cells (Supplementary Figure S3J-K). In addition to the findings in murine CD8+ T cells, repeated activation of CD8+ T cells from human PBMCs in the presence of hTSH markedly inhibited the production of TNFα and IFNγ (Figure 2E) as well as elevated PD-1 and TIM3 expression (Figure 2F), thereby highlighting the evolutionarily conserved function of TSH/TSHR signaling in promoting CD8+ T cell exhaustion. Together, these data indicated that activation of TSH/TSHR signaling contributed to the functional exhaustion of CD8+ T cells.
3.3 TSH/TSHR signaling promoted CD8+ T cell exhaustion through the PKA/CREB signaling pathway
To systemically identify the target genes of TSH/TSHR signaling in CD8+ T cells, we performed RNA-seq analysis comparing Tshr-KO and Tshr-WT murine CD8+ T cells. A total of 573 and 846 DEGs were significantly up-regulated and down-regulated, respectively, in Tshr-KO CD8+ T cells compared with Tshr-WT CD8+ T cells (Figure 3A). Of these, 354 DGEs regulated by TSH/TSHR signaling were relevant to the T cell effector and exhaustion signature. More than 26% (151/573) of the up-regulated genes in Tshr-KO CD8+ T cells overlapped with the genes expressed by the effector PD-1−TIM3−CD8+ T cells, whereas approximately 24% (203/846) of the down-regulated genes overlapped with the exhausted PD-1+TIM3+CD8+ T cells (Supplementary Figure S4A-D). Many of the up-regulated genes encoded proinflammatory cytokine, including Il-2 and Tnf, whereas classical T cell exhaustion-associated genes (Il-10, Ctla4, Pdcd1, Havcr2) showed marked down-regulation (Figure 3B). We also confirmed that the mRNA levels of those exhaustion-associated genes were regulated by TSH/TSHR signaling on the basis of real-time PCR (Figure 3C-D).
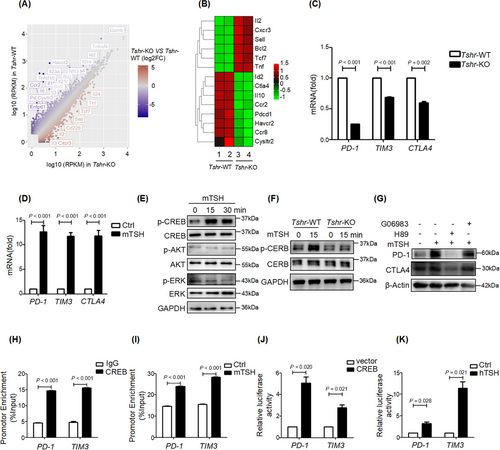
TSH/TSHR exerts its action through various signaling cascades, such as the cyclic adenosine monophosphate (cAMP)/PKA, protein kinase C/extracellular signal-regulated kinase (PKC/ERK) and phosphatidylinositol-3-hydroxykinase/protein kinase B (PI3K/AKT) signaling pathways, in different types of cells [22]. The phosphorylation of CREB substantially increased immediately after TSH treatment in both primary CD8+ T cells and Jurkat cells (Figure 3E, Supplementary Figure S4E). Although TSH treatment induced the instantaneous phosphorylation of ERK in Jurkat cells, it had little effect on the phosphorylation of ERK and AKT in murine CD8+ T cells (Figure 3E). Moreover, Tshr-KO CD8+ T cells showed diminished CREB phosphorylation levels under quiescent conditions and did not respond to TSH treatment (Figure 3F), indicating that TSH-induced phosphorylation of CREB in CD8+ T cells was TSHR-dependent. We next investigated whether TSH induced immunosuppressive molecules via PKA/CREB signaling and found that TSH-induced expression of PD-1 and CTLA4 was abolished by a PKA inhibitor (H89), but not by a PKC inhibitor (GO6983) in primary CD8+ T cells and Jurkat cells (Figure 3G, Supplementary Figure S4F). These results confirmed that TSH/TSHR signaling activated the PKA/CREB pathway to regulate the expression of immunosuppressive molecules in CD8+ T cells.
Phosphorylated CREB translocates to the nucleus and interacts with a number of coactivators to regulate gene transcription [23]. To determine whether TSH/PKA/CREB signaling directly regulates the expression of checkpoint receptors, we searched the target gene database of CREB and found that CREB binds directly to PDCD1, HAVCR2, CTLA4, lymphocyte activation gene 3 (LAG3), and IL-10. The results of ChIP-qPCR confirmed that CREB bound directly to the promoter regions of PD-1 and TIM3 (Figure 3H), and CREB occupancy on the promoters of PD-1 and TIM3 increased in the presence of mTSH stimulation (Figure 3I). To further verify the regulation model, we cloned the well-studied functional promoter of human PD-1 (-2kb) and TIM3 (-2kb) with a luciferase reporter, and tested the activity of those cis-regulatory elements in response to TSH/CREB signaling. In line with the ChIP-qPCR results, both CREB overexpression and hTSH treatment transactivated PD-1 and TIM3 expression (Figure 3J-K). These results suggested that TSH/TSHR signaling promoted PD-1 and TIM3 transcription through the PKA/CREB pathway in CD8+ T cells.
3.4 TSHR deficiency in CD8+ T cells increased antitumor activity
We next examined the importance of TSH/TSHR signaling in antitumor immunity. OT-1 mice-derived CD8+ T cells were differentiated into CTLs with or without TSH and then co-cultured with OVA257-264 loaded EL4 cells. The killing efficiency of CTLs was significantly diminished in the presence of TSH, particularly when CTLs were more abundant than target cells (Supplementary Figure S5A). However, mTSH treatment had little effect on the regulation of tumor growth in the MC38 cell subcutaneous inoculation model (Supplementary Figure S5B-C). This finding might be due to the opposite function of TSH/TSHR signaling in other types of cells in the TME, thus counteracting the anti-tumor effects of CD8+ T cells. Using an orthotopic tumor formation assay with MC38 cells, we found that Tshr-KO mice had diminished orthotopic tumor volumes (Figure 4A-D), whereas this effect was abolished after depletion of CD8+ T cells (Supplementary Figure S5D-G). Moreover, we subcutaneously implanted either MC38 or CMT93 CRC cells into Tshr-WT and Tshr-KO mice and found that Tshr-KO mice exhibited inhibited tumor growth (Figure 4E-J). Loss of Tshr in T cells also restricted tumor progression of subcutaneous MC38 tumor even when CD4+ T cells were depleted (Supplementary Figure S5H-K), thus indicating that the antitumor activity of TSHR was CD4+ T cell-independent. Moreover, the TSH concentration in the blood of Tshr-KO mice did not significantly differ from that in Tshr-WT mice (Supplementary Figure S5L).
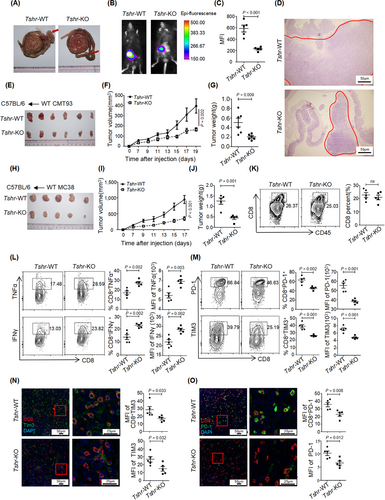
Immunofluorescence and flow cytometry analyses showed that the frequency and spatial distribution of CD8+ TILs in the TME were similar (Figure 4K and Supplementary Figure S5M), thereby suggesting that the recruitment or proliferation of CD8+ TILs was independent of TSH/TSHR signaling. The infiltration of CD4+ T cells, neutrophils, monocytes and macrophages also remained unchanged in the subcutaneous tumors of Tshr-KO mice (Supplementary Figure S5N-Q). In line with the In vitro results, CD8+ TILs from Tshr-KO mice showed a markedly elevated ability to produce inflammatory cytokines TNFα and IFNγ (Figure 4L), while the expression of the checkpoint receptors PD-1 and TIM3 was dramatically diminished (Figure 4M). In addition, PD-1 and TIM3 were down-regulated in non-CD8+ T cells in the TME of Tshr-KO mice (Figure 4N-O), indicating that blocking TSHR signaling in CD8+ T cells was sufficient to shape the immunosuppressive TME.
3.5 TSHR in CRC cells inhibited antitumor immunity via CD8+ T cells
Given the high protein expression of TSHR in CRC cells (Figure 1A-B, 1E-F), we wondered whether TSHR in CRC cells might affect tumor growth. Knockdown (KD) of TSHR had no significant effect on the proliferation of MC38 cells In vitro (Supplementary Figure S6A-B) or tumor growth in subcutaneous models in nude mice (Figure 5A-C). TSHR overexpression also had no significant effect on tumor growth in subcutaneous models in nude mice (Supplementary Figure S6C-D). However, TSHR-KD in MC38 cells significantly inhibited tumor growth in subcutaneous models in syngeneic immunocompetent C57BL/6 mice (Figure 5D-F), suggesting that TSHR in CRC decreased the antitumor effect through the immune microenvironment. When CD8+ T cells were depleted in C57BL/6 mice, TSHR-KD had no significant effect on tumor growth in subcutaneous models (Figure 5G-I), thus indicating that TSHR in CRC cells inhibited antitumor immunity via CD8+ T cells. Interestingly, although knockdown of TSHR in CRC cells did not change the number of CD8+ TILs (Supplementary Figure S6E-F), it significantly decreased the expression of the immunosuppressive molecules PD-1 and Tim3 and promoted the production of TNFα and IFNγ (Figure 5J-K). Taken together, these results suggested that TSHR in CRC cells also promoted the functional failure of CD8+ TILs, resembling the function of TSHR in CD8+ T cells.
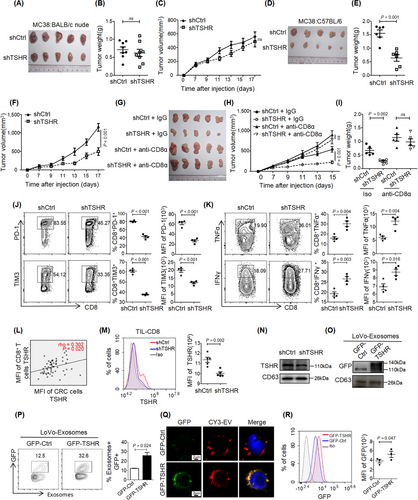
3.6 CRC cells delivered the TSHR protein to CD8+ TILs through exosomes
The similar expression pattern and homologous function of THSR in CD8+ T cells and CRC cells prompted us to explore the relationship between the two kinds of cells. The expression level of TSHR was markedly higher in tumor cells than in CD8+ TILs (Figure 1E-F), and the expression of TSHR in CD8+ TILs was positively correlated with that in tumor cells (Figure 5L). TSHR-KD in CRC cells down-regulated the expression of TSHR in CD8+ TILs in vivo (Figure 5M), suggesting a potential flow of TSHR protein from tumor cells to CD8+ TILs. Furthermore, TSHR protein was detected in the culture supernatant of normal intestinal epithelium and tumor cells, and was present at higher levels in the culture supernatant of CRC cells (Supplementary Figure S7A). The culture supernatant of CRC cells increased the expression of TSHR in T cells In vitro (Supplementary Figure S7B), thereby indicating that CRC cells may secrete TSHR protein to accumulate in CD8+ TILs.
Exosomes are small extracellular vesicles secreted by cells to communicate with other cells, and they are involved in regulating the TME. The TSHR protein was present in the exosomes from MC38 cells, and its levels were decreased in the exosomes from TSHR-KD MC38 cells (Figure 5N), suggesting that the TSHR protein was secreted via exosomes. For dynamic tracking of TSHR protein, the GFP-CD28 (GFP-Ctrl) fusion protein expressed on the cell membrane, and GFP-TSHR fusion protein were overexpressed in CRC cells (Supplementary Figure S7C). Ectopic overexpression of GFP-TSHR had little effect on exosome formation (Supplementary Figure S7D-E). The GFP-TSHR fusion protein, but not GFP-Ctrl, was enriched in the exosomes (Figure 5O-P) and secreted into the culture supernatant (Supplementary Figure S7F). Addition of exosomes from CRC cells with GFP-TSHR led to the translocation of GFP-TSHR in exosomes to the cell membrane of CD8+ T cells or Jurkat cells (Figure 5Q-R, Supplementary Figure S7G-H), and this effect peaked at 3 h and stabilized after 6 h in CD8+ T cells (Supplementary Figure S7I), thus providing direct evidence that the TSHR protein secreted by CRC cells via exosomes was taken up by CD8+ T cells. On the basis of these results, we investigated the delivery pathway of GFP-TSHR-containing exosomes in CD8+ T cells in the presence of endocytosis inhibitors. The results showed that the GFP-TSHR-containing exosomes proceeded with clathrin-mediated endocytosis and caveolae-mediated endocytosis, but not macropinocytosis (Supplementary Figure S7J-K).
However, because Tshr-KO CD8+ T cells took up the same amount of GFP-TSHR-containing exosomes as Tshr-WT CD8+ T cells (Supplementary Figure S7L), tumor cell exosomes did not restore the TSHR expression level of Tshr-KO CD8+ T cells (Supplementary Figure S7M). Low TSHR expression was also found in tumor-infiltrating CD8+ T cells from Tshr KO mice (Supplementary Figure S7N). In agreement with the immunofluorescence results (Supplementary Figure S1B), GFP-TSHR-containing exosomes could also translocate to the cell membrane of macrophages and neutrophils, but did not affect their functions (Supplementary Figure S8). These results suggested that CRC cells delivered the TSHR protein to CD8+ TILs through exosomes.
3.7 TSHR desensitized tumors to PD-1 blockade therapy
We investigated the effects of TSHR exosomes on CD8+ T cell function. The TSHR protein level increased in CD8+ T cells treated with MC38 exosomes, and expression of PD-1 and CTLA4 was also induced (Figure 6A). Tshr-KO CD8+ T cells showed re-expression of TSHR (Figure 6B) and subsequently respond to TSH stimulation (Figure 6C, Supplementary Figure S9A) after the treatment of MC38 exosomes. Inhibition of exosome secretion did not affect TSHR expression in MC38 cells (Supplementary Figure S9B), but CD8+ T cells took up less TSHR-containing exosomes (Supplementary Figure S9C). Furthermore, TSHR-KD restrained subcutaneous MC38 tumor growth, and this effect was abolished by GW4869 (Figure 6D-G). TSHR expression was also diminished in CD8+ TILs after GW4869 treatment (Supplementary Figure S9D-E). Thus these data indicated that TSHR in CRC cells regulated antitumor immunity via exosomes. Because ICB reverses the terminal exhaustion in T cells, we examined the expression of TSHR in tissue samples from 50 CRC immunotherapy cases. The expression of TSHR in the PR + SD group was lower than that in the PD group (Figure 6H). Although ICB therapy for CRC is suitable for patients with gene mismatch repair or high microsatellite instability (MSI-H), this group of patients accounts for approximately 5% of the population with advanced CRC, and many patients with MSS receive immunotherapy. TSHR expression was lower in the PR + SD group than that in the PD group in 28 cases of MSS (Figure 6I). Therefore, the expression of TSHR in CRC patients may serve as a marker to evaluate the efficacy of immunotherapy.
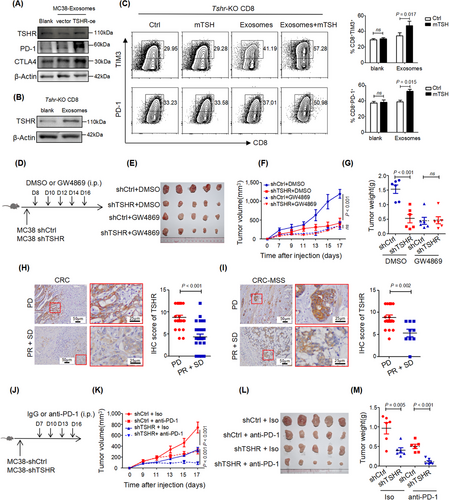
Finally, we tested whether TSH/TSHR signaling affected the response to ICB in mice. We treated C57BL/6 mice bearing TSHR-WT MC38 and TSHR-KD MC38 tumors with anti-PD-1 antibody. The results showed that TSHR-KD MC38 tumors were sensitized to anti-PD-1 antibody treatment, with some TSHR-KD MC38 tumors nearly completely diminished (Figure 6J-M). Together, these data suggested robust sensitization to PD-1 blocking therapy by decreasing TSHR levels in the TME.
3.8 TSH released by MDSCs promoted the immunosuppressive phenotype of CD8+ TILs
Although the anterior pituitary is the primary source of TSH, TSH could also be produced by other types of cells, including T cells, B cells, splenic DCs, bone marrow hematopoietic cells, and intestinal epithelial cells. The expression level of TSH was higher in CRC tumor tissue than in the normal intestinal epithelium, and similar results were also obtained from the TCGA datasets GSE21510 and GSE87211 (Figure 7A-B, Supplementary Figure S10A-B). Although TSH protein was expressed in CRC cells, the secreted TSH protein was not detected in the culture supernatant (Supplementary Figure S10C), suggesting stromal cells as the main source of TSH in the TME.
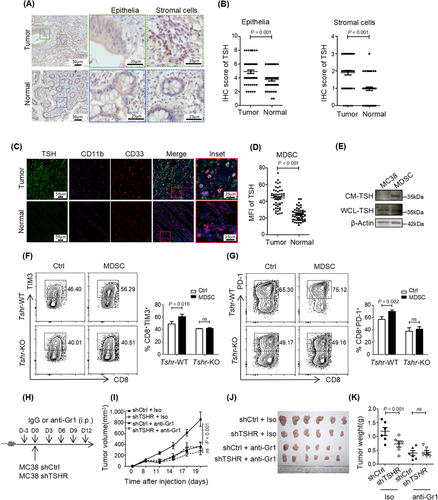
To identify the cell types responsible for TSH production in the TME, we detected the expression of TSH in different immune cell types by immunofluorescence in human CRC tissues. TSH was nearly undetectable in macrophages (CD68+ cells), B cells (CD19+ cells), T cells (CD3+ cells), DCs (CD11c+ cells), neutrophils (CD66b+ cells) and monocytes (CD14+ cells) (Supplementary Figure S10D). Unexpectedly, TSH was highly expressed in MDSCs, which were marked by CD33 and CD11b in human (Figure 7C-D). Similar results were obtained by flow cytometry (Supplementary Figure S10E). TSH expression was higher in MDSCs from CRC than in those from the normal intestinal mucosa (Figure 7C-D), and secreted TSH protein was detected in the culture supernatant of MDSCs (Figure 7E). These results indicated that MDSCs were the primary source of TSH in the TME.
To explore whether TSH/TSHR signaling is essential to the immunosuppressive function of MDSCs, we co-cultured differentiated MDSCs with CD8+ T cells from Tshr-WT and Tshr-KO mice. MDSCs upregulated the expression of TIM3 and PD-1 in Tshr-WT CD8+ T cells, whereas they had no such effect in Tshr-KO CD8+ T cells (Figure 7F-G). As reported previously, anti-Gr1 antibody eliminated MDSCs in vivo [24]. Depletion of MDSCs by an anti-Gr1 antibody abolished the function of TSHR-KD restrained subcutaneous MC38 tumor growth (Figure 7H-K). In conclusion, these data suggested that MDSCs induced CD8+ T cell dysfunction and antitumor immunity through TSH/TSHR signaling in the TME.
4 DISCUSSION
Here, we identified a novel mechanism of CD8+ T cell function exhaustion in the CRC microenvironment. We found that CRC cells up-regulated TSHR expression in CD8+ T cells by secreting TSHR, which bound to TSH produced by tumor-associated MDSCs and activated the TSH/TSHR signaling pathway to induce effector differentiation and dysfunction in CD8+ TILs. TSH/TSHR signaling induced PD-1 and TIM3 transcription by promoting the binding of CREB to the PD-1 and TIM3 promoter regions, thus revealing the mechanism by which TSH/TSHR signaling suppresses the immune response. Activation of TSHR signaling was associated with failure to respond to ICB in CRC patients and could predict the treatment outcome in patients with MSS-type CRC.
The expression and functional role of TSHR have been reported in various nonthyroidal adenocarcinoma tissues, including melanoma, glioma, lung, breast, ovarian, and liver cancers. However, there are no studies of TSHR in CRC. TSH could be synthesized in the subvillous crypt region and local areas of the epithelium in the mouse small intestine. During acute rotavirus infection, TSH staining was markedly increased in epithelial cells, especially in the virus-infected region, suggesting that TSH was involved in intestinal immune regulation, although the underlying mechanism remains unknown. In this study, we found that TSH/TSHR signaling promoted immune evasion in CRC by inducing the function exhaustion of CD8+ T cells.
The TSH/TSHR pathway mainly functions through the PKA/CREB, PKC/ERK, and PI3K/AKT signaling pathways in tumor cells, whereas cAMP could promote the binding of CREB to the promoter region of CTLA4 and IL10 to promote the transcription of CTLA4 and IL10 [25-27]. cAMP could also induce the expression of TIM3 in T cells [28], although the underlying mechanism remains unclear. We found that TSH/TSHR signaling activated the cAMP/PKA signaling pathway in T cells, promoted CREB phosphorylation, enhanced CREB binding to the PD-1 and TIM3 promoter regions, and increased the transcriptional levels of PD-1 and TIM3. Consistent with the results in the literature, the results of RNA sequencing indicated that TSHR signaling affects the expression of CTLA4 and IL10. These findings suggest that the transcription factor CREB can activate the expression of multiple anti-inflammatory factors and suppress T cell function.
Tumor cells secrete various molecules that affect the function of T cells in the microenvironment (exosomes, microRNAs, and inflammatory factors, among others), whereas exosomes secreted by tumor cells play an essential role in the formation of the tumor immunosuppressive microenvironment. Exosomal PD-L1 from the tumor suppresses T-cell activation in the draining lymph node [29]. The present data indicated that CRC cells secreted TSHR into the tumor microenvironment, CD8+ T cells took up TSHR secreted by CRC cells, and TSHR expression in TIL-CD8+ T cells was decreased when TSHR was knocked down in CRC cells. Thyroid cancer cells expressed high levels of TSHR and secreted TSHR into the microenvironment via the exosomal pathway. We speculated that CRC cells may also transfer TSHR to CD8+ T cells through the exosomal pathway, resulting in high expression of TSHR in CD8+ T cells.
This study focused on the effect of endogenous TSH in the TME. However, whether exogenous factors influence the progression of CRC remains unclear. Current CRC screening guidelines do not consider thyroid dysfunction as a risk factor for disease progression. The results of a recent study show that untreated hypothyroidism (TSH > 4 mg/dl) is associated with a mildly elevated risk of CRC [30]. Although thyroid hormone replacement therapy has been found to have a protective effect against CRC risk [31], this finding does not contradict our results. During thyroid replacement therapy, negative feedback suppresses TSH secretion when the blood thyroid hormone concentration reaches a certain level, thus decreasing the risk of CRC. The present findings provide guidance for patients treated with checkpoint blockade. In particular, TSHR expression was identified as an important indicator to predict the efficacy of immunotherapy in patients with MSS-type CRC due to the poor effect of immunotherapy. Because of the lack of TSHR-specific inhibitors and TSHR antibodies in mice, we were not able to validate the effect of TSHR with PD-1 treatment.
5 CONCLUSIONS
Herein, we found that local TSH/TSHR signaling promoted the exhaustion of CD8+ T cells and immune evasion in CRC through the PKA/CREB signaling pathway. CRC cells secreted TSHR via extracellular vesicles, thereby increasing TSHR levels in CD8+ T cells and resulting in immunosuppression in the TME. MDSCs were the main source of TSH within the TME. Most importantly, TSHR insensitized tumors to PD-1 blockade therapy and TSHR expression was correlated with failure to respond to ICB in CRC patients.
AUTHOR CONTRIBUTIONS
Sisi Zeng, Huiling Hu, Zhiyang Li, Qi Hu performed most of the experiments and data analysis; Rong Shen analyzed the bulk RNA-seq data; Mingzhou Li, Yunshi Liang, Zuokang Mao, Yandong Zhang, Wanqi Zhan, Qin Zhu. Feifei Wang, Jianbiao Xiao, Bohan Xu, Guanglong Liu assisted in the in vivo or in vitro experiments; Yanan Wang, Bingsong Li, Shaowan Xu, Zhaowen Zhang, Ceng Zhang led the collection of blood and CRC tumor specimens and related clinical data. Sisi Zeng and ZhiZhang Wang wrote the original draft. Zhizhang Wang and Li Liang contributed in experiments guidance. Sisi Zeng and Huiling Hu revised and edited the manuscript.
ACKNOWLEDGMENTS
This work was supported by the National Key R&D Program of China (Grant No. 2021YFF1201004), the National Natural Science Foundation of China (Grant No. 82273358; No. 81802306; No. 81903002; No. 81672821; No. 82071742; No. 32270926); Natural Science Foundation of Guangdong Province of China (Grant No. 2019A1515012196; No. 2022A1515012059).
CONFLICT OF INTEREST STATEMENT
All authors declare no competing interest.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study was approved by the Clinical Research Ethics Committee of Southern Medical University (NFEC-2024-127). Each participant signed an informed consent before participating to this study. The animal study was carried out in compliance with the guidance suggestion of Animal Care Committee of Southern Medical University (No. 00301532).
Open Research
DATA AVAILABILITY STATEMENT
The datasets generated and/or analyzed in the current study are available from the corresponding author on reasonable request. The sequencing datasets have been deposited in National Center for Biotechnology Information (PRJNA943102).




