Metabolic landscape of head and neck squamous cell carcinoma informs a novel kynurenine/Siglec-15 axis in immune escape
Xin-Yu Zhang, Jian-Bo Shi, and Shu-Fang Jin contributed equally to this work.
Abstract
Background
Metabolic reprograming and immune escape are two hallmarks of cancer. However, how metabolic disorders drive immune escape in head and neck squamous cell carcinoma (HNSCC) remains unclear. Therefore, the aim of the present study was to investigate the metabolic landscape of HNSCC and its mechanism of driving immune escape.
Methods
Analysis of paired tumor tissues and adjacent normal tissues from 69 HNSCC patients was performed using liquid/gas chromatography-mass spectrometry and RNA-sequencing. The tumor-promoting function of kynurenine (Kyn) was explored in vitro and in vivo. The downstream target of Kyn was investigated in CD8+ T cells. The regulation of CD8+ T cells was investigated after Siglec-15 overexpression in vivo. An engineering nanoparticle was established to deliver Siglec-15 small interfering RNA (siS15), and its association with immunotherapy response were investigated. The association between Siglec-15 and CD8+ programmed cell death 1 (PD-1)+ T cells was analyzed in a HNSCC patient cohort.
Results
A total of 178 metabolites showed significant dysregulation in HNSCC, including carbohydrates, lipids and lipid-like molecules, and amino acids. Among these, amino acid metabolism was the most significantly altered, especially Kyn, which promoted tumor proliferation and metastasis. In addition, most immune checkpoint molecules were upregulated in Kyn-high patients based on RNA-sequencing. Furthermore, tumor-derived Kyn was transferred into CD8+ T cells and induced T cell functional exhaustion, and blocking Kyn transporters restored its killing activity. Accroding to the results, mechanistically, Kyn transcriptionally regulated the expression of Siglec-15 via aryl hydrocarbon receptor (AhR), and overexpression of Siglec-15 promoted immune escape by suppressing T cell infiltration and activation. Targeting AhR in vivo reduced Kyn-mediated Siglec-15 expression and promoted intratumoral CD8+ T cell infiltration and killing capacity. Finally, a NH2-modified mesoporous silica nanoparticle was designed to deliver siS15, which restored CD8+ T cell function status and enhanced anti-PD-1 efficacy in tumor-bearing immunocompetent mice. Clinically, Siglec-15 was positively correlated with AhR expression and CD8+PD-1+ T cell infiltration in HNSCC tissues.
Conclusions
The findings describe the metabolic landscape of HNSCC comprehensively and reveal that the Kyn/Siglec-15 axis may be a novel potential immunometabolism mechanism, providing a promising therapeutic strategy for cancers.
List of abbreviations
-
- HNSCC
-
- head and neck squamous cell carcinoma
-
- Kyn
-
- kynurenine
-
- siS15
-
- Siglec-15 small interfering RNA
-
- PD-1
-
- programmed cell death protein 1
-
- AhR
-
- aryl hydrocarbon receptor
-
- TME
-
- tumor microenvironment
-
- TIL
-
- Tumor-infiltrating lymphocyte
-
- Treg
-
- regulatory T cells
-
- GC-MS
-
- gas chromatography-mass spectrometry
-
- LC-MS
-
- liquid chromatography-mass spectrometry
-
- RNA-seq
-
- RNA-sequencing
-
- TMA
-
- tissue microarray
-
- DMEM
-
- Dulbecco's modified Eagle's medium
-
- FBS
-
- fetal bovine serum
-
- RPMI
-
- Roswell Park Memorial Institute
-
- STR
-
- short tandem repeat
-
- BCH
-
- 2-Aminobicyclo-(2,2,1)-heptane-2-carboxylic acid
-
- PHA
-
- polyhydroxyalkanoate
-
- DMSO
-
- dimethyl sulfoxide
-
- PCA
-
- principal component analysis
-
- OPLS-DA
-
- orthogonal partial least-squares discriminant analysis
-
- RPT
-
- response permutation testing
-
- VIP
-
- variable importance in the projection
-
- KEGG
-
- Kyoto Encyclopedia of Gene and Genome
-
- AUC
-
- Area under the curve
-
- ROC
-
- receiver operating characteristic
-
- GSEA
-
- gene set enrichment analysis
-
- GEPIA
-
- Gene Expression Profiling Interactive Analysis
-
- TCGA
-
- The Cancer Genome Atlas
-
- GTEx
-
- The Genotype-Tissue Expression
-
- Trp
-
- tryptophan
-
- CCK8
-
- Cell Counting Kit-8
-
- EdU
-
- 5-ethynyl-2′-deoxyuridine
-
- SDS
-
- sodium dodecyl sulfate
-
- PVDF
-
- polyvinylidene fluoride
-
- PD-L1
-
- programmed cell death 1 ligand 1
-
- PBS
-
- phosphate buffered saline
-
- HRP
-
- horseradish peroxidase
-
- ECL
-
- enhanced chemiluminescence
-
- NH2-MSN
-
- NH2-modified mesoporous silica nanoparticle
-
- NP-siRNA
-
- NH2-MSN-siRNA
-
- CTAB
-
- cetyltrimethyl ammonium bromide
-
- siScr
-
- siScramble
-
- H&E
-
- hematoxylin and eosin
-
- DAPI
-
- 4',6-diamidino-2-phenylindole
-
- Cy
-
- cyanine
-
- FITC
-
- fluorescein isothiocyanate
-
- ChIP
-
- chromatin immunoprecipitation
-
- RT-qPCR
-
- real-time quantitative polymerase chain reaction
-
- IL-2
-
- interleukin 2
-
- APC
-
- allophycocyanin
-
- PE
-
- phycoerythrin
-
- IFN-γ
-
- interferon-gamma
-
- LAG-3
-
- lymphocyte activation gene 3
-
- CFSE
-
- carboxyfluorescein succinimidyl ester
-
- ELISA
-
- enzyme-linked immunosorbent assay
-
- TNF-α
-
- tumor necrosis factor-alpha
-
- IC50
-
- half maximal inhibitory concentration
-
- IDO1
-
- indoleamine 2,3-dioxygenase 1
-
- Fa
-
- fraction affected
-
- CI
-
- combination index
-
- IHC
-
- immunohistochemistry
-
- TUNEL
-
- TdT-mediated dUTP-biotin nick end labeling
-
- IRS
-
- immunoreactive score
-
- ANOVA
-
- analysis of variance
-
- HPV
-
- human papillomavirus
-
- KMO
-
- kynurenine 3-monooxygenase
-
- KYNU
-
- kynureninase
-
- IL4I1
-
- interleukin-4-induced-1
-
- TDO2
-
- tryptophan 2,3-dioxygenase 2
-
- SEM
-
- tandard error of the mean
-
- ns
-
- not significant
-
- ROI
-
- region of interest
-
- CTL
-
- cytotoxic T lymphocyte
1 BACKGROUND
Metabolism is involved extensively in pathological and physiological processes, such as growth, development, and aging, and is regulated finely by an organism. Once metabolism becomes dysregulated, severe illnesses will emerge, such as gout and cardiovascular disease [1, 2]. Metabolism reprogramming, a key hallmark of cancer, endows tumors with adaptation to continuous metabolic stress, thereby facilitating cancer progression [3, 4]. Cancer cells can suppress antitumor immunity by reducing the metabolic fitness of tumor-infiltrating immune cells [5]. In addition, altered metabolites could convey information into the nucleus and remodel the epigenetic landscape [6]. For example, several metabolites, such as methionine [7], acetyl coenzyme A [8], and lactate [9], are sources of epigenetic modifications that regulate the expression of tumor-associated genes. Targeting tumor and immune cell metabolism, for example, via glutaminase and glucose uptake inhibitors, is an emerging cancer therapy field [10, 11]. However, more ideal and precise metabolic targets are required urgently. Elucidating aberrant metabolic profiles is vital for understanding the mechanisms of tumor development and for the development of novel targeted therapies.
Immune escape, another hallmark of cancer, allows cancer cells to escape detection and attack by the body's immune system through recruitment of inhibitory immune cells and alteration of the tumor microenvironment (TME) [12, 13]. Accumulating evidence indicates that metabolic reprogramming in the TME is a key driver of tumor immune escape [4, 14], which alters immune cell activation, differentiation, and killing [15]. Several metabolites, such as lactate [16], potassium [17], itaconate [18], and L-arginine [19], in the TME impair the function of tumor-infiltrating lymphocytes (TILs) and support energy supply of regulatory T (Treg) cells to promote immune escape. Therefore, exploring the key metabolites driving immune escape is crucial for understanding immune escape mechanisms and target identification. Targeting metabolic fragility can remold the anti-tumor TME and enhance the efficiency of immune checkpoint inhibitors.
Regarding patients with head and neck squamous cell carcinoma (HNSCC), their objective response rate for immunotherapy is less than 20% [20]. Therefore, the mechanisms of immunotherapy failure in most patients with HNSCC should be explored. Our previous study demonstrated that endogenous interferon-alpha [21] and lactate [22] could induce the expression of immune checkpoint molecules in HNSCC. However, the study of immune checkpoint molecules regulating the function of immune cells is still a long way off. Currently, studies have focused on the mechanism of immune escape in tumors such as breast cancer [23], renal cell carcinoma [24] and melanoma [25]. Unveiling the mechanism driving immune escape in HNSCC is essential for improving immunotherapy efficacy.
The aim of the present study was to reveal the landscape of metabolic reprogramming in HNSCC and elucidate the mechanisms via which the metabolites mediate immune escape. Metabolomics analysis of patients with HNSCC and functional profiling of TILs in vivo after metabolite stimulation were performed. We further explored how Kynurenine (Kyn) regulated the downsteam target of Siglec-15 involving in immune escape in HNSCC, shining a new light on better cancer treatment strategies for HNSCC.
2 MATERIALS AND METHODS
2.1 Patient information and tissue specimens
HNSCC diagnosis was based on histopathological features, including depth of invasion, pathological stage, and lymph node metastasis. Tumor tissues and adjacent normal tissues from 69 patients with HNSCC were collected and frozen immediately in liquid nitrogen within 30 min after surgery and stored at -80°C between June 2020 and January 2021. Analyses were performed on these samples using gas chromatography-mass spectrometry (GC-MS), liquid chromatography-mass spectrometry (LC-MS), and RNA-sequencing (RNA-seq). A tissue microarray (TMA) was prepared using 70 HNSCC and 18 adjacent normal tissues from patients who had undergone surgery between January 2007 and December 2008 and was used for immunohistochemical analysis. All enrolled primary HNSCC patients met our inclusion criteria: (1) treatment with surgical resection; (2) pathologically confirmed by two independent experts; and (3) no local or systemic treatment prior to the surgery. Exclusion criteria were as follows: (1) patients with history of previous malignancies; (2) patients refused to utilize their surgical specimens for research purposes. The clinical characteristics of HNSCC patients for metabolomics and RNA-seq analyses are listed in Supplementary Table S1, and those for TMA immunohistochemical analysis are listed in Supplementary Table S2. All patients had signed informed consent prior to surgery. The use of human specimens in the present study was approved by the Institutional Research Ethics Committee of Shanghai Ninth People's Hospital, Shanghai, China (approval ID: SH9H-2021-T135).
2.2 Cell lines and cell cultures
Human tongue squamous cell carcinoma cell lines (SCC9, SCC25, and Cal27) were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Human tongue squamous cell carcinoma cell lines (HN4 and HN6) and human pharyngeal squamous cell carcinoma cell line HN30 were obtained from the University of Maryland Dental School (Baltimore, MD, USA). Mouse squamous cell carcinoma cell line SCCVII was donated by Prof. Zhuang Liu (Soochow University, Suzhou, Jiangsu, China). The human embryonic kidney cell line 293T and human T-cell leukemia cell line Jurkat were purchased from the American Type Culture Collection (Manassas, VA, USA). All cultures were tested routinely using MycoAlertTM PLUS Mycoplasma Detection Kit (Lonza, Basel, Switzerland) and found to be negative for mycoplasma. All cancer cells were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM, Gibco, Grand Island, NY, USA) or DMEM/F-12 (Gibco) comprised of 0.584 g/L glutamine and supplemented with 10% fetal bovine serum (FBS, Gibco). Jurkat and SCCVII cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 (Gibco), supplemented with 10% FBS and 1% penicillin/streptomycin (Gibco). All cell lines were maintained at 37°C in 5% CO2. Short tandem repeat (STR) profiling was used to test the authenticity of the cell lines. Aryl hydrocarbon receptor (AhR) inhibitors, BAY-218 (S8842) and CH-223191 (S7711), were purchased from SelleckChem (Houston, TX, USA). System L amino acid transporter (Kyn transporter) inhibitor 2-aminobicyclo-(2,2,1)-heptane-2-carboxylic acid (BCH, S6894) and polyhydroxyalkanoate (PHA, HY-N7038) were purchased from MedChemExpress (Monmouth Junction, NJ, USA). Dimethyl sulfoxide (DMSO, 0.1%, 67-68-5, Sigma, St. Louis, MO, USA) was used as solvent control.
2.3 Mice
All animal experiments were approved by the Animal Care and Use Committee of Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China (approval ID: SH9H-2022-A031-SB). Female 4-6 weeks old C3H/He mice and BALB/c nude mice were purchased from Shanghai Bikai Keyi Biotechnology Co., Ltd (Shanghai, China). All mice were maintained under pathogen-free conditions in the animal care facilities of the Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine. All animals were maintained at room temperature (22-25°C) and with free access to food and water with a 12-h light/dark cycle. All mice used in experiments throughout the study exhibited normal health. All mice were used after 1 week of acclimatization to the facility. All animal studies were performed in accordance with the Guide for Care and Use of Laboratory Animals (The Ministry of Science and Technology of China, 2006).
2.4 LC-MS analysis
The metabolic profiles of HNSCC and paired adjacent normal tissues were analyzed using LC (Dionex U3000 UHPLC, Thermo Fisher, Waltham, MA, USA)-tandem MS (Q-Exactive, Thermo Fisher). Briefly, prepared samples were loaded separately on an Acquity UPLC BEH C18 column (1.7 µm, 2.1 mm × 100 mm, Waters, Milford, MA, USA). Each sample was eluted using a gradient elution system consisting of mobile phase A (99.9% H2O, 0.1% formic acid, v/v) and mobile phase B (99.9% acetonitrile, 0.1% formic acid, v/v). Elution gradient solutions were added as follows: from 100% to 95% mobile phase A in 2 min; from 95% to 70% mobile phase A in 2 min; from 70% to 50% mobile phase A in 4 min; from 50% to 20% mobile phase A in 2 min; from 20% to 0% mobile phase A in 4 min; holding at 0% mobile phase A for 1 min; from 0% to 95% mobile phase A in 5 s; holding at 95% mobile phase A for 55 s. The flow rate was 0.35 mL/min, column temperature was 45°C, and injection volume was 2 µL. Sample signals were collected continuously in the mass spectrometer at a capillary temperature of 320°C and a spray voltage of 3,800 V. Principal component analysis (PCA) and orthogonal partial least-squares discriminant analysis (OPLS-DA) were carried out to visualize the metabolic alterations among experimental groups, after mean centering and Pareto variance scaling, respectively. The Hotelling's T2 region, shown as an ellipse in score plots of the models, defines the 95% confidence interval of the modeled variation. Seven-fold cross-validation was applied with 1/7 of the samples being excluded from the mathematical model in each round, in order to guard against overfitting. In addition, 200 response permutation testing (RPT) was performed to evaluate the risk of model overfitting. Linear regression was conducted with R2Y and Q2Y of the original model, and the obtained intercept values between the regression line and Y-axis were R2 and Q2, respectively, evaluating whether the OPLS-DA model was overfitting. Variable importance in the projection (VIP) ranks the overall contribution of each variable to the OPLS-DA model, and those variables with VIP > 1 are considered relevant for group discrimination. Metabolics was annotated and analyzed using the Kyoto Encyclopedia of Gene and Genome (KEGG) database (https://www.genome.jp/kegg). Area under the curve (AUC) was calculated by summing the area under the receiver operating characteristic (ROC) curve, which shows the classifier's performance at varying decision thresholds.
2.5 GC-MS analysis
All the prepared samples were analyzed using an Agilent gas chromatograph (7890B, Agilent Technologies, Santa Clara, CA, USA) and an Agilent mass spectrometer (5977A, Agilent Technologies). A DB-5MS (30 m × 0.25 mm × 0.25 µm, Agilent Technologies) fused-silica capillary column (Agilent Technologies) was utilized for the GC-MS analysis. The column operating conditions were as follows: helium (> 99.999%) was used as carrier gas with a flow rate of 1.0 mL/min, inlet volume of 1 µL, and split-less inlet temperature of 260°C. The initial oven temperature was 60°C and held for 0.5 min; increased to 125°C at a rate of 8°C/min, 210°C at a rate of 5°C/min, 270°C at a rate of 10°C/min, and 305°C at a rate of 20°C/min; and finally held at 305°C for 5 min. The temperature of the MS quadrupole was set to 150°C, and electron impact was set to 230°C. The collision energy was 70 eV, and data were acquired in full-scan mode (m/z: 50-500). Data processing was the same in LC-MS.
2.6 RNA-seq and data analysis
Total RNA extraction was performed using a TRIzol reagent according to the manufacturer's protocol (Takara, Dalian, Liaoning, China). mRNA library construction was performed on an Illumina HiSeq X Ten platform (OE Biotech Co., Ltd., Shanghai, China). Raw data (raw reads) in fastq format for each sample were generated and processed using Trimmomatic [26]. Clean reads for each sample were retained by removing low-quality reads and mapped to the human genome (GRCh38) using HISAT2 for subsequent analyses [27]. The patients were separated into two groups based on the median values of the Kyn level (Kyn-high or Kyn-low) or CD274 expression (CD274-high or CD274-low). Gene set enrichment analysis (GSEA) was performed with a gene list ordered based on the log2 (Ratio) value, the absolute value of the log2 (Ratio), and the network-based score, respectively, using the GSEA-preranked function, from which we computed enrichment scores, normalized enrichment scores, P values, and false discovery rates for each gene set based on a modified K–S test. Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.pku.cn/) is a newly developed interactive web server for analyzing the bulk gene expression datasets in The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) projects. In the present study, SIGLEC15 expression and survival analysis were performed using the GEPIA HNSCC dataset.
2.7 LC-MS targeted metabolomics analysis
Samples were ground in 100% methanol for 10 min and centrifuged (16,000 ×g) at 4°C for 10 min. The supernatant was filtered through a 0.22-µm membrane filter and analyzed using a liquid chromatograph (UltiMate 3000 RS, Thermo Fisher). The samples were loaded separately on a Thermo Hypersil GOLD HILIC column (1.9 µm, 2.1 mm × 100 mm, Thermo Fisher). Samples were eluted with an elution including phase water (containing 0.1% formic acid, v/v) and phase acetonitrile (containing 0.1% formic acid, v/v). Elution gradient solutions were added as follows: from 5% to 95% water in 2.5 min, holding at 95% water for 1 min, from 95% to 5% water in 18 s, holding at 5% water for 1 min. The flow rate was 0.40 mL/min, column temperature was 40°C, and inlet volume was 2 µL. Tryptophan (Trp), Kyn, kynurenic acid, indole-3-carboxaldehyde and serotonin standards (Sigma) were used to construct a standard curves for quantifying the concentrations. Briefly, all standard samples were dissolved in 0.1% formic acid and methanol for a stock solution of 2 mg/mL, then a series of standard curve solutions were diluted with pure methanol in concentration gradients of 20,000, 5,000, 2,000, 500, 200, 50, 20, and 5 ng/mL in turn. The compounds’ chromatogram collection and integration were processed using Xcalibur 3.0 software (Thermo Fisher). Linear regression was performed with a weighting coefficient of 1/x2 to calculate the content of metabolites in the samples.
2.8 Cell counting kit-8 (CCK8), colony formation assay, 5-ethynyl-2’-deoxyuridine (EdU) assay
Cells were seeded at an initial density of 1,000 cells/well in 96-well plates and treated with 0.1% DMSO, 50 or 100 µmol/L of Kyn for 5 days to detect cell proliferation using a CCK8 kit (Dojindo, Kumamoto, Japan).
Cells were seeded in 6-well plates at 1,000 cells/well and treated with 0.1% DMSO, 50 or 100 µmol/L of Kyn for 10 days. Subsequently, cells were prefixed with 4% paraformaldehyde (P0099, Beyotime, Shanghai, China) for 10 min at room temperature and stained by crystal violet staining solution (C0121, Beyotime) for 15 min to analyze colony formation.
Cells were seeded in 6-well plates at 5,000 cells/well and treated with 0.1% DMSO, 50 or 100 µmol/L of Kyn for 24 h. Subsequently, cells were stained using a Cell-Light EdU Apollo567 In Vitro Kit (C10371-1, RiboBio, Guangzhou, Guangdong, China) according to the manufacturer's protocol. Cells were viewed under a Zeiss inverted light microscope (Axio vert A1, Zeiss, Oberkochen, Germany), and the percentage of EdU-positive cells was analyzed using ImageJ software (NIH, Bethesda, ML, USA).
2.9 Cell migration and invasion assay
Wound healing and transwell assays using uncoated polycarbonate inserts (Millipore, Billerica, MA, USA) were performed to determine cell migration. Transwell invasion assays were performed using Bio-Coat™ inserts (BD Biosciences, San Jose, CA, USA). Cells were pretreated with 1 µg/mL mitomycin C (Sigma) for 1 h to exclude the effect of cell proliferation. Medium without FBS (2 × 104 cells/200 µL) were added into the upper portion of a migration (uncoated insert) or invasion (matrigel-coated insert) chamber, with 500 µL DMEM containing 10% FBS added into the lower chamber. After staining with crystal violet, the number of colonies was counted and analyzed statistically. The experiment was performed in triplicate.
2.10 Western blotting analysis
Western blotting was performed as described in our previous study [28]. The cell protein was extracted in sodium dodecyl sulfate (SDS) lysis buffer (Beyotime) with protease inhibitor cocktail (4693132001, Roche, Basel, Switzerland). The protein samples were electrophoresed on 4%-20% polyacrylamide gels (Genscript, Nanjing, Jiangsu, China) and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore). Membranes were then incubated overnight at 4°C with primary antibody. The antibodies used in this study included anti-E-cadherin Ab (1:1,000 dilution, 3195, Cell Signaling Technology, Danvers, MA, USA), anti-Vimentin Ab (1:1,000 dilution, 5741, Cell Signaling Technology), anti-Snail Ab (1:1,000 dilution, 3879, Cell Signaling Technology), anti-programmed death-ligand 1 (PD-L1) Ab (1:1,000 dilution, 85164, Cell Signaling Technology), anti-Siglec-15 Ab (1:1,000 dilution, 37925, Signalway Antibody, Greenbelt, ML, USA), primary rabbit anti-human AhR Ab (1:1,000 dilution, 83200, Cell Signaling Technology), and anti-programmed cell death protein 1 (PD-1) Ab (1:5,000 dilution, 66220-1-Ig, Proteintech, Rosemont, IL, USA). An anti-β-actin antibody (1:2,000 dilution, 8457, Cell Signaling Technology) was used as an internal control. After washing with phosphate buffered saline (PBS), the membrane was probed with horseradish peroxidase (HRP)-conjugated secondary antibody (Cell Signaling Technology) at 1:2,000 dilution for 1 h, and the signals were detected with enhanced chemiluminescence (ECL) detection reagents (Millipore). Immunoreactive bands were analyzed using ImageJ software.
2.11 RNA interference and lentiviral transfection
Lipofectamine 2000 (11668019, Thermo Fisher) was used for transfection with siRNAs (siAhR #1-3 for homo sapiens and siSIGLEC15 [siS15] #1-3 for mouse, RiboBio), at a final concentration of 100 nmol/L. Mouse SIGLEC15 cDNA was cloned into the PGMLV-CMV-MCS-EF1-ZsGreen1-T2A-Puro vector (Genomeditech, Shanghai, China). Polybrene was used for the lentiviral transfection. Puromycin (1 µg/mL) was used for selection for 7 days to establish a stable SCCVIISiglec-15 cell line. Vector lentivirus was used to establish SCCVIIVector cell line. The sequences are listed in Supplementary Table S3.
2.12 Preparation and characterization of NH2-modified mesoporous silica (NH2-MSN) nanoparticles and NH2-MSN-siRNA (NP-siRNA)
NH2-MSNs were synthesized according to previous reports [29, 30]. Briefly, 1.5 g of cetyltrimethyl ammonium bromide (CTAB, H5882, Sigma) was dissolved and stirred, and different amounts of diethanolamine (D8885, Sigma) were added and stirred. Subsequently, 12 mL of tetraethyl orthosilicate (131903, Sigma) was added dropwise to the mixture under intense stirring. The products were dried, collected, and refluxed in methanol/HCl. The steps were repeated to remove CTAB and obtain MSNs. Subsequently, the MSNs were redispersed and stirred with 3-aminopropyltriethoxysilane (A3648, Sigma). After refluxing, the obtained products were cleaned and dried successively, as described above. The amino-functionalized samples were named NH2-MSNs. The positively charged NH2-MSNs (10 µg/mL) were mixed with the siRNA (siS15 or siScramble [siScr]) solution in PBS at pH 6.0 and shaken on a bench-top shaker for 1 h to complete the binding between siRNA and NH2-MSNs via electrostatic interactions. The prepared samples were termed NP-siRNA (NP-siS15 or NP-siScr) herein. The morphologies of NH2-MSNs and NP-siS15 were examined under transmission electron microscopy (JEM-1400, JEOL, Tokyo, Japan). Zeta potential measurements of NH2-MSNs and NP-siS15 were performed using the Zetasizer Nanoseries (Nano ZS90, Malvern Instrument Ltd., Worcestershire, UK).
2.13 in vivo experiments
Orthotopic transplants were performed as described [31]. Required number of SCCVII cells were collected by centrifuging (300 ×g) at 4°C for 3 min, and supernatant was discarded before washed twice by pre-cold PBS. Subsequently, cell pellets were resuspended in Matrigel (356234, Corning, NY, USA) at concentration of 2,000 cells/µL and mixed gently on ice. C3H/He mice were anesthetized, and the tongues were gently grasped and pulled out from the mouth using forceps. Using a 1-mL syringe attached to a 29 gauge needle, 25 µL cells (5 × 104) were injected slowly into the right lateral side of each tongue (∼1.5 mm from the tip of the tongue) so that a bulbous mass formed in the center of the tongue. Small white tumors were visible 10 days after injection. The mice received intraperitoneal injections of either PBS or Kyn (50 mg/kg) every 3 days for 3 cycles. The mice were euthanized at 20 days and analyzed. For tongue samples taken thereafter, the entire tongue was removed by cutting the tongue at the back of the mouth and bisecting it down the middle from tip to back. Generally, the entire tongue was used for immunohistology analysis. For the Siglec-15 overexpression experiment, SCCVIIVector or SCCVIISiglec-15 cells were injected into the tongue as orthotopic transplants.
Female BALB/c nude mice were used to establish lung metastasis model. A total of 5 × 105 HN30 cells in 100 µL PBS were injected intravenously into the lateral tail vein. After 10 days of tumor cell injection, 4 mg/100 µL Kyn or 100 µL PBS was administered by gavage twice a week for 4 weeks. Mice were euthanized 5 weeks after treatment. Whole lung tissues were removed and fixed in Bouin's fixative (HT10132, Sigma) diluted 1:5 with neutral-buffered formalin for metastasis nodule observation.
Subcutaneous tumorigenesis models were established using C3H/He mice. A total of 1 × 105 SCCVIIVector or SCCVII Siglec-15 cells in 100 µL PBS were inoculated subcutaneously into both flanks, and small tumors were visible 10 days after injection. The length (L) and width (W) of the tumors were measured every 2 days from the 10th day after injection, and the volume was calculated as (L × W2 × π/6). On day 20, the mice were euthanized, and the tumors were extracted carefully for flow cytometry and immunohistochemical analyses. For the Kyn stimulation experiment, SCCVII cells (1 × 105) were inoculated subcutaneously into C3H/He mice as before. After 10 days of tumor transplantation, mice received oral gavage of PBS, Kyn (100 mg/kg), and/or CH-223191 (10 mg/kg) once every 2 days. The mice were euthanized at day 20 and analyzed as previously described. The vital organs were fixed, embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E), and blood samples were obtained for hepatic and renal function testing. For anti-PD-1 and anti-Siglec-15 experiments, SCCVII subcutaneous tumorigenesis models were established as before and separated randomly into 5 groups. After 10 days of tumor transplantation, the anti-PD-1 Ab (10 mg/kg, BE0146, BioXCell, West Lebanon, NH, USA) or isotype IgG (10 mg/kg, BE0089, BioXCell) were administered by intraperitoneal injection, while NP-siScr (0.5 nmol siScr:4 µL NH2-MSNs) and NP-siS15 (0.5 nmol siS15:4 µL NH2-MSNs) were administered by intravenous injection. Tumor volume was measured every 2 days, and mice were euthanized at the end of the experiment. Overall survival probability curves were used to evaluate treatment efficacy. For anti-PD-L1 and anti-Siglec-15 experiments, SCCVII subcutaneous tumorigenesis models were established as before and separated randomly into 4 groups. After 10 days of tumor transplantation, the anti-PD-L1 (5 mg/kg, 2115, Selleck) or isotype IgG (5 mg/kg, A2116, Selleck) were administered by intraperitoneal injection, whereas NP-siS15 (0.5 nmol siS15: 4 µL NH2-MSNs) were administered by intravenous injection. Tumor volume measurement and analysis were performed as previously described.
Tribromethyl alcohol (HY-B1372, MedChemExpress) was used to anesthetize mice. Mice were euthanized by cervical dislocation after anesthesia in the following conditions: near death or immobility, significant weight loss, inability to feed or drink, at the end of experiment, or tumor volume > 2,000 mm3.
2.14 Immunofluorescence
Cells with the indicated treatments were fixed for 20 min with 4% paraformaldehyde and blocked for 1 h with 10% goat serum at 37°C. Subsequently, they were incubated with primary anti-AhR antibody (1:200, 67785-1-Ig, Proteintech, USA) overnight at 4°C. The cells were stained with Alexa Fluor 647-conjugated secondary antibody (4410, Cell Signaling Technology), and nuclei were counterstained with 4’,6-diamidino-2-phenylindole (DAPI) (D21490, Thermo Fisher). SCCVII cells were transfected with NH2-MSNs that contained siS15 (RiboBio) labeled with Cyanine (Cy) 5 (NP-siS15-Cy5). The cytoskeleton and nuclei were counterstained using fluorescein isothiocyanate (FITC)-phalloidin (40735ES75, Yeasen, Shanghai, China) and DAPI. The cellular uptake of NP-siS15-Cy5 was observed using Cytation 5 (Biotek, Winooski, VT, USA).
2.15 Chromatin immunoprecipitation (ChIP) assay
The SimpleChIP® Enzymatic Chromatin IP kit (9003, Cell Signaling Technology) was used for the ChIP assay, as previously described [21]. Briefly, after the indicated treatment, Cal27 and HN30 cells were cross-linked by formaldehyde (Millipore) at 1% final concentration. Chromatin was digested with micrococcal nuclease and sonicated into 150-900 bp fragments. An aliquot of the cross-linked protein complexes was immunoprecipitated by incubation with either primary rabbit anti-human AhR antibody or IgG antibody (2729, Cell Signaling Technology) overnight at 4°C with rotation. Chromatin-antibody complexes were isolated from solution by incubation with ChIP-Grade Protein G Magnetic Beads (9003, Cell Signaling Technology) for 1 h at 4°C with rotation. The bead-bound immune complexes were then washed and eluted from the beads with elution buffer. Eluates were heated at 65°C overnight to reverse the formaldehyde cross-linking, and DNA was extracted. DNA samples from chromatin immunoprecipitation preparations were analyzed by real-time quantitative polymerase chain reaction (RT-qPCR). The promoter primers for SIGLEC15 and CD274 were synthesized by Sangon Biotech (Shanghai, China) and are listed in Supplementary Table S4.
2.16 Fluorescent multiplex immunohistochemistry staining and analysis
For multiple-immunofluorescence staining, paraffin-embedded tissues were sequentially stained with indicated primary antibodies overnight at 4°C and HRP-conjugated secondary antibodies for 1 h at room temperature, followed by microwave treatment (700 w for 5 min and then 100 w for 30 min) and another round of staining with three different primary antibodies. Anti-CD3 rabbit antibody (1:200 dilution, ab16669, Abcam, Cambridge, MA, USA) and anti-CD8 rabbit antibody (1:200 dilution, ab209775, Abcam) for SCCVII tumors, anti-CD8 (D8A8Y) rabbit antibody (1:200 dilution, 85336, Cell Signaling Technology) for HNSCC tissues, and anti-PD-1 rabbit antibody (1:200 dilution, 66220-1-Ig, Proteintech) were used as primary antibodies. Cy3, FITC, Cy5, and DAPI dyes were used for staining. The slides were mounted with the nuclear dye DAPI. Images of the samples were captured and collected using a slice scanner (Pannoramic MIDI, 3DHISTECH Ltd., Budapest, Hungary). Analyses of the different immunofluorescence images differed in each experiment and are indicated in the figure legends.
2.17 RT-qPCR assay
The total RNA of related cells was isolated using a TRIzol reagent (15596018, Invitrogen, Carlsbad, CA, USA). Total RNA (500 ng) was reversely transcribed into first-strand cDNA using PrimeScript™ RT Master Mix (RR036A, TaKaRa, Tokyo, Japan). RT-qPCR was conducted to quantify mRNA expression using a StepOnePlus Real-time PCR system (Applied Biosystems, Foster City, CA, USA). The reaction conditions were denaturation at 95°C for 30 s, followed by 40 cycles of 5 s of denaturation at 95°C, and annealing and extension for 30 s at 60°C. The primer sequences used in the present study are listed in Supplementary Table S5. Relative quantification of mRNA levels compared to the internal control gene was performed using the 2-ΔΔct method. All samples were assayed in triplicate.
2.18 Luciferase reporter assay
293T cells were transfected with luciferase reporter (firefly and renilla luciferase) plasmids containing the promoter of SIGLEC15 or CD274. Transfected 293T cells were then treated with Kyn at a concentration or time gradient. To examine the transcription of AhR for SIGLEC15 or CD274, 293T cells were co-transfected with siAhR or siScr and luciferase reporter (firefly and renilla luciferase) plasmids, including the SIGLEC15 or CD274 promoter. After 48 h of incubation, a dual-luciferase reporter assay kit (RG027, Beyotime) was used to detect luciferase activity as previously described [32].
2.19 TIL analysis
Tumors established using the indicated treatments were removed from C3H/He mice, dissociated, and digested, as previously described [32]. The digested tissues were washed twice with PBS and filtered through a 70-µm strainer. The obtained single-cell suspensions were stained with anti-mouse CD45 PerCP-Cy5.5 (BioLegend, San Diego, CA, USA) to label the immune cells and stained with viability dye 450 (Peprotech, Cranbury, NJ, USA) at 4°C in dark for 30 min to distinguish between live and dead cells.
2.20 Isolation of CD8+ T cells
Human CD8+ T cells were isolated from peripheral blood of 4 healthy donors at physical examination using a human CD8 MicroBeads kit (130-045-201, Miltenyi Biotec, Bergisch Gladbach, Germany), according to the manufacturer's recommendations. All donors gave informed consent. Enriched CD8+ T cells (1 × 105) were stimulated in 6-well plates precoated with 2 µg/mL anti-human CD3 (01121-25, BioGems, Westlake Village, CA, USA) and cultured in RPMI-1640 with 2 µg/mL anti-human CD28 (10311-25, BioGems) and 20 ng/mL recombinant human interleukin-2 (IL-2, 200-02, Pepro Tech, Rocky Hill, NJ, USA) for 48 h. Cells were then transferred to new wells with a new culture medium for follow-up experiments. PHA (1 µg/mL) was used to sitmulate Jurkat T cells for 24 h, and BCH (5 mmol/L) was applied to block the transport of kyn to cytoplasm in CD8+ T and Jurkat T cells for 48 h.
2.21 Flow cytometry assay
For TIL analysis, samples were incubated with membrane antibodies containing anti-mouse CD3 allophycocyanin (APC)-Cy7, anti-mouse CD8-FITC, anti-mouse PD-1 phycoerythrin (PE)-Cy7, and incubated with transcription factor fixation/permeabilization working solution (92550-00, BioGems) for 30 min at 4°C, and washed with permeabilization buffer (BioGems). Samples were then stained with intracellular or intranuclear antibodies, including anti-mouse interferon-gamma (IFN-γ) APC, anti-mouse lymphocyte activation gene 3 (LAG-3) PE, anti-mouse Ki-67 BV605, anti-mouse granzyme PE, anti-mouse perforin APC for 30 min at 4°C in the dark. The samples were then washed with PBS and analyzed using a BD Fortessa cytometer (BD Biosciences). For T cell analysis, human CD8+ T and Jurkat cells were detected using EdU Cell Proliferation Kit with 10 µmol/L Alexa Fluor 555 (Beyotime) for 2 h, 1 µmol/L carboxyfluorescein succinimidyl ester (CFSE, 65-0850-84, Thermo Fisher) for 10 min, and surface PD-1 expression (Anti-Human PD-1 PE, 31831-60, BioGems) assays, with the indicated treatments. All antibodies against immune cells were purchased from PeproTech and analyzed using a BD Fortessa cytometer (BD Biosciences). Flow cytometry data were analyzed using FlowJo 10.8.1 (BD Biosciences).
2.22 Enzyme-linked immunosorbent assay (ELISA)
The concentration of cytokines in C3H/He mice were detected by ELISA. Mouse serum was collected at the experimental endpoint and analyzed using a mouse tumor necrosis factor-alpha (TNF-α) ELISA kit (YC113, Shycbio, Shanghai, China) and an IFN-γ ELISA kit (YC111, Shycbio) according to the manufacturer's instructions.
2.23 Drug combination studies
In vitro drug combination studies were conducted similar to the steps in our previous study using the CCK8 assay [33]. The molar ratio of equipotent doses of the two agents (at the ratio of their half maximal inhibitory concentration [IC50] values) was applied. Briefly, cells were seeded at 3,000 cells/well in 96-well plates, and then treated with serial dilutions of epacadostat (an indoleamine 2,3-dioxygenase 1 [IDO1] inhibitor, S7910, SelleckChem) and CH-223191 for 72 h. The cell viability was measured using the CCK8 assay. The dimensionless value at fraction affected (Fa) = 0.5 was calculated from Fa–CI curve. The combination index (CI) was used to analyze the synergistic inhibitory effects of drug combinations using CompuSyn software 1.0 (CompuSyn Inc., Paramus, NJ, USA) according to the Chou–Talalay equation [34].
2.24 Immunohistochemistry (IHC)
Tumor tissues were stained with H&E, Ki-67 (AF1738, Beyotime), and TdT-mediated dUTP-biotin nick end labeling (TUNEL, C1098, Beyotime), according to the recommended protocols. The tissues of the patient were embedded and sectioned into 4-µm slides. Subsequently, slides were deparaffinized, rehydrated, antigen repaired, and incubated with primary rabbit anti-human Siglec-15 Ab (dilution 1:100; 37925, Signalway Antibody), primary rabbit anti-human AhR Ab (dilution 1:100) overnight at 4°C. The immunoreactive score (IRS) was calculated by multiplying the intensity score (0 = negative, 1 = weak, 2 = intermediate, 3 = strong) and the fraction score (percentage of positive tumor cells, range = 0-100) [32]. Images were acquired and analyzed using the CaseViewer software 2.4 (3DHISTECH Ltd., Budapest, Hungary).
2.25 Statistical analysis
A power analysis to determine sufficient sample size was not conducted. Samples were randomized before LC-MS, GC-MS, and RNA-seq analyses. R sofeware 4.2.0 (https://www.r-project.org/) was used for the analysis of metabolomic and transcriptomic data. The R packages pheatmap v1.0.12 (https://www.rdocumentation.org/packages/pheatmap/versions/1.0.12/topics/pheatmap) and ggplot2 v3.4.2 (https://ggplot2.tidyverse.org/index.html) were used for data visualization. For animal experiments, mice injected with tumor cells were randomized before allocation to cages. All other experiments were not randomized, and the investigators were not blinded to the allocation during the experiments or outcome assessment. Statistical analysis and plotting were performed using the GraphPad Prism 9.0 (GraphPad Software, San Diego, CA, USA). Two-sided Student's t-test or one-way analysis of variance (ANOVA) was used to compare two or more groups. Tukey's test was used as the post-hoc after ANOVA. The relationship between Siglec-15 or AhR expression and clinicopathological characteristics was determined using two-sided χ2 tests. Data are expressed as mean ± standard error of mean. Statistical significance was set at P < 0.05.
3 RESULTS
3.1 Untargeted metabolomics revealed metabolic landscape and Trp/Kyn pathway alterations in HNSCC
Metabolic reprogramming is a critical characteristic of malignant tumors, which plays an important role in tumor progression, metastasis, and immune escape [3]. However, a comprehensive analysis of metabolic reprogramming in HNSCC remains elusive. To reveal the metabolic profile of HNSCC, 69 pairs of tumor and adjacent normal tissues were collected (Supplementary Table S1), and their metabolic and transcriptional alterations were analyzed using GC/LC-MS and RNA-seq (Figure 1A). Nine human papillomavirus (HPV)-negative oropharyngeal samples were included in this cohort. As HPV-negative HNSCCs have similar biological characteristics [35, 36], our research conclusions would not be affected by the diversity of sites. PCA and OPLS-DA model score plots showed a perfect separation between HNSCC and adjacent normal tissues (Supplementary Figure S1A-B). Seven-fold cross-validation and 200 RPT indicated the accuracy of the OPLS-DA model (Supplementary Figure S1C). Collectively, 178 differential metabolites were identified using LC-MS, out of which 121 had elevated levels, and 57 had decreased levels (Figure 1B, Suppementary Figure S1D). They were further classified as carbohydrates (n = 22), lipids and lipid-like molecules (n = 42), amino acids (n = 56), and others (n = 58) (Figure 1C, Supplementary Figure S1E). Amino acids were the major category of differential metabolites, indicating that altered amino acid metabolism may be involved in HNSCC development.
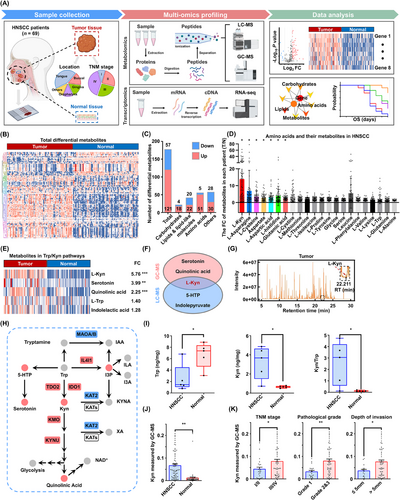
GC/LC-MS analysis revealed metabolic reprograming in HNSCC and alterations of the Trp/Kyn pathway. (A) Flowchart of the study design. Tumor tissues and adjacent normal tissues from HNSCC patients (n = 69) were analyzed using multi-omics profiling, including GC-MS, LC-MS, and RNA-seq, to unveil the metabolic landscape of HNSCC. (B) Metabolomics heatmap based on LC-MS results. Rows show the different metabolites (n = 178), and columns show the different tissues (n = 69). The log-transformed metabolite intensities were Z scored/standardized. (C) The numbers of differential metabolites. (D) The expression of human amino acids and their metabolites were measured in HNSCC. Each point represents the fold change of metabolites in each patient (T/N). (E) Heatmap depicts the relative intensities of the metabolites (n = 138) in the Trp pathway, analyzed by GC-MS. (F) Venn diagram shows the overlap of the metabolites in Trp pathways identified by GC-MS and LC-MS. (G) A representative chromatogram of Kyn in GC-MS. (H) Schematic representation of key enzymes and metabolites involved in the Trp/Kyn pathway. Red and blue indicated high and low expression/levels, respectively. (I) LC-MS targeted metabolomics analysis shows the Trp and Kyn contents in 5 HNSCC patients. (J) Levels of Kyn, as measured using GC-MS, in HNSCC patients (n = 69). (K) Levels of Kyn and their association with TNM stage, pathological grade, and depth of invasion in HNSCC (n = 69). Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. Abbreviations: HNSCC, head and neck squamous cell carcinoma; LC-MS, liquid chromatography-mass spectrometry; GC-MS, gas chromatography-mass spectrometry; FC, fold change; OS, overall survival; Kyn, kynurenine; Trp, tryptophan; 5-HTP, 5-hyroxy-L-tryptophan; RT, retention time; I3P, indole-3-pyruvate; IAA, indole-3-acetic acid; ILA, indole-3-lactic acid; I3A, indole-3-aldehyde; KYNA, kynurenic acid; XA, xanthurenic acid; IL4I1, interleukin-4-induced-1; IDO1, indoleamine-2,3-dioxygenase 1; TDO2, tryptophan-2,3-dioxgenase; MAOA/B, monoamine oxidase A/B; KATs, kynurenine aminotransferases; KMO, kynurenine-3-monooxidase, KYNU, kynureninase; NAD+, nicotinamide adenine dinucleotide; SEM, tandard error of the mean.
Similarly, KEGG metabolic pathway enrichment analysis revealed that several amino acid pathways, such as those of glycine, serine, threonine, and Trp, were enriched significantly (Supplementary Figure S1F). We identified the key metabolites or molecules in the metabolic and gene networks. The most significant hub network had 59 nodes and 771 interactions, containing 25 genes and 34 metabolites (Supplementary Figure S1G). Top 20 altered metabolites were obtained using GC-MS (Supplementary Table S6) and LC-MS analyses (Supplementary Table S7). Furthermore, we analyzed the contents of all human amino acids and their metabolites and found that Kyn had the most significant fold change (Figure 1D, Supplementary Figure S1H), indicating a potential association between Kyn and HNSCC progression. In addition, Kyn is the only molecule included in the top 10 altered metabolites and the hub network, suggesting that its reprograming may associated with HNSCC progression.
Trp is an essential amino acid involved in the biosynthesis of cellular proteins and formation of the cytoskeleton, and Kyn is a major metabolite of Trp catabolism [37]. Consequently, we focused on the Trp degradation trends, and Kyn, serotonin, quinolinic acid, 5-hydroxy-L-tryptophan, and N-methyltryptamine were identified by GC-MS (Figure 1E) and LC-MS analyses (Supplementary Figure S2A) as immediate products. The most significantly altered molecule in the Trp pathway was Kyn (Supplementary Figure S2B-C). Indeed, Kyn was the only differential metabolite involved in the Trp pathway that was detected by both GC-MS and LC-MS analyses (Figure 1F), and the mass spectrogram of Kyn is shown in Figure 1G. Correlation analysis showed good concordance between LC-MS and GC-MS data with regard to Kyn content (Supplementary Figure S2D).
The gene expression of metabolic enzymes involved in the Trp metabolic pathway was analyzed, and 13 differentially expressed genes were observed, among which kynurenine 3-monooxygenase (KMO), kynureninase (KYNU), interleukin-4-induced-1 (IL4I1), IDO1, and tryptophan 2,3-dioxygenase 2 (TDO2) were increased significantly in HNSCC (Supplementary Figure S2E-F). A schematic representation of the dysregulated enzymes and metabolites in the Trp pathway was drawn according to the metabolomics and transcriptomics data (Figure 1H). Considering the impact interconnected systems between metabolites, we found that blocking the Kyn pathway by epacadostat resulted in Trp accumulation and serotonin generation (Supplementary Figure S2G).
To confirm the metabolomics results, 5 pairs of fresh HNSCC tissues were collected for determination of Trp and Kyn contents. The HNSCC tissues had lower Trp levels and higher Kyn levels and Kyn/Trp ratios than the adjacent normal tissues (Figure 1I). Furthermore, the HNSCC cell lines had higher Kyn content than normal primary oral mucosal epithelia (Supplementary Figure S2H). Indeed, the Kyn content was increased in 69 patients with HNSCC (Figure 1J, Supplementary Figure S2I). Increased levels of Kyn were associated with advanced TNM stage, poor pathological grade, and greater depth of invasion (Figure 1K, Supplementary Figure S2J) but were not associated with age or sex (Supplementary Figure S2K-L).
ROC analysis was performed to further characterize the predictive value of Kyn. In GC-MS, Kyn had a high AUC value of 0.8895, indicating a good diagnostic efficiency in HNSCC (Supplementary Figure S2M). Similar results were obtained for LC-MS, with an AUC of 0.9053 (Supplementary Figure S2N). The results indicate that upregulated Kyn levels may play a role in HNSCC progression.
3.2 Kyn promoted HNSCC proliferation and metastasis
To investigate the biological role of Kyn in HNSCC, a series of in vitro and in vivo assays were conducted. The CCK8 and colony formation assays indicated that 50 and 100 µmol/L Kyn enhanced proliferation, colony formation ability and the percentage of EdU-positive cells compared with the control in Cal27 and HN30 cells (Figure 2A-C). Cells after Kyn stimulation migrated more quickly than DMSO control in the would healing assay(Figure 2D), and the number of migratory and invasive cells increased in the transwell assay (Figure 2E-F). Moreover, Kyn decreased the expression of E-cadherin significantly but increased that of Vimentin and Snail (Figure 2G, Supplementary Figure S3A), indicating that Kyn promotes epithelial-to-mesenchymal transition in HNSCC cell lines. The metastasis model showed that Kyn induced lung metastasis significantly, as shown by H&E staining. The number of metastatic nodules increased after Kyn treatment (Supplementary Figure S3B). Considering the limitations of the metastasis model, SCCVII orthotopic transplant models were established in C3H/He immune-competent mice (Figure 2H). Tumor volume and Ki-67 analysis showed that Kyn accelerated tumor progression (Figure 2I, Supplementary Figure S3C-D). More importantly, Kyn promoted neck lymph node metastases after tongue injection (Figure 2J).
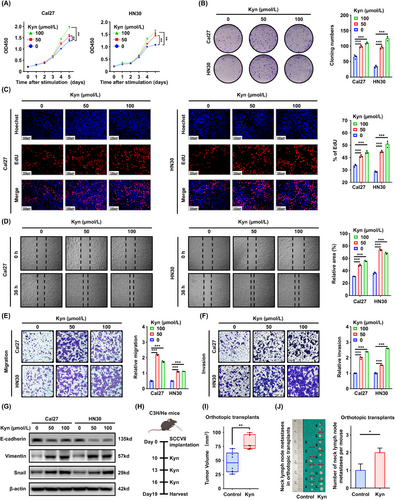
Kyn promoted HNSCC aggressive progression both in vivo and in vitro. (A) CCK8 assay following indicated Kyn treatments in Cal27 and HN30 cells. (B) Colony formation assay was performed using Cal27 and HN30 cells following stimulation with Kyn. (C) EdU assay was performed to detect cell proliferation after Kyn stimulation for 24 h. (D) Wound healing assay was conducted to detect the migration ability of HNSCC cells after Kyn stimulation for 36 h. (E-F) Migration and invasion capacities were detected using the transwell assay after Kyn stimulation for 24 h. (G) E-cadherin, Vimentin, and Snail were detected after Kyn stimulation for 24 h. (H) Schematic diagram of the Kyn administration strategy in C3H/He tongue orthotopic transplant models. (I) The volume of tumor was compared between the two groups (n = 6 mice per group). (J) Neck lymph node metastasis analysis of the orthotopic transplant models. Data are represented as the mean ± SEM (A-G and I-J) based on three independent experiments (A–G). *P < 0.05, **P < 0.01, ***P < 0.001. Abbreviations: Kyn, kynurenine; HNSCC, head and neck squamous cell carcinoma, CCK8, Cell Counting Kit-8; EdU, 5-ethynyl-2′-deoxyuridine; SEM, tandard error of the mean.
3.3 Kyn promoted CD8+ T cell dysfunction and Siglec-15 expression in tumor cells
To explore the effects of Kyn for HNSCC immunity, the expression of immunecheckpoint molecules was detected by RNA-seq in the Kyn-high group and Kyn-low group. Notably, most classical immune checkpoint molecules were significantly higher in the Kyn-high group than in the Kyn-low group (Figure 3A). In addition, GSEA revealed that the T cell receptor signaling pathway was enriched significantly in Kyn-high patients (Figure 3B). The data suggested that Kyn may impede HNSCC immunity by affecting T cell signaling. To further investigate the role of Kyn in T cells, the primary human CD8+ T cells and Jurkat T cell line were used for follow-up experiments. EdU and CFSE staining showed that Kyn suppressed T cell proliferation (Figure 3C, Supplementary Figure S4). PD-1 is well-known as a T cell-exhaustion marker [38]. Therefore, PD-1 in T cells was detected after stimulation with Kyn, and PD-1 was upregulated significantly in CD8+ T and Jurkat T cells (Figure 3D). Western blotting analysis also confirmed that Kyn induced PD-1 expression in primary CD8+ T and Jurkat T cells, with or without PHA stimulation (Figure 3E), indicating that Kyn could promote T cell dysfunction.
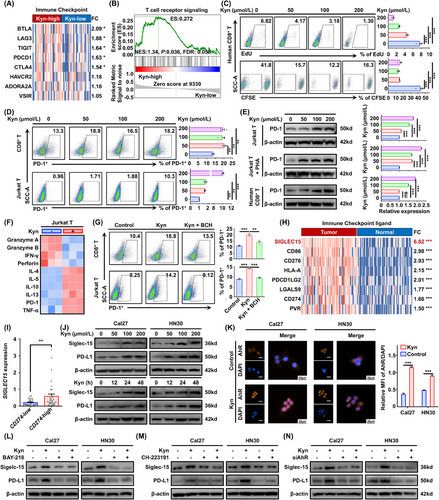
Kyn promoted CD8+ T cell dysfunction and Siglec-15 expression in tumor cells. (A) Heatmap analysis showing classical immune checkpoint gene expression in HNSCC, grouped by Kyn levels (n = 69). (B) Gene set enrichment analysis (GSEA) of mRNAs in the Kyn-high group. (C) The percentage of EdU+ (48 h, upper) and CFSE+ (96 h, lower) cells among human primary CD8+ T cells, isolated from the peripheral blood of healthy controls using a human CD8 MicroBeads kit and measured using flow cytometry, after the indicated Kyn stimulation. (D) The percentage of PD-1+ cells in primary CD8+ T (upper) and Jurkat cell line (lower), as measured using flow cytometry, after the indicated Kyn stimulation for 48 h. (E) Western blotting analysis of PD-1 expression after the indicated Kyn stimulation for 48 h in Jurkat cell line with or without PHA stimulation and in primary CD8+ T cells. Representative images (left) and three experiment replicates (right) are displayed. (F) Comparative heatmap depicted differential gene expression after Kyn stimulation (200 µmol/L, 48 h) using RT-qPCR data. (G) Kyn (100 µmol/L) and the system L inhibitor, BCH (5 mmol/L) were used as the indicated treatments to analyze the functions of Kyn on the dysfunction of primary CD8+ T cells and Jurkat cells (48 h). (H) Heatmap analysis shows classical common immune checkpoint ligand gene expression in HNSCC and adjacent normal tissues from 69 patients. (I) Analysis of SIGLEC15 expression in HNSCC stratified by high or low CD274 expression. (J) PD-L1 and Siglec-15 expression after treatment with the indicated Kyn concentrations for 48 h (upper) or 200 µmol/L Kyn for 0, 12, 24, and 48 h (lower) by Western blotting. (K) Nuclear expression of AhR was detected by immunofluorescence staining after 200 µmol/L Kyn stimulation for 1 h. (L) Cal27 and HN30 cell lines were treated with PBS or Kyn (200 µmol/L) and/or BAY-218 (10 µmol/L) for 48 h, and PD-L1 and Siglec-15 expression was detected by Western blotting. (M) Cal27 and HN30 cell lines were treated with PBS or Kyn (200 µmol/L) and/or CH-223191 (10 µmol/L) for 48 h, and PD-L1 and Siglec-15 expression was detected by Western blotting. (N) PD-L1 and Siglec-15 expression was detected using Western blotting after siAhR transfection for 48 h and 200 µmol/L Kyn treatment for 48 h. Data are represented as the mean ± SEM based on three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ns: not significant. Abbreviations: Kyn, kynurenine; HNSCC, head and neck squamous cell carcinoma, FC, fold change; GSEA, gene set enrichment analysis; ES, enrichment score; NES, normalized enrichment score; FDR, false discovery rate; EdU, 5-ethynyl-2′-deoxyuridine; CFSE, carboxyfluorescein succinimidyl ester; PD-1, programmed cell death protein 1; PHA, polyhydroxyalkanoate; IFN-γ, interferon-gamma; TNF-α, tumor necrosis factor-alpha; RT-qPCR, real-time quantitative polymerase chain reaction; BCH, 2-Aminobicyclo-(2,2,1)-heptane-2-carboxylic acid; FC, fold change; PD-L1, programmed death-ligand 1; DAPI, 4',6-diamidino-2-phenylindole; AhR, aryl hydrocarbon receptor; MFI, mean fluorescence intensity; PBS, phosphate buffered saline; SEM, tandard error of the mean; ns, not significant.
We further examined the mRNA expression of effector molecules in Jurkat T cells and found that the cytotoxic effector molecules were down-regulated while the inflammatory molecules were up-regulated after Kyn stimulation (Figure 3F), indicating that Kyn may reduce the secretion of cytotoxic effector molecules and killing function of T cells. To test whether the process depends on Kyn intake, Kyn transport was blocked using the system L transporter SLC7A5 inhibitor BCH [39]. BCH reversed Kyn-mediated T cell dysfunction in both human CD8+ T cells and Jurkat T cells (Figure 3G). The data confirmed that Kyn could inhibit the T cell vitality and promote the PD-1 expression.
To investigate whether Kyn affects HNSCC immunity through targeting tumor cells, the expression of immune checkpoint ligand in tumor and adjacent normal tissues was analyzed by RNA-seq. Strikingly, SIGLEC15, a freshly reported immune checkpoint ligand molecule [40], exhibited the maximum fold change of expression (Figure 3H). Subsequently, the levels of SIGLEC15 in the CD274-low (a classical immune checkpoint ligand, encoding PD-L1) and CD274-high groups were detected via RNA-seq data. SIGLEC15 had higher levels in the CD274-high group than in the CD274-low group (Figure 3I). Based on the above findings, SIGLEC15 and CD274 mRNA and protein were measured by RT-qPCR and Western blotting after Kyn treatment. The mRNA levels of SIGLEC15 and CD274 were elevated after Kyn stimulation (Supplementary Figure S5A). As expected, Western blotting analysis verified that Kyn increased the expression of Siglec-15 and PD-L1 (Figure 3J, Supplementary Figure S5B-C). In addition, based on immunofluorescence staining, Kyn promoted the nuclear translocation of AhR (Figure 3K), a well-known feature of AhR activation by Kyn [41]. Furthermore, two AhR inhibitors, BAY-218 and CH-223191, reversed Kyn-induced Siglec-15 and PD-L1 expression upregulation (Figure 3L-M, Supplementary Figure S5D-E), and similar results were obtained following knockdown of AhR via siRNA transfection (Figure 3N, Supplementary Figure S5F-H). Therefore, we demonstrated that Kyn could induce CD8+ T cell dysfunction and promote Siglec-15 and PD-L1 expression through AhR in HNSCC.
3.4 Kyn induced Siglec-15-mediated immune escape
To test whether Kyn-induced expression of Siglec-15 depends on AhR transcriptional activity, ChIP-PCR assays were performed using two specific primers. The AhR antibody was bound to the promoter region of SIGLEC15 or CD274 after Kyn stimulation in the Cal27 and HN30 cell lines (Figure 4A). Dual-luciferase assays showed that Kyn increased the promoter activity of SIGLEC15 and CD274 significantly (Figure 4B-C). Conversely, forced knockdown of AhR in 293T cells reduced the luciferase activity of SIGLEC15 or CD274 (Figure 4D), which was also confirmed in the Cal27 cell line (Supplementary Figure S6). The data suggested that Kyn can transcriptionally activate the expression of Siglec-15 and PD-L1 through AhR in HNSCC.
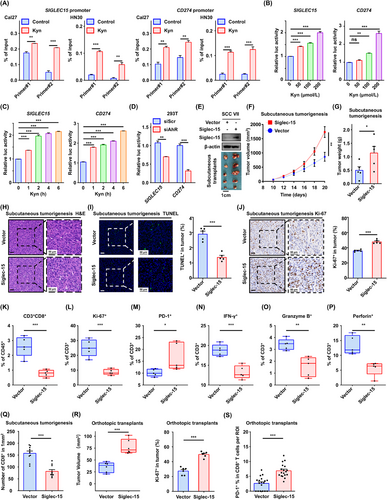
Kyn induced Siglec-15-mediated immune escape. (A) The chromatin immunoprecipitation (ChIP)-PCR assay was performed using an IgG or AhR antibody after treatment with 200 µmol/L Kyn for 1 h. Two primers targeting the promoter region of SIGLEC15 and CD274 mRNA were used for RT-qPCR analysis. (B) 293T cells were co-transfected with SIGLEC15/CD274 promoter-luciferase reporter PGL3 for 24 h and treated with the indicated concentration of Kyn for another 6 h, followed by an analysis of luciferase activity. (C) 293T cells were co-transfected with SIGLEC15/CD274 promoter-luciferase reporter PGL3 for 24 h and treated with 200 µmol/L Kyn for the indicated time, followed by an analysis of luciferase activity. (D) 293T cells were co-transfected with promoter-luciferase reporter plasmids and siAhR /siScr for 24 h and treated with 200 µmol/L Kyn for 6 h. (E) SCCVII cells were transfected with lentivirus-SIGLEC15, and transfection was confirmed using Western blotting analysis. (F) Vector or SIGLEC15-overexpressing SCCVII cells were subcutaneously injected into C3H/He mice (n = 5 per group). Tumor volumes were measured once every two days. (G) Tumor weights were measured after mice were euthanized. (H-J) H&E (H), TUNEL (I), and Ki-67 (J) staining analyses of tumor tissues in each group. (K-P) Tumor-infiltrating lymphocytes (TIL) harvested from xenograft tumors, and the percentages of CD3+CD8+ (K), Ki-67+ (L), PD-1+ (M), IFN-γ + (N), granzyme B+ (O), and perforin+ cells (P) were analyzed using flow cytometry. (Q) The percentages of CD8+ in the indicated tumors were analyzed by multiplex immunofluorescence staining. (R) The volume and Ki-67 expression of tumor were compared in tongue orthotopic transplant models established by vector or SIGLEC15-overexpressing SCCVII cells (n = 6 mice per group). (S) PD-1+CD8+ T cells were compared in tongue orthotopic transplant models (n = 6 mice per group), based on multiplex immunofluorescence staining. Three regions of interest (ROIs) in each tumor were analyzed and measured. Data are represented as mean ± SEM (A-D, F-G and I-S) based on three independent experiments (A-D). *P < 0.05, **P < 0.01, ***P < 0.001, ns: not significant. Abbreviations: Kyn, kynurenine; ChIP, chromatin immunoprecipitation; RT-qPCR, real-time quantitative polymerase chain reaction; AhR, aryl hydrocarbon receptor; siScr, siScramble; H&E, hematoxylin and eosin; TUNEL, TdT-mediated dUTP-biotin nick end labeling; PD-1, programmed cell death protein 1; TIL, Tumor-infiltrating lymphocyte; IFN-γ, interferon-gamma; ROI, region of interest; SEM, tandard error of the mean; ns, not significant.
To investigate the immunoregulatory functions of Siglec-15 in HNSCC, SIGLEC15-overexpressing subcutaneous tumorigenesis models were established in C3H/He mice (Figure 4E, Supplementary Figure S7A-B). Tumor growth curves and weight analysis showed that the tumors grew faster in the Siglec-15 group than in the vector group (Figure 4F-G). SCCVII tumors were confirmed using H&E staining (Figure 4H). TUNEL and Ki-67 staining analyses showed that Siglec-15 overexpression reduced apoptosis and promoted tumor cell proliferation (Figure 4I-J).
We further analyzed immune cell infiltration within the TME and observed that Siglec-15 reduced CD3+CD8+ T cell infiltration significantly (Figure 4K, Supplementary Figure S7C). The percentage of Ki-67+ T cells was reduced significantly in the Siglec-15 group (Figure 4L, Supplementary Figure S7D), indicating that Siglec-15 suppresses T cell proliferation. Regarding immune-exhaustion markers on T cells, there was a significant increase in the percentage of PD-1+ T cells in the xenograft TME (Figure 4M, Supplementary Figure S7E), whereas LAG-3+ showed no difference (Supplementary Figure S7F).
The effector molecules were analyzed, and the positive expression of IFN-γ, granzyme B, and perforin in tumor-infiltrating T cells was decreased notably in the Siglec-15 group (Figure 4N-P, Supplementary Figure S7G-I). Fluorescent multiplex immunohistochemistry showed that Siglec-15 decreased CD8+ T cell infiltration in xenograft tumors significantly (Figure 4Q, Supplementary Figure S7J). Furthermore, SIGLEC15-overexpressing orthotopic transplant models were established and showed that Siglec-15 promoted tumor progression and neck lymph node metastases (Figure 4R, Supplementary Figure S8A-B). In addition, the proportions of PD-1+CD8+ T cells in the Siglec-15 group were increased significantly based on fluorescent multiplex immunohistochemistry analysis results (Figure 4S, Supplementary Figure S8C). Furthermore, Kyn promoted PD-1+CD8+ T cell infiltration in orthotopic transplant models (Supplementary Figure S8C-D). These results suggested that Kyn promotes HNSCC progression through Siglec-15-mediated immune escape.
3.5 Targeting AhR reversed Kyn-Siglec-15-mediated immune escape in vivo
To confirm the role of Kyn and AhR in CD8+ T cell dysfunction, subcutaneous tumorigenesis C3H/He immunocompetent mouse models were established. Kyn promoted tumor growth, whereas the AhR inhibitor CH-223191 reversed the effect (Figure 5A), which was verified by tumor weight analysis (Figure 5B). Western blotting analysis also confirmed that CH-223191 could block Kyn-induced Siglec-15 expression in fresh tumor tissues (Figure 5C). SCCVII tumors were examined using H&E staining to confirm squamous cell carcinoma (Figure 5D), and TUNEL and Ki-67 staining analyses showed that Kyn reduced apoptosis and promoted tumor proliferation, whereas CH-223191 effectively reversed the effect (Figure 5E-F).
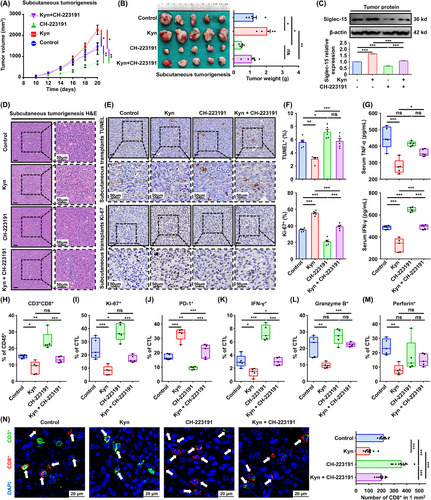
Targeting AhR reversed Kyn-Siglec-15-mediated immune escape in vivo. (A) Mice with SCCVII were treated with Kyn (100 mg/kg) or/and CH-223191 (10 µmol/L) once every two days for three times (n = 5 per group). Tumor volumes were measured once every two days. (B) Tumor weights were measured after mice euthanized on day 20 (n = 5 per group). (C) Western blotting analysis of Siglec-15 expression after Kyn stimulation in mice. (D-F) H&E (D), TUNEL (E and F upper), and Ki-67 (E and F lower) staining analyses of tumor tissues in each group. (G) Concentration of serum TNF-α and IFN-γ in mice were measured by ELISA. (H-M) The percentage of CD3+CD8+ (H), Ki-67+ (I), PD-1+ (J), IFN-γ+ (K), granzyme B+ (L), and perforin+ cells (M) in cytotoxic T lymphocytes (CTLs) isolated from the indicated tumors were analyzed. (N) Representative images of multiplex immunofluorescence staining of CD3+ (green) and CD8+ (red) are shown, and quantification analysis were performed in tumor tissues (n = 10 fields of 5 mice per group). Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ns: not significant. Abbreviations: AhR, aryl hydrocarbon receptor; Kyn, kynurenine; H&E, hematoxylin and eosin; TUNEL, TdT-mediated dUTP-biotin nick end labeling; TNF-α, tumor necrosis factor-alpha; IFN-γ, interferon-gamma; ELISA, enzyme-linked immunosorbent assay; PD-1, programmed cell death protein 1; CTL, cytotoxic T lymphocyte; DAPI, 4',6-diamidino-2-phenylindole; SEM, tandard error of the mean; ns, not significant.
Serum TNF-α and IFN-γ levels were decreased in the Kyn treatment group but were increased in the CH-223191 group (Figure 5G). Flow cytometry analysis showed that the proportion of CD3+CD8+ T cells in the TME was reduced following Kyn treatment, and this was reversed by treatment with CH-223191 (Figure 5H, Supplementary Figure S9A), similar findings were observed with regard to Ki-67+ T cell percentage (Figure 5I, Supplementary Figure S9B).
The PD-1+ T cells induced by Kyn were blocked partially by CH-223191 (Figure 5J, Supplementary Figure S9C), indicating that Kyn can exhaust tumor-infiltrating CD8+ T cells by activating AhR. Moreover, IFN-γ, granzyme B, and perforin analyses showed that Kyn reduced the secretion of cytotoxic effector molecules in CD8+ T cells, whereas CH-223191 reversed IFN-γ and granzyme B expression, but not perforin (Figure 5K-M, Supplementary Figure S9D-F). Fluorescent multiplex immunohistochemistry observed that Kyn reduced CD8+ T cell infiltration, whereas CH-223191 reversed the effects (Figure 5N).
Lastly, in vivo drug toxicity was analyzed, and no significant alterations in the levels of alanine transaminase, aspartate aminotransferase, blood urine nitrogen, and creatinine were observed, as well as no pathohistological changes in the main organs (Supplementary Figure S9G-H). Moreover, we investigated the effect of a combination of epacadostat and CH-223191, and the CI value was < 0.5 (Cal27: 0.010; HN30: 0.400), which suggested a strong synergistic effect between epacadostat and CH-223191 (Supplementary Figure S10).
The results indicate that Kyn can promote HNSCC progression by inducing CD8+ T cell dysfunction, and the AhR inhibitor CH-223191 can reverse this effect.
3.6 Siglec-15 specific siRNA delivered by NH2-MSN nanoparticles enhanced immunotherapy efficacy in vivo
Owing to a lack of Siglec-15 inhibitor, siRNA delivery through nanoparticles for mRNA interference is a potential solution for enhanced immunotherapy efficacy. Therefore, a nanoparticle delivery system (herein, NH2-MSNs) was established to encapsulate siS15 for targeted delivery (Figure 6A). NH2-MSNs loaded with siS15 were near transparent liquid (Figure 6B), and the morphology and zeta potential of a NH2-MSNs and NP-siS15 were determined. The average size of NH2-MSNs was 81 nm, with an electric potential of 31 mV. After siS15 loading, the diameters of the nanoparticles increased, and their edges became blurred, with a rapid decrease in zeta potential (Figure 6C-E). These findings indicate that siS15 was adsorbed on the surface of NH2-MSNs.
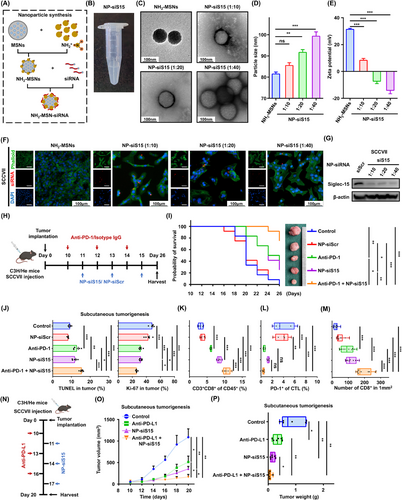
Siglec-15 specific siRNA delivered by NH2-MSN nanoparticles enhances immunotherapy efficacy in vivo. (A) Synthesis routes of NH2-MSN nanoparticles loaded with siRNA. (B) Macroscopy characterization of NH2-MSNs with siS15. (C) Transmission electron microscopy images of NH2-MSNs and NP-siS15 with indicated ratios. (D-E) The morphology and Zeta potential of NH2-MSNs and NP-siS15. (F) Scanning microscopy analysis of the cellular uptake of NH2-MSNs with or without the indicated siS15-Cy5 by SCCVII cells after in vitro treatment for 24 h. (G) Western blotting analysis confirming Siglec-15 gene-silencing effect by siRNA released from nanoparticles in SCCVII cells. (H) Schematic diagram of the treatment strategy in SCCVII mouse model. (I) Probability of survival analysis for each group was performed. (J) Analysis of TUNEL and Ki-67 staining of tumor tissues in each group. (K-L) The percentage of CD3+CD8+ (K) and PD-1+cells in CTLs (L) of the indicated treatment groups. (M) Analysis of multiplex immunofluorescence staining of CD3+ (green) and CD8+ (red) were shown and quantification analysis were performed in tumor tissues (n = 10 fields of five mice per group). (N) Schematic diagram of the anti-PD-L1 and NP-siS15 combination treatment strategy in C3H/He subcutaneous tumorigenesis models. (O-P) Tumor volume and weights were measured and analyzed in the indicated groups. Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ns: not significant. Abbreviations: siS15, Siglec-15 small interfering RNA; DAPI, 4',6-diamidino-2-phenylindole; AhR, aryl hydrocarbon receptor; NP-siS15, NH2-MSN-siSIGLEC15; DAPI, 4',6-diamidino-2-phenylindole; siScr, siScramble; anti-PD-1, anti-programmed cell death protein 1 antibody; TUNEL, TdT-mediated dUTP-biotin nick end labeling; CTL, cytotoxic T lymphocyte; anti-PD-L1, anti-programmed death-ligand 1 antibody; SEM, tandard error of the mean; ns, not significant.
Immunofluorescence analysis showed that NP-siS15 labeled with Cy5 could be effectively internalized and released in the cytoplasm of SCCVII cells at different mixing ratios (Figure 6F). Western blotting analysis confirmed that NP-siS15 effectively degraded the Siglec-15 protein in SCCVII compared with NP-siScr (Figure 6G). The results confirmed the successful establishment of NP-siS15, which can achieve efficient siRNA delivery into cancer cells and degrade the immune checkpoint molecule Siglec-15.
To investigate the effect of NP-siS15 treatment in vivo, subcutaneous tumorigenesis C3H/He mouse models were established (Figure 6H). The anti-tumor effect between the NP-siS15 and anti-PD-1 groups was not significantly different. More importantly, the mice treated with both anti-PD-1 and NP-siS15 exhibited prolonged survival compared with those treated with a single agent (Figure 6I, Supplementary Figure S11A), indicating a superior antitumor effect of anti-PD-1 and NP-siS15. The combination treatment group had significantly increased tumor cell apoptosis and decreased proliferation capacity, as indicated by TUNEL and Ki-67 staining (Figure 6J, Supplementary Figure S11B-C). TIL analysis showed that the NP-siS15 group could more significantly promote CD8+ T cell infiltration than the anti-PD-1 group, and the number of CD8+ T cells were the highest in the combination group (Figure 6K, Supplementary Figure S11D). Interestingly, the percentage of PD-1+CD8+ T cells decreased notably in the combination treatment group (Figure 6L, Supplementary Figure S11E). Fluorescent multiplex immunohistochemistry confirmed that NP-siS15 or anti-PD-1 promoted CD8+ T-cell infiltration, while combined NP-siS15 and anti-PD-1 had a better effect on immunotherapy efficacy (Figure 6M, Supplementary Figure S11F). Additionally, we applied the combination therapy strategy and observed that NP-siS15 enhanced the efficiency of anti-PD-L1 therapy (Figure 6N-P, Supplementary Figure S11G).
Overall, our results indicated that the newly-established NP-siS15 can effectively restore the killing function of CD8+ T cells and enhance anti-PD-1/PD-L1 therapy efficacy.
3.7 Siglec-15 expression positively correlated with CD8+ T cell exhaustion in HNSCC patients
An HNSCC tissue microarray was performed to determine the clinical correlation between AhR and Siglec-15 in CD8+ T cells. Siglec-15 expression was positively correlated with AhR expression (Figure 7A-B). Furthermore, high levels of AhR and Siglec-15 expression were associated with advanced TNM stage, while only AhR expression was associated with tumor location, which may be due to the large variation in the location of TMA samples (Supplementary Table S2). In particular, the percentage of Siglec-15-positive cases was 44.3% in HNSCC tissues, which was significantly higher than that of PD-L1-positive cases, at 22.8% (Figure 7C). Higher SIGLEC15 levels indicated poorer prognosis in the GEPIA HNSCC dataset (Figure 7D). The results indicated that Siglec-15 may play a more important immunosuppressive role than PD-L1 in HNSCC. More importantly, high Siglec-15 expression was closely associated with a high infiltration of exhausted CD8+PD-1+ T cells (Figure 7E). The results suggested that tumor-derived Kyn promoted CD8+ T cell exhaustion via the AhR/Siglec-15 pathway in HNSCC (Figure 7F).
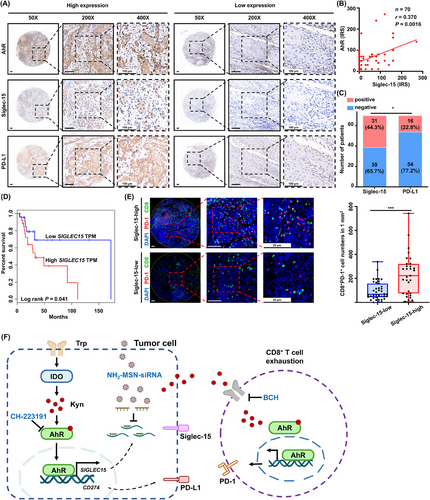
Siglec-15 expression positively correlates with CD8+ T cell exhaustion in HNSCC patients. (A) Representative images of immunohistochemical staining for AhR, Siglec-15, and PD-L1 on a TMA of 70 HNSCC patients. (B) The correlation between Siglec-15 expression and AhR expression using immunoreactive score (IRS) (n = 70). (C) Comparison of positive percentage of Siglec-15 and PD-L1 expression in HNSCC (n = 70). (D) GEPIA HNSCC dataset shows SIGLEC15 expression with the corresponding survival rates (n = 50) (E) Representative images show CD8+PD-1+ T cells using multiplex immunofluorescence staining in TMA (n = 70). (F) A schematic showing the mechanism via which Kyn/AhR/Siglec-15 signaling promotes CD8+ T cell exhaustion. Data are represented as mean ± SEM. *P < 0.05, ***P < 0.001. Abbreviations: HNSCC, head and neck squamous cell carcinoma; AhR, aryl hydrocarbon receptor; PD-L1, programmed death-ligand 1; TMA, tissue microarray; IRS, immunoreactive score; GEPIA, Gene Expression Profiling Interactive Analysis; TPM, transcripts per million; PD-1, programmed cell death protein 1; DAPI, 4',6-diamidino-2-phenylindole; Trp, tryptophan; IDO, indoleamine 2,3-dioxygenase 1; Kyn, kynurenine; BCH, 2-Aminobicyclo-(2,2,1)-heptane-2-carboxylic acid; SEM, tandard error of the mean.
4 DISCUSSION
Metabolic reprogramming plays a vital role in most cancers [42]. However, how it drives tumor metastasis and immune escape in HNSCC remains obscure. To address the knowledge gap, the present study revealed comprehensive and significant metabolic reprogramming in HNSCC and observed that the Trp-Kyn pathway is a key driving factor in tumor immune escape. Kyn, a metabolite of Trp, was identified as the key bridge that could promote Siglec-15 expression and ultimately lead to CD8+ T exhaustion. In addition, we developed NH2-MSN nanoparticles to deliver siS15 and observed that it could enhance the efficacy of immunotherapy significantly.
Currently, LC-MS and GC-MS are commonly used for untargeted metabolomics analysis. LC-MS has a high sensitivity and resolution but poor separation efficiency [43]. Meanwhile, GC-MS has a relatively robust database and good separation efficiency but limited detection range [44]. The metabolic landscape of several malignancies, such as lung cancer [45] and breast cancers [46], has been revealed by LC/GC-MS. However, that of HNSCC remains lacking. To minimize errors and omissions, metabolomic analysis was conducted using both LC-MS and GC-MS in the present study, which provided comprehensive information on the HNSCC metabolic landscape.
Song et al. [47] reported metabolic alterations in saliva and tumors which could be used to non-invasively diagnose HNSCC. In addition, Paul et al. [48] reported that all metabolites, excluding lipids, were upregulated in cancer and could predict the prognosis of HNSCC. However, the effect of HNSCC metabolic heterogeneity on tumor progression remains unclear. Therefore, in the present study, metabolomics and transcriptomics analysis was coducted, and multiple significantly differential metabolites, such as ascorbic acid, galacturonic acid, and Kyn, were revealed in HNSCC. A total of 56 differential metabolites were screened and classified as amino acids. In addition to the 20 standard amino acids, the others were called non-standard amino acids, which were either occured in living organisms but not in proteins, or were chemically modified after being incorporated into a protein [49]. Notably, amino acid metabolism was severely imbalanced in HNSCC patients, highlighting that amino acid metabolism plays a crucial role in tumor progression, and Kyn was identified as a key metabolite. In the present study, Kyn was observed to promote tumor metastasis. We speculated that Kyn could promote the expression of the metastasis-related transcription factor Snail, which regulated the expression of genes involved in multiple metastasis-related processes. The migration capacity induced by 100 µmol/L Kyn was weaker than by 50 µmol/L Kyn (Figure 2D-E), which may be due to the system L transporters not being exclusive for Kyn transport [39]. Excessive concentrations of Kyn can lead to the inability of the transporter to transport other essential amino acids, thereby affecting tumor physiological functions.
Metabolic abnormalities, such as adenosine [50] and epinephrine [51], are important drivers of tumor immune escape [52]. The present study had identified an immune driver Kyn, which could promote tumor PD-L1 and T cell PD-1 expression by activating AhR. However, the anti-PD-1/PD-L1 response rate in HNSCC was only 15%-20% [20], indicating that multiple mechanisms of immune escape may exist. Siglec-15, a novel immune checkpoint molecule, was rarely expressed in normal tissues, but was mainly expressed in tumor cells and macrophages [40]. Siglec-15 can facilitate immune escape independently of PD-1, and anti-Siglec-15 could improve prognosis in mice with melanoma and colon cancer [40, 53]. Moreover, Siglec-15 was overexpressed in bladder cancer [54] and lung adenocarcinoma [55], which were associated with its prognosis. However, there is still no literature reporting the upstream regulatory mechanism of Siglec-15. In the present study, Siglec-15 was the most significantly altered immune checkpoint molecule in HNSCC patients, and it could be promoted by Kyn. Furthermore, targeting Siglec-15 in combination with anti-PD-1/PD-L1 achieved greater anti-tumor efficacy, suggesting that Kyn/Siglec-15 signaling may be another key signaling pathway in immune evasion in addition to PD-1. The findings suggest that Kyn/Siglec-15 may be a new target for cancer immunotherapy and that targeting Kyn/Siglec-15 may provide a new strategy for ameliorating the low response rate to immunotherapy. Moreover, the immunoregulatory function of Siglec-15 was investigated, and the mechanism of immune escape elucidated from an amino acid metabolism perspective.
Kyn had been previously reported to induce Foxp3+ Treg-mediated immunosuppression through activation of AhR [56]. Here, Kyn was observed to activate Siglec-15 expression through AhR, which in turn directly suppressed CD8+ T cell function to promote immune escape. Due to the unclear binding molecules of Siglec-15 on T cells [40], flow cytometry was employed to detect CD8+ T cell proliferation, exhaustion, and killing factors. The results confirmed that Siglec-15 overexpression inhibited CD8+ T proliferation significantly, leading to functional exhaustion. Several preclinical and clinical trials are currently exploring the inhibitors of the Kyn signaling pathway. A phase III study of epacadostat in combination with Keytruda did not meet the expected results [57], indicating that simply targeting Trp degradation makes it challenging to block its promotion in tumor progression. Thus, further investigations on Kyn pathways may improve therapeutic efficacy. Kenison et al. [31] found that AhR could promote tumor progression by enhancing PD-L1-mediated immunosuppression. However, the functional status data of CD8+ T cells were lacking. The present study revealed that the Kyn/AhR/Siglec-15 axis played a critical role in CD8+ T cell exhaustion in HNSCC, providing a target for immunotherapy. More interestingly, the positive rate of Siglec-15 was higher than that of PD-L1 (44.3% vs 22.8%) in HNSCC patients, suggesting that it might be a more promising target for future immunotherapy. The clinical application of Siglec-15 blockade may improve the low response rate of immunotherapy. Furthermore, innovative nanoparticle carriers were designed to deliver siS15, and targeting Siglec-15 cound enhance anti-PD-1/PD-L1 therapeutic efficacy significantly, thus providing a new therapeutic strategy for subsequent clinical treatment.
However, the present study had some limitations. First, metabolonomics studies are based on bulk analysis, which cannot identify the source of the differential metabolites. Single-cell analysis techniques could provide additional information in the future. Second, Siglec-15 is expressed not only on tumor cells but also on macrophages, and the function of Siglec-15 on macrophages was not investigated owing to the COVID-19 pandemic and limited time and resources. Lastly, the ligand of Siglec-15 on immune cells and therapeutic antibodies remains unknown, suggesting that further studies are required before its clinical application.
5 CONCLUSIONS
In summary, the present study clearly delineated the metabolic landscape of HNSCC and reports that Kyn induced CD8+ T dysfunction significantly through AhR, which transcriptionally activated Siglec-15 expression. Targeting AhR or Siglec-15 could reverse CD8+ T dysfunction in the TME, thereby improving the efficacy of immunotherapy. The present study reveals a novel mechanism of immune escape from a metabolic pathway perspective and provides novel therapeutic targets for tumor immunotherapy.
DECLARATIONS
AUTHOR CONTRIBUTIONS
Hai-Long Ma, Xi Yang, and Zhi-Yuan Zhang conceived of the study. Xin-Yu Zhang, Jian-Bo Shi, and Shu-Fang Jin performed animal and in vitro experiments. Ming-Yu Li, Rui-Jie Wang, and Xi Yang provided and analyzed patient clinical samples. Xin-Yu Zhang, and Hai-Long Ma analyzed histological data. Hai-Long Ma, Xi Yang, Zhi-Yuan Zhang, Shu-Fang Jin, and Jian-Bo Shi provided funding. Xin-Yu Zhang and Jian-Bo Shi wrote the paper with assistance from Hai-Long Ma. Shu-Fang Jin, Zhi-Yuan Zhang, Xi Yang, and Hai-Long Ma supervised the study. This study was supported by the National Natural Science Foundation of China (82303280 to Xinyu Zhang, 82072980 and 82272831 to Xi Yang, 82272983 to Hai-Long Ma, 82172897 to Shu-Fang Jin, 82203614 to Jian-Bo Shi), Science and Technology Daystar Program of Shanghai (22QA1405300 to Hai-Long Ma), Natural Science Foundation of Shanghai (22ZR1436800 to Shu-Fang Jin, 20ZR1447300 to Xi Yang), Young Talent Lift Project by the China Association for Science and Technology (2020QNRC001 to Hai-Long Ma), the Shanghai Sailing Program (22YF1421600 to Jian-Bo Shi), Young physicians collaborative innovation team of Shanghai Ninth People's Hospital (QC202004 to Hai-Long Ma), The Innovative Research Team of High-level Local Universities in Shanghai (SHSMU-ZDCX20212500 to Shu-Fang Jin and SHSMU-ZLCX20212300 to Zhi-Yuan Zhang)
ACKNOWLEDGMENTS
The authors thank Prof. Zhen Tian (Department of Oral Pathology, Shanghai Jiao Tong University School of Medicine, Shanghai, China) for providing histological examination and analysis.
CONFLICTS OF INTERESTS STATEMENT
The authors declare no potential conflicts of interest.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
All animal experiments were approved by the Animal Care and Use Committee of Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine (approval ID: SH9H-2022-A031-SB, SH9H-2023-A408-SB). All patients had signed informed consent prior to surgery. The use of human specimens in this study was approved by the Institutional Research Ethics Committee of Shanghai Ninth People's Hospital (approval ID: SH9H-2021-T135, SH9H-2023-TK245-1).
Open Research
DATA AVAILABILITY STATEMENTS
The RNA-seq data have been deposited in the NCBI Gene Expression Omnibus under accession number GEO: GSE222673.
No new code was used in the preparation of this manuscript.




