Enhancing sensitivity of triple-negative breast cancer to DNA-damaging therapy through chemical inhibition of the m6A methyltransferase METTL3
Abbreviations
-
- AML
-
- Acute Myeloid Leukemia
-
- BCO
-
- breast cancer organoid
-
- BRCA1
-
- Breast cancer gene 1
-
- BRCA2
-
- Breast cancer gene 2
-
- CI
-
- combination index
-
- GFP
-
- green fluorescent protein
-
- GO
-
- gene ontology
-
- H2AX
-
- H2A Histone Family Member X
-
- IC50
-
- half maximal inhibitory concentration
-
- m6A
-
- N6-methyladenosine
-
- MeRIP-seq
-
- Methylated RNA Immunoprecipitation Sequencing
-
- METTL3
-
- Methyltransferase-like 3
-
- PARP1
-
- Poly (ADP-ribose) polymerase 1
-
- PARP2
-
- Poly (ADP-ribose) polymerase 2
-
- PVS
-
- perivitelline space
-
- qPCR
-
- quantitative real-time polymerase chain reaction
-
- RAD51
-
- RecA-related protein 51
-
- STM
-
- STM2457
-
- TNBC
-
- Triple negative breast cancer
Dear Editor,
N6-methyladenosine (m6A) is a critical mRNA modification catalyzed by the enzyme methyltransferase-like 3 (METTL3), with implications in RNA metabolism. METTL3 upregulation is associated with cancer progression, metastasis, and drug resistance, making it a potential therapeutic target [1]. The small-molecule METTL3 inhibitor, STM2457, has shown promise in treating acute myeloid leukemia (AML) and has demonstrated good tolerance in mice [2, 3]. However, the specific cancer types where METTL3 inhibitors are most effective remain unknown.
In breast cancer, METTL3 knockdown markedly suppresses proliferation, invasiveness, and metastasis [4]. Therefore, METTL3 inhibition is proposed as a therapeutic approach for breast cancer. Triple-negative breast cancer (TNBC), the most aggressive subtype, lacks targeted therapies, and its primary treatments involve conventional chemotherapy and DNA-damaging agents [5]. Homologous recombination deficiency, such as mutations in the breast cancer gene 1 (BRCA1) and BRCA2, serves as a predictive biomarker for identifying patients who would benefit from genotoxic chemotherapy and poly(ADP-ribose) polymerase (PARP) inhibitors. Notably, METTL3 is recruited to DNA-damaged sites and is crucial for subsequent DNA repair [6, 7]. Consequently, METTL3 knockdown reduces DNA repair activity and sensitizes cancer cells to genotoxic drugs [7, 8]. However, while TNBC exhibits elevated METTL3 levels, and its nuclear catalytic activity associates with invasiveness and metastasis [9], it remains uncertain whether METTL3 inhibition enhances chemotherapy response in TNBC.
Here, we aimed to explore the potential of METTL3 catalytic inhibition by STM2457 as a valuable treatment option for TNBC. Furthermore, we assessed the impact of STM2457 on the sensitivity of TNBC cells and a TNBC patient-derived organoid line to clinical DNA-damaging therapies, like platinum-based chemotherapy and the PARP inhibitor olaparib (Supplementary file of methods).
STM2457 significantly reduced the proliferation and viability of TNBC cells, including both BRCA1/2 wild-type (MDA-MB-231 and MDA-MB-468) and BRCA1-mutated (MDA-MB-436, HCC1395, and HCC1937) cell lines. STM2457 exhibited negligible effects on the proliferation of non-tumoral breast epithelial cells (MCF-10A), with significant reduction observed only at the highest concentration tested (100 μmol/L) (Figure 1A, Supplementary Figure S1A-B). The treatment with 10 μmol/L STM2457 for 48 h decreased the global m6A levels in mRNA by approximately 50% in both MDA-MB-231 and MCF-10A cells (Supplementary Figure S1C). Colony formation assays further confirmed the anti-proliferative impact of STM2457 on TNBC cell lines (Figure 1B, Supplementary Figure S2). Moreover, wound healing assays indicated that the inhibition of METTL3 activity reduced TNBC cell migration (Figure 1C). The specificity of the effects exerted by STM2457 were confirmed by METTL3 knockdown (Supplementary Figure S3). To identify the molecular pathways regulated by METTL3 inhibition, we investigated its effect on MDA-MB-231 cells, one of the most aggressive TNBC cell line. Cells were treated with 10 μmol/L STM2457 for 48 h and subjected to RNA-sequencing. Bioinformatics analysis revealed 3,182 genes downregulated upon STM2457 treatment, with 3,237 genes upregulated by the drug (Supplementary Figure S4A, Supplementary Table S1). Gene ontology (GO) analysis of differentially expressed genes showed a significant downregulation of biological processes involved in cell proliferation, translation, and DNA repair (Supplementary Figure S4B). This result is consistent with previous analyses performed in breast cancer cells knocked down for METTL3 expression [8, 9] and in AML cell lines treated with STM2457 [2]. Comparison of our RNA-sequencing dataset with a methylated RNA immunoprecipitation (MeRIP)-sequencing dataset performed in MDA-MB-231 cells [9] showed a substantial overlap between the transcripts regulated by STM2457 treatment and those that are m6A methylated (Supplementary Figure S5A). GO analysis of the m6A-methylated genes, which exhibit differential expression upon METLL3 inhibition, revealed a significant downregulation of biological processes related to the cell cycle and DNA damage repair (Supplementary Figure S5B). Consistent with global m6A level observations, individual MeRIP-qPCR analysis on highly methylated transcripts confirmed around a 50% reduction due to STM2457 treatment (Supplementary Figure S6). We focused on the DNA damage repair pathway because of its relevance for TNBC therapy [8].
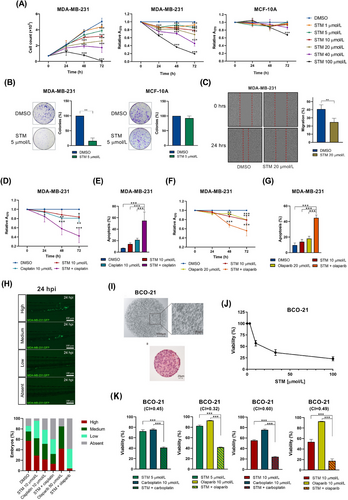
First, we investigated the effect of METTL3 inhibition in combination with DNA-damaging agents. Platinum-based chemotherapeutic agents and PARP inhibitors, such as olaparib, showed mostly favourable responses in TNBC harboring BRCA1/2 mutations [5]. Since STM2457 downregulated genes involved in the DNA repair pathway, we hypothesized that it may reduce homologous recombination (HR) proficiency also in TNBC cells that are wild-type for BRCA1/2. To test this hypothesis, we examined the effect of METTL3 inhibition on the response to cisplatin and olaparib in MDA-MB-231 cells, which are BRCA1/2 wild-type and characterized by lower sensitivity to DNA-damaging agents. MDA-MB-231 cells were treated with doses of STM2457, cisplatin, and olaparib that were below their half maximal inhibitory concentration (IC50) either as single agents or in combination. Combined treatment with STM2457 and either cisplatin or olaparib significantly reduced MDA-MB-231 cell proliferation and increased apoptosis compared to single-drug treatments (Figure 1D-G). Notably, treatment with STM2457 alone was sufficient to induce DNA damage, as indicated by the elevated phosphorylation of the H2A histone family member X (γH2AX) (Supplementary Figure S7A-B). Additionally, we noticed a global reduction in RecA-related protein 51 (RAD51) protein levels and RAD51 foci upon DNA damage (Supplementary Figure S7C-D). These findings are in line with those obtained in cells knocked down for METTL3 expression [7, 8]. Moreover, we observed substantial and consistent increase in DNA damage in response to combined treatments (Supplementary Figure S7A-B). Together, these data indicate that STM2457 strongly sensitizes HR-proficient TNBC cells with wild-type BRCA1/2 to DNA damage induced by genotoxic chemotherapy or PARP inhibitors.
As STM2457 treatment reduced TNBC cell migration, we further explored its impact on cancer cell metastasis. We chose the metastatic TNBC cell line MDA-MB-231 for injection into zebrafish larvae, a well-established in vivo xenograft model for studying metastasis [10]. MDA-MB-231 cells expressing green fluorescent protein (GFP) were microinjected into the perivitelline space of zebrafish embryos at 48 h post-fertilization. Embryos were then exposed to compounds, either independently or in combinations, as employed in the in vitro assays. We observed no toxicity during the treatment with STM2457 in the larvae. While some extravasated cells were scattered throughout the larva, GFP-positive clusters were enriched in the caudal hematopoietic tissue region. We quantified micrometastases in this area (Figure 1H). Intriguingly, while each drug alone reduced cell metastasis, the combined STM2457 + cisplatin and STM2457 + olaparib treatments exhibited a notable mutually reinforcing trend, leading to a substantial reduction in both the number and size of micrometastases (Figure 1H).
Finally, we assessed the effect of STM2457 in a patient-derived organoid (BCO-21) from a TNBC patient with wild-type BRCA1/2 genes (Figure 1I). This patient was ineligible for olaparib treatment. Initially, we determined the IC50 for single-drug treatments with STM2457, olaparib, and carboplatin, a platinum-based chemotherapy used in TNBC. In line with the results from MDA-MB-231 cells, STM2457 decreased BCO-21 cell viability (Figure 1J, Supplementary Figure S8). We then investigated whether STM2457 enhances the BCO-21 sensitivity to DNA-damaging therapy. Notably, combination treatment with both optimal (10 μmol/L) and suboptimal (5 μmol/L) doses of STM2457 synergistically increased the cytotoxic effect of carboplatin and olaparib (combination index < 1; Figure 1K, Supplementary Figure S8).
In conclusion, this study showed that STM2457 displayed anti-tumor activity in TNBC and enhanced chemotherapy response while sensitizing BRCA1/2 wild-type patients to olaparib. Given the observed upregulation of METTL3 activity across various cancer types, METTL3 inhibitors may offer a novel approach to improve the efficacy of DNA-damaging agents, overcome inherent resistance, and mitigate genotoxic drug adverse effects.
DECLARATIONS
AUTHORS' CONTRIBUTIONS
BC, AI, FP, MT, SM, GF conducted investigations in triple negative breast cancer cell lines; BC, FP, AI performed statistical analyses and generated figures; DR, AM synthesized chemical compounds; AD, ML, LDG conducted zebrafish xenograft experiments; EC, RM, ADL, CN, CS produced and performed experiments in breast cancer organoids; CS assisted with manuscript review and editing; FF, CS, LDG provided funding and supervision; AF contributed to conceptualization, funding acquisition, supervision, and original draft writing. All authors approved the final revised manuscript.
ACKNOWLEDGEMENTS
We thank Marcella Marchioni (Institute of Biology, Molecular Medicine and Nanobiotechnology, Italian National Research Council) for technical assistance, Dr. Davide Mariani (Center for Human Technologies, Italian Institute of Technology) for the ePB-PGK-BSD plasmid, Prof. Giuseppe Giannini (Department of Molecular Medicine, Sapienza University of Rome) for the HCC1937 cell line, and Dr Pietro Pichierri (Mechanisms, Biomarkers and Models Unit, Department of Environment and Health, Istituto Superiore di Sanità) for the anti-RAD51 antibody.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no competing interests.
FUNDING INFORMATION
This work was supported by NextGenerationEU-PNRR M4C2-Investment 1.4- CN00000041 to A.F., F.F., and L.D.G., “Progetti Ateneo Sapienza” RP1201729D714976 to A.F. Work in the laboratory of C.S. was supported by Breast Cancer now (project 2018NovPCC1283) and AIRC (project IG23416).
CONSENT FOR PUBLICATION
Not applicable.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Zebrafish larvae were obtained through natural spawning of wild type adult fish of the AB strain. Our facility strictly complies with the relevant European (EU Directive 2010/63/EU for animal experiments) and Italian (Legislative Decree No. 26/2014) laws, rules, and regulations (Auth. Min. 24/2019-UT), as also confirmed by the authorization issued by the municipality of Milan (PG 198283/2019). The procedures were carried out in accordance with the relevant guidelines and regulations.
Patient-derived organoids were derived from tumor biopsies from patients treated at “Fondazione Policlinico Universitario A. Gemelli IRCCS” (FPG), Rome, Italy. The protocol was approved by the Institutional Review Board (Protocol ID: 3642) and conducted in accordance with the Helsinki Declaration. All patients enrolled gave their written informed consent for participation.
Open Research
DATA AVAILABILITY STATEMENT
All data generated or analyzed during the current study are available from the corresponding author upon reasonable request. RNA-sequencing data are deposited at the ArrayExpress database under the accession number E-MTAB-13184.




