AP4 induces JNK1 and a miR-22-3p/FOSL1 feed-forward loop to activate AP-1 and promote colorectal cancer metastasis
List of abbreviations
-
- 3’-UTR
-
- 3’-untranslated region
-
- AP-1
-
- activator protein-1
-
- Cas9
-
- clustered regularly interspaced short palindromic repeats-associated protein 9
-
- CDH1
-
- E-cadherin 1
-
- MYC
-
- c-MYC proto-oncogene
-
- CRC
-
- colorectal cancer
-
- CRISPR
-
- clustered regularly interspaced short palindromic repeats
-
- DOX
-
- doxycycline
-
- EMT
-
- epithelial-mesenchymal transition
-
- FOSL1/FRA1
-
- FOS-like 1/FOS-related antigen 1
-
- JNK1
-
- c-Jun N-terminal protein kinase 1
-
- JUN
-
- c-Jun proto-oncogene
-
- JUNB
-
- c-JunB proto-oncogene
-
- KO
-
- knock-out
-
- MAP2K7
-
- mitogen-activated protein kinase kinase 7
-
- MAP3K13
-
- mitogen-activated protein kinase kinase kinase 13
-
- MAPK
-
- mitogen-activated protein kinase
-
- mCRC
-
- metastatic colorectal cancer
-
- MDC1
-
- mediator of DNA damage checkpoint 1
-
- MIR22HG
-
- MIR22 host gene
-
- MUT
-
- mutant
-
- NOD/SCID
-
- non-obese diabetic/severe combined immunodeficiency
-
- qChIP
-
- quantitative real-time polymerase chain reaction-chromatin immunoprecipitation
-
- qPCR
-
- quantitative real-time polymerase chain reaction
-
- SMS
-
- seed-matching sequence
-
- t1/2
-
- half-life time
-
- TCGA-COAD
-
- The Cancer Genome Atlas Colorectal Adenocarcinoma
-
- TF
-
- transcription factor
-
- TFAP4/AP4
-
- transcription factor AP4/activating enhancer binding protein 4
-
- VIM
-
- vimentin
-
- WT
-
- wild-type
-
- ΔSMS
-
- seed-matching sequence deletion
Dear Editor,
Colorectal cancer (CRC) is the third most deadly cancer worldwide [1]. The mortality of CRC has remained high due to limited treatment options for metastatic CRC (mCRC) [2]. Epithelial-mesenchymal transition (EMT) is an important contributor to mCRC [2]. The c-MYC proto-oncogene (MYC)-induced transcription factor AP4 (TFAP4/AP4) is a driver of EMT, thereby presumably facilitates mCRC [3, 4]. The mitogen-activated protein kinase (MAPK)/c-Jun N-terminal kinase (JNK)/activator protein-1 (AP-1) pathway has been implicated in the regulation of EMT and mCRC [5].
Here, we analyzed whether AP4 regulates components of the MAPK/JNK/AP-1 pathway after MYC activation using CRC cells rendered AP4-deficient by a clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) approach. The detailed methods are shown in the Supplementary file. First, we grouped MYC-induced changes in mRNA expression observed in the CRC cell line DLD-1 AP4 wild-type 1/pRTR-c-MYC-VSV (AP4-WT1 DLD-1/pRTR-c-MYC-VSV) into 6 non-overlapping expression clusters (Supplementary Figure S1A, left), with cluster 1 representing mRNAs down-regulated, and clusters 2-6 representing different patterns of mRNA up-regulated after MYC activation. MAPK signaling pathway components were strongly over-represented in cluster 6 (Supplementary Figure S1A, right). The AP4 targets MIR22 host gene (MIR22HG) and E-cadherin 1 (CDH1) were down-regulated after MYC activation in AP4-WT1 DLD-1/pRTR-c-MYC-VSV cells (Supplementary Figure S1B). MAPK signaling effectors, including c-Fos proto-oncogene (FOS), c-Jun proto-oncogene (JUN) and c-JunB proto-oncogene (JUNB), were over-represented in cluster 6. Additional MAPK signaling pathway components. such as c-Jun N-terminal kinase 1 (JNK1), mitogen-activated protein kinase kinase kinase 1 (MAP3K1), mitogen-activated protein kinase kinase kinase 13 (MAP3K13), mitogen-activated protein kinase kinase 3 (MAP2K3), mitogen-activated protein kinase kinase 7 (MAP2K7) and FOS-like 1 (FOSL1) were found in clusters 3-5 (Supplementary Figure S1B). Interestingly, MAP3K13, MAP2K7, JNK1 and FOSL1 were induced by MYC in an AP4-dependent manner (Supplementary Figure S1C-D).
Notably, MAP3K13, FOSL1, JNK1 and MAP2K7 were also up-regulated after activating AP4 for 48 or 72 hours (Figure 1A) and showed AP4-binding sites (CAGCTG) and AP4 occupancy (Supplementary Figure S2A). Therefore, these genes presumably represent direct AP4 targets. MAP3K13 and FOSL1 protein and phosphorylated JNK1 were up-regulated after AP4 activation, whereas JNK1 protein levels remained unchanged (Figure 1B). Furthermore, AP4 occupancy at the MAP3K13 and FOSL1 promoters was confirmed by quantitative real-time polymerase chain reaction-chromatin immunoprecipitation (qChIP) analysis (Figure 1C). The nascent mRNA of MAP3K13 and FOSL1 was decreased in AP4-deficient DLD-1 and SW480 cells (Supplementary Figure S2B). Ectopic AP4 induced nascent MAP3K13 and FOSL1 mRNA in CRC cells (Supplementary Figure S2C). Therefore, AP4 activates the JNK1 pathway through a MAP3K13/MAP2K7/JNK1 cascade. CRCs with liver metastasis are significantly more often FOSL1-positive [6], and MAP3K13 is up-regulated in colon cancer tissues [7]. Thus, AP4-induced FOSL1 and MAP3K13 are presumably clinically relevant in CRC. FOSL1 showed a trend towards a positive correlation with AP4 in 471 primary CRC samples of The Cancer Genome Atlas Colon Adenocarcinoma (TCGA-COAD) cohort and in an independent patient cohort (GSE13294) (Supplementary Figure S2D). The dimerization of FOSL1 and c-Jun proto-oncogene (JUN) results in the formation of AP-1 [8]. Consistently, AP-1 activity was induced by ectopic AP4 (Figure 1D, Supplementary Figure S2E). In addition, AP-1 activity was significantly lower in AP4-deficient DLD-1 cells (Supplementary Figure S2F). Taken together, these results show that AP4 activates JNK1 and AP-1 by directly inducing MAP3K13 and FOSL1.
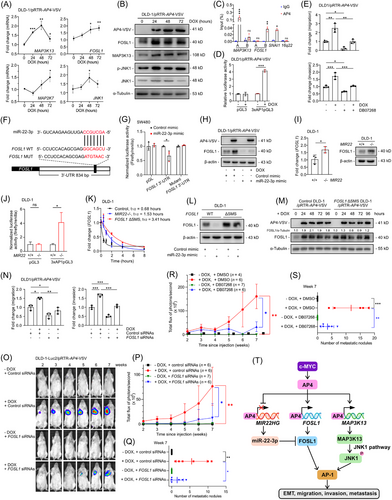
Next, we analyzed the relevance of JNK1 for AP4-induced EMT. The JNK1 inhibitor DB07268 partially abrogated the induction of vimentin (VIM) and the repression of CDH1 by AP4 in CRC cell lines (Supplementary Figure S3A), indicating that AP4-mediated JNK1 activation contributes to the induction of VIM and the CDH1 repression by AP4 [3]. In addition, DB07268 significantly suppressed AP4-induced migration and invasion (Figure 1E, Supplementary Figure S3B-C). Furthermore, DB07268 largely suppressed the AP4-induced proliferation of DLD-1 cells (Supplementary Figure S3D). Therefore, JNK1 activation mediates the induction of EMT, migration, invasion and proliferation by AP4.
We have previously shown that AP4 represses the tumor-suppressive miRNA miR-22-3p, which contributes to the up-regulation of mediator of DNA damage checkpoint 1 (MDC1), a miR-22-3p target, by AP4 [9]. Since we identified a miR-22-3p seed-match sequence (SMS) in the FOSL1 3’-untranslated region (3’-UTR), we investigated whether FOSL1 is also regulated by AP4 via repression of miR-22-3p (Figure 1F). Indeed, a FOSL1 3’-UTR reporter was repressed by ectopic miR-22-3p, and mutation of the SMS rendered the reporter refractory to miR-22-3p (Figure 1G). Ectopic miR-22-3p repressed FOSL1 (Supplementary Figure S4A) and effectively decreased basal and AP4-induced FOSL1 protein levels (Figure 1H, Supplementary Figure S4B). Next, we generated MIR22-/- (MIR22-knockout [KO]) and FOSL1 ΔSMS CRC cell lines (Supplementary Figure S4C). FOSL1 mRNA and protein expression were elevated in MIR22-deficient CRC cells (Figure 1I, Supplementary Figure S4D). MIR22-deficiency increased the activity of the FOSL1 3’-UTR reporter in a miR-22-3p SMS-dependent manner (Supplementary Figure S4E) and enhanced migration and invasion (Supplementary Figure S4F). Furthermore, the intrinsic AP-1 activity was significantly higher in MIR22-deficient CRC cells (Figure 1J, Supplementary Figure S4G). The half-life (t1/2) of FOSL1 mRNA was prolonged in MIR22-deficient and FOSL1 ΔSMS DLD-1 cells (Figure 1K). Ectopic expression of miR-22-3p failed to repress FOSL1 in FOSL1 ΔSMS CRC cells (Figure 1L, Supplementary Figure S4H), which showed increased basal FOSL1 expression, and attenuated the up-regulation of FOSL1 by AP4 (Figure 1M). Therefore, the direct targeting of FOSL1 by miR-22-3p is essential for maintaining low levels of FOSL1 expression, which allows FOSL1 to be induced by AP4 directly and indirectly via repression of MIR22.
Next, we determined whether FOSL1 mediates functional effects of AP4 on CRC cells. Ectopic AP4 repressed CDH1 and induced VIM in CRC cells, and silencing FOSL1 partially compromised these effects (Supplementary Figure S5A). The positive effect of ectopic AP4 on migration and invasion was largely abrogated by FOSL1 silencing (Figure 1N, Supplementary Figure S5B-C), indicating that FOSL1 mediates these effects of AP4. Silencing FOSL1 significantly repressed AP-1 activity and compromised the effects of AP4 on AP-1 activity (Supplementary Figure S5D). In addition, ectopic AP4 induced the proliferation of DLD-1 cells, which was repressed by concomitant siRNA-mediated silencing of FOSL1 (Supplementary Figure S5E). Taken together, these results demonstrate that the FOSL1-mediated activation of AP-1 by AP4 is required for the effects of AP4 on EMT, migration, invasion and proliferation.
miR-22-3p was shown to be significantly down-regulated in primary CRCs, which displayed lymph node metastasis, and ectopic miR-22-3p repressed lung metastasis formation in a xenograft model [10]. Therefore, deregulation of the miR-22-3p/FOSL1 axis by AP4 may be clinically relevant for mCRC. Notably, AP4 activation in non-metastatic DLD-1 cells allowed these to form lung metastases in non-obese diabetic/severe combined immunodeficiency (NOD/SCID) mice as determined by non-invasive bioluminescence imaging (Figure 1O-P) and histological analysis (Supplementary Figure S6A, right). Therefore, AP4 activation is sufficient to convert CRC cells from a non-metastatic to a metastatic state. Silencing FOSL1 efficiently prevented the AP4-induced formation of lung metastases (Figure 1Q, Supplementary Figure S6A left). Therefore, FOSL1 mediates and is required for the formation of AP4-induced metastases. Similar results were obtained after the inhibition of JNK1 using DB07268 (Figure 1R-S, Supplementary Figure S6B-C).
In conclusion, AP4 activates JNK1 and induces FOSL1 directly, as well as indirectly, via repression of miR-22-3p. AP4, miR-22-3p and FOSL1 form a coherent, regulatory feed-forward loop. These regulations converge in the activation of AP-1 (Figure 1T), which ultimately promotes EMT, migration and invasion, thereby enhancing metastasis formation. In the future, these findings may be exploited for the prevention and/or treatment of mCRC.
DECLARATIONS AUTHOR CONTRIBUTIONS
Heiko Hermeking conceived, planned and supervised the project; Heiko Hermeking, Jinjiang Chou and Markus Kaller designed experiments; Jinjiang Chou performed experiments and analyzed results; Markus Kaller performed bioinformatics analysis; Matjaz Rokavec performed and analyzed mouse experiments. Fangteng Liu histochemically evaluated lung metastases in mice. Heiko Hermeking, Jinjiang Chou and Markus Kaller wrote the manuscript. All authors read and approved the final manuscript.
ACKNOWLEDGMENTS
We are grateful to Raffaele Conca (Dr. von Haunersches Children's Hospital, Munich) for fluorescence-activated cell sorting and to Chunfeng Liu and Ursula Götz for technical assistance.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no competing interests.
FUNDING INFORMATION
This work was supported by German Cancer Aid/Deutsche Krebshilfe grants (70114235 and 70112245) to Heiko Hermeking.
ETHICS APPROVAL
Animal experimentations and analyses were approved by the Government of Upper Bavaria, Germany (55.2-2532.vet_02-18-57).
CONSENT FOR PUBLICATION
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
All data generated or analyzed during this study are included in this article and its supplementary information files.




