Residual circulating tumor DNA after adjuvant chemotherapy effectively predicts recurrence of stage II-III gastric cancer
Shu-Qiang Yuan, Run-Cong Nie, and You-Sheng Huang contributed equally to this work.
Trial registration: ClinicalTrials.gov Identifier: NCT02887612
Abstract
Background
Circulating tumor DNA (ctDNA) is a promising biomarker for predicting relapse in multiple solid cancers. However, the predictive value of ctDNA for disease recurrence remains indefinite in locoregional gastric cancer (GC). Here, we aimed to evaluate the predictive value of ctDNA in this context.
Methods
From 2016 to 2019, 100 patients with stage II/III resectable GC were recruited in this prospective cohort study (NCT02887612). Primary tumors were collected during surgical resection, and plasma samples were collected perioperatively and within 3 months after adjuvant chemotherapy (ACT). Somatic variants were captured via a targeted sequencing panel of 425 cancer-related genes. The plasma was defined as ctDNA-positive only if one or more variants detected in the plasma were presented in at least 2% of the primary tumors.
Results
Compared with ctDNA-negative patients, patients with positive postoperative ctDNA had moderately higher risk of recurrence [hazard ratio (HR) = 2.74, 95% confidence interval (CI) = 1.37–5.48; P = 0.003], while patients with positive post-ACT ctDNA showed remarkably higher risk (HR = 14.99, 95% CI = 3.08-72.96; P < 0.001). Multivariate analyses indicated that both postoperative and post-ACT ctDNA positivity were independent predictors of recurrence-free survival (RFS). Moreover, post-ACT ctDNA achieved better predictive performance (sensitivity, 77.8%; specificity, 90.6%) than both postoperative ctDNA and serial cancer antigen. A comprehensive model incorporating ctDNA for recurrence risk prediction showed a higher C-index (0.78; 95% CI = 0.71–0.84) than the model without ctDNA (0.71; 95% CI = 0.64–0.79; P = 0.009).
Conclusions
Residual ctDNA after ACT effectively predicts high recurrence risk in stage II/III GC, and the combination of tissue-based and circulating tumor features could achieve better risk prediction.
List of abbreviations
-
- GC
-
- gastric cancer
-
- ACT
-
- adjuvant chemotherapy
-
- MRD
-
- molecular residual disease
-
- ctDNA
-
- circulating tumor DNA
-
- cfDNA
-
- cell-free DNA
-
- NGS
-
- next-generation sequencing
-
- VAF
-
- variant allele frequency
-
- SOX
-
- S-1 plus oxaliplatin
-
- CapOX
-
- capecitabine plus oxaliplatin
-
- CEA
-
- carcinoembryonic antigen
-
- CA199
-
- carbohydrate antigen 199
-
- CA72-4
-
- carbohydrate antigen 72-4
-
- TCGA
-
- The Cancer Genome Atlas
-
- STAD
-
- stomach adenocarcinoma
-
- ERBB4
-
- Erb-B2 receptor tyrosine kinase 4
-
- RFS
-
- recurrence-free survival
-
- OS
-
- overall survival
-
- DFS
-
- disease-free survival
-
- LASSO
-
- Least Absolute Shrinkage and Selection Operator
-
- PPV
-
- positive predictive value
-
- NPV
-
- negative predictive value
-
- ROC
-
- receiver operating characteristic
-
- AUC
-
- area under the curve
-
- HR
-
- hazard ratio
-
- CI
-
- confidence interval
-
- HAS
-
- hepatoid adenocarcinoma of the stomach
-
- GASC
-
- gastric adenosquamous carcinoma
-
- SRCC
-
- signet ring cell carcinoma
-
- AJCC
-
- American Joint Committee on Cancer
-
- TP53
-
- tumor protein p53
-
- ARID1A
-
- AT-rich interactive domain-containing protein 1A
-
- CDH1
-
- Cadherin 1
-
- ESMO
-
- European Society for Medical Oncology.
1 BACKGROUND
Gastric cancer (GC) has become the sixth most common cancer and the fourth leading cause of cancer-related deaths worldwide [1]. Nearly 40% of newly diagnosed cases are from China and have an unsatisfactory 5-year overall survival (OS) rate of <50% [2]. Surgical resection with adjuvant chemotherapy (ACT) or perioperative chemotherapy has become the standard treatment for localized GC [3]. However, despite multimodal therapy, the incidence of regional recurrence and distant metastasis in patients with resectable GC remains high [4].
Although conventional histopathologic and radiographic examinations being widely used for the diagnosis and assessment of GC spread, solely depending on them to determine which patients have molecular residual disease (MRD) is insufficient, especially in earlier stage of treatment and in those who have achieved complete disease remission after curative treatment. Further, even though circulating serum cancer antigen detection contributes to the dynamic observation of treatment efficacy and prognosis evaluation, their predictive sensitivity and specificity are far from satisfactory [5].
As such, liquid biopsy, the detection of circulating tumor DNA (ctDNA) from the peripheral blood, is being rapidly implemented in clinical settings as a non-invasive, real-time and more reliable approach across multiple cancers, such as colorectal [6], breast [7], hepatocellular [8] and lung cancers [9]. In recent years, the intra- and inter-tumoral heterogeneity of GC has been investigated [10, 11], suggesting the potential prospects of utilizing ctDNA for providing a more general profile of genomic alterations in GC.
To date, most data only support ctDNA's clinical utility in the metastatic setting, with emerging utility in the early-stage MRD setting in ongoing clinical trials. Emerging studies have shown that residual ctDNA after surgery or during surveillance could reflect the existence of MRD and risk of relapse in various solid tumors [12-16], with early changes in ctDNA as a promising marker of clinical response [9, 17]. Recently, several groups applied next-generation sequencing (NGS) assays targeting somatic variants of ctDNA in GC patients and indicated the relation between postoperative ctDNA detection and disease relapse [18-20]. However, considering the relatively limited size (n = ∼ 50) and technical complexity in previous studies, clinical implementation of subsequent adjuvant therapies would be hindered.
In this prospective observational study, a 425-gene NGS panel-based approach was used to accurately capture tumor-specific somatic mutations in ctDNA perioperatively and after ACT in 100 patients with stage II/III resectable GC. The current study aimed to determine whether perioperative or post-ACT ctDNA detection could predict disease recurrence and to explore the additional value of ctDNA as a biomarker for response evaluation in the postoperative setting.
2 MATERIALS AND METHODS
2.1 Study design and patient enrollment
This prospective and observational study (ClinicalTrials.gov identifier: NCT02887612) was designed and implemented in the Sun Yat-sen University Cancer Center, Guangzhou, Guangdong, China. Patients diagnosed with resectable stage II/III gastric cancer who underwent R0 D2 gastrectomy between October 2016 and June 2019 and met the eligibility criteria (Figure 1) were enrolled for this study. Patients were excluded if they had previously received neoadjuvant anti-tumor therapy. The study was conducted in accordance with the Declaration of Helsinki and Good Clinical Practice Guidelines after approval by the institutional independent ethics committee of Sun Yat-sen University Cancer Center (ethics approval ID: B2016-050-01).

The primary tumor tissues of each patient were collected during surgery. The peripheral blood samples of each patient were collected at the following time points: (i) within 1 week prior to the surgical resection; (ii) within 1 week after the surgical resection; and (iii) within 3 months after ACT. All participants provided written informed consent.
2.2 Somatic variant detection in primary tumors and ctDNA
Genomic DNA from tumor tissues and peripheral blood leukocytes was extracted using QIAamp DNA FFPE Tissue Kit and DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany), respectively. At each sampling point, 8-10 mL of peripheral blood was drawn into Cell-Free DNA BCT® tubes (Streck, La Vista, NE, USA) or K2-EDTA tubes (BD Biosciences, Oxord, UK), the plasma fraction was prepared within 2 h after blood collection by centrifugation at 1800 ×g for 10 min and transferred to the central testing laboratory within 72 h. Cell-free DNA (cfDNA) was extracted using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany), and then quantified using Qubit 3.0 Fluorometer with the dsDNA High Sensitivity Assay Kit (Life Technologies, Eugene, OR, USA) to determine the yield of cfDNA. Then, we detected cfDNA and matched normal DNA from the control whole-blood samples of patient to determine somatic alterations of ctDNA. Sequencing libraries were constructed using the KAPA Hyper Library Prep Kit (KAPA Biosystems, Cape Town, South Africa). Hybridization capture-based targeted enrichment was performed using the Geneseeq Prime™ 425-gene panel (Nanjing Geneseeq Technology Inc., Nanjing, Jiangsu, China) (Supplementary Table S1). Libraries were quantified by quantative PCR (qPCR) using KAPA Library Quantification kit (KAPA Biosystems). Library fragment size was determined by Bioanalyzer 2100 (Agilent Technologies, Waldbronn, Germany). The target-enriched library was then sequenced on the HiSeq4000 NGS platforms (Illumina, San Diego, CA, USA).
The peripheral blood leukocytes, primary tumors and plasma samples were all target-sequenced using the Geneseeq Prime™ 425-gene panel, separately yielding a mean sequencing depth of 104×, 856× , and 2630× after removing duplicate reads. Fastp [21] (v0.20.0) was used for quality control and adapter removal of the FastQ files. Leading/trailing low quality (quality score below 30) or N bases were removed. Sequencing reads were mapped to the hg19 reference genome using bwa-mem [22] (v0.7.17-r1188). PCR duplicates were marked using sambamba [23] (v0.8.0). Single nucleotide variants and small insertions/deletions were identified by VarScan2 [24] with tumor and matched normal DNA.
Further filtering criteria were applied to the variants identified in primary tumors, including (i) the variants not presenting in the internal database of normal peripheral blood leukocyte samples from ∼500 healthy donors; (ii) the variants presenting in <1% of the population in the 1000 Genomes Project [25], the Exome Aggregation Consortium [26], and the Genome Aggregation Database [27]; (iii) variant allele frequency (VAF) ≥ 0.5%, supporting reads ≥ 3, depth ≥ 30× for recurrent variants (≥20 mentions in COSMIC [28] v92), and VAF ≥ 1%, supporting reads ≥ 6, depth ≥ 30× for non-recurrent variants. All qualified variants identified in the primary tumor of each patient were regarded as patient-specific somatic variants for further ctDNA tracking (tracking variants hereafter).
As to plasma variant calling, a set of cfDNA samples from 33 healthy donors were pre-processed to estimate the background VAF distribution for background polishing as previously described [29]. Briefly, for each tracking variant, a one-dimensional vector v was generated containing all observed allele frequencies (AFs) at that position and base substitution in the healthy cfDNA controls. A Gaussian distribution was used to model vector (v) with mean (μ) and standard deviation (σ) calculated from the remaining AFs if the number of non-zero AFs was less than 5. In cases where the number of non-zero AFs was 5 or more, a Weibull distribution was fitted to the non-zero AFs, and the resulting shape and scale parameters were saved. The fraction of non-zero AFs in v was saved to encounter zero-inflation and incorporate the frequency of zero-valued observations into the final model. The fractional abundance (f) of each tracking variant background allele was assessed using the corresponding background model in the resultant background database (Φ). A one-sided z-test was used to yield P value when the distribution was assigned as Gaussian. While for Weibull distribution, shape and scale parameters were used to calculate the cumulative probability (P*). Zero-inflation of dataset were accounted by the following formula: P value = 1 – [(1 – δ) + (δ × P*)]. False discovery rate correction was achieved using Bonferroni Correction with significance threshold at α = 0.01 yielding the final adjusted P value. Based on our understanding, a false discovery rate at a level of 0.01 carries higher stringency compared to the commonly used level of 0.05 in similar publications [29]. Indeed, during assay development of a mutation analysis, detecting limit of blank (LoB) among donor controls is often emphasized to establish background noise level in the absence of the target mutation [30]. Therefore, we opted for determining the LoB and selecting a fixed cutoff value instead of testing various P value thresholds. Herein, we performed leave-one-out cross-validation within our 33 healthy donors and tested various adjusted P value thresholds, including 0.001, 0.005, 0.01, 0.05, and 0.1. We found that false positives were detected only when the threshold reached 0.05. Consequently, we selected the next maximum threshold (0.01) to ensure sufficient sensitivity while maintaining a high confidence level. It is worth mentioning that the same cutoff has been utilized in previous published reports on colorectal cancer [14, 31] and non-small cell lung cancer [14, 31]. For each tracking variant, the read depth of mutant and reference alleles in the plasma was used to calculate the P value against the background VAF distribution, and variant with a corresponding FDR-adjusted P value < 0.01 was retained as a true variant; we further required a minimum supporting reads of 3, a minimum depth of 100× for recurrent variants and a minimum supporting reads of 6, a minimum depth of 100× for non-recurrent variants. In addition, for ctDNA variants not presenting in the corresponding primary tumor, they were rescued as true variants to account for tumor heterogeneity and clonal evolution if the following stringent criteria were met: (i) VAF ≥ 1%, supporting reads ≥ 6, depth ≥ 100×; (ii) not present in our previously published database of clonal hematopoiesis variants [32]; (iii) VAF = 0% in the paired peripheral blood leukocyte sample. To account for the intra- and inter-tumoral heterogeneity of different patients’ tumors and reduce false-positive rates, a plasma sample was prospectively defined as ctDNA-positive only if one or more variants in the plasma were detected in at least 2% of the primary tumors. The ctDNA levels of each ctDNA-positive plasma was calculated by the number of tumor-specific mutations per ng of cfDNA and per mL of plasma.
2.3 Treatment and follow-up
All patients were treated and followed up according to the Chinese Society of Clinical Oncology guidelines [3]. The adjuvant chemotherapy was standardized, consisting of oral fluoropyrimidine (S-1 or capecitabine), S-1 plus oxaliplatin (SOX), capecitabine plus oxaliplatin (CapOX). The implementation of ACT after surgery was at the discretion of the clinicians and patients, but both were blinded to the ctDNA results. Clinical follow-up included clinical review and serum cancer antigen test (i.e., carcinoembryonic antigen [CEA], carbohydrate antigen 199 [CA199], and carbohydrate antigen 72-4 [CA72-4]) for every 3 months, and standard-of-care radiologic imaging for every 6 months. Follow-up was recorded until November 16, 2021.
2.4 Validation cohort information
We included the stomach adenocarcinoma (STAD) cohort of The Cancer Genome Atlas (TCGA) to determine the prognostic value of Erb-B2 receptor tyrosine kinase 4 (ERBB4) mutational status. The TCGA STAD cohorts [33, 34] were collected with complete clinical and genomic data from the cBioPortal database (https://www.cbioportal.org/).
2.5 Nomogram construction
Univariate Cox regression analysis was performed to identify clinicopathological risk factors that were significantly associated with recurrence-free survival (RFS). To select recurrent mutations significantly associated with RFS, we used the Least Absolute Shrinkage and Selection Operator (LASSO) method in the Cox regression model, as we previously described [35]. Genes with a mutational frequency above 5% were subjected to the LASSO Cox regression model, and we required selected genes to appear over 125 times out of a total of 500 repetitions [8].
2.6 Statistical analyses
The primary outcomes were RFS and OS. RFS was calculated from the date of surgical resection to the date of verified radiological recurrence or death as a result of GC for patients who relapsed and was censored at last follow-up or non-GC-related death for patients who were not documented as recurrence. OS was measured from the date of inclusion until the date of death or last follow-up. The secondary outcomes were sensitivity, specificity, accuracy, positive predictive value (PPV) and negative predictive value (NPV), which were measured to assess the performance of each predictive biomarker. RFS and OS analyses and univariate and multivariate analysis were performed using the Kaplan–Meier estimator and Cox proportional hazard regression analysis. Variables with P < 0.05 in the univariate Cox model were entered into multivariate Cox model. We plotted the time-dependent receiver operating characteristic (ROC) curves and compared area under the curve (AUC) using the timeROC package, and comparison between two C-indices was performed using compareC package. Associations between predictive biomarker status (i.e., postoperative ctDNA, post-ACT ctDNA, post-treatment serum cancer antigen and ERBB4 mutation) and recurrence status were measured using the Fisher's exact test. Statistical analyses were performed using the R software v4.0.1 (https://www.r-project.org/), and a two-sided P value < 0.05 was considered significant.
3 RESULTS
3.1 Clinicopathological characteristics and preoperative ctDNA detection
A total of 100 patients with pathological stage II/III resectable GC were enrolled (Figure 1). The patients’ baseline characteristics are summarized in Supplementary Table S2. Sixty-three patients were diagnosed with stage III GC, and 90 patients received at least one course of ACT. The median duration of ACT was 6.2 months (range: 0.8-13.1 months). At a median follow-up of 52.2 (95% CI = 50.5-54.3) months, 33 patients ultimately relapsed, and 27 patients died from any cause.
In the primary tumors, tumor protein p53 (TP53), AT-rich interactive domain-containing protein 1A (ARID1A), and Cadherin 1 (CDH1) were the three most frequently mutated genes (Figure 2). A total of 855 variants were detected, and 83 (9.7%) variants presented in only one tumor. More importantly, only 4 of the 83 variants were detected in 3 patients’ corresponding preoperative plasma samples.
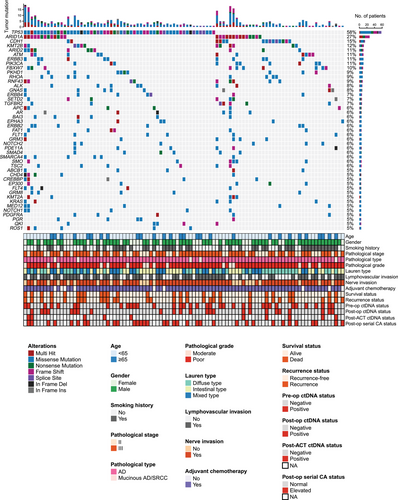
Preoperatively, 223 variants were detected in 42 of the 100 patients, 28 (12.6%) variants called in plasma were presented in corresponding primary tumors, and only 33 preoperative plasma samples were considered positive for ctDNA. Besides, preoperative ctDNA status of the patients was not associated with baseline clinical features (Supplementary Table S2), and no significant RFS or OS difference was observed between patients with and without detectable preoperative ctDNA (RFS: HR = 1.10, 95% CI = 0.55–2.22, P = 0.785; OS: HR = 1.13, 95% CI = 0.53–2.43, P = 0.754; Supplementary Figure S1).
3.2 Postoperative ctDNA detection and its association with recurrence risk
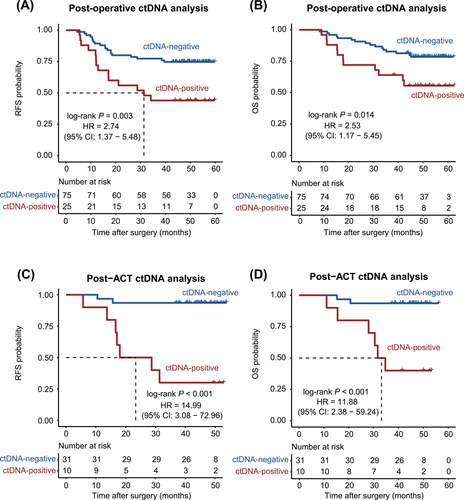
After surgical resection (median, 4 days; range, 1-7 days), 25.0% (25/100) of patients were positive for ctDNA. Patients with postoperative positive ctDNA had a higher proportion of preoperative elevated CEA compared with those with postoperative negative ctDNA (P = 0.046; Supplementary Table S2). No difference in other clinicopathological characteristics was observed between patients with detectable versus undetectable postoperative ctDNA (Supplementary Table S2). Patients with postoperative residual ctDNA had moderately higher risk of recurrence compared to those with undetectable ctDNA (Figure 3A). Patients with positive ctDNA also had inferior OS compared to those without (Figure 3B). Multivariable Cox analysis confirmed postoperative ctDNA as an independent factor for RFS (Supplementary Table S3). Higher cutoffs for circulating tumor-specific mutations per ng of cfDNA and per mL of plasma were not associated with significantly higher risk of relapse (Supplementary Figure S2), suggesting that presence of ctDNA rather than the total load of ctDNA is more related to high recurrence risk.
3.3 Post-ACT ctDNA predicted significantly higher recurrence and death risks
Among 41 patients who had plasma samples collected within 3 months after ACT, 10 (24.4%) patients were positive for ctDNA. Patients with preoperative elevated CEA had a higher proportion of post-ACT ctDNA positivity compared to those with preoperative normal CEA (P = 0.036; Supplementary Table S2). No difference in other clinicopathological characteristics was observed between patients with positive versus negative post-ACT ctDNA (Supplementary Table S2). After ACT, ctDNA-positive patients had remarkably higher risk of recurrence than ctDNA-negative patients (Figure 3C). Similarly, ctDNA-positive patients had significantly reduced OS compared to ctDNA-negative patients (Figure 3D). Multivariate Cox regression analysis showed that post-ACT ctDNA status was still the strongest prognostic factor associated with recurrence and death risk (Supplementary Table S4).
Next, we evaluated the dynamic ctDNA changes from preoperative to postoperative status and from postoperative to post-ACT. We found that those patients with both preoperative and postoperative ctDNA negative experienced the least disease relapse (Supplementary Table S5). In addition, compared with postoperative ctDNA status, both the 2 patients who experienced ctDNA clearance and 27 (93.1%) of 29 patients who remained ctDNA-negative were disease-free at last follow-up; 3 of 5 patients who were initially ctDNA-negative but turned positive and 4 of 5 patients who remained ctDNA-positive ultimately had disease relapse (Supplementary Table S6).
3.4 Association of recurrence site and ctDNA status
We also evaluated the association between recurrence site and ctDNA status. Of note, the peritoneum was the most common recurrence site among patients with negative postoperative or post-ACT ctDNA (Supplementary Table S7), which indicated that ctDNA detection rate is rather low in patients with peritoneal disease.
3.5 Post-ACT ctDNA outperformed other biomarkers in predicting recurrence
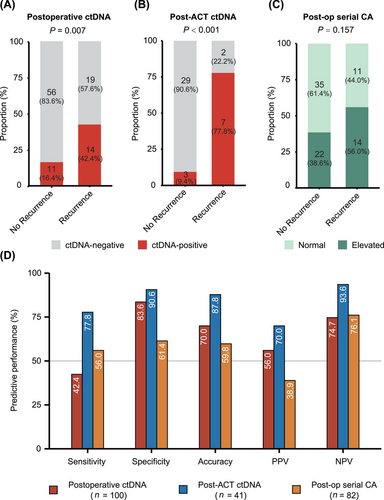
Next, we tested the predictive power of postoperative ctDNA, post-ACT ctDNA and postoperative serial cancer antigen (Figure 4A-C). We found that post-ACT ctDNA was the most related biomarker to disease relapse (P < 0.001). Further, post-ACT ctDNA consistently achieved better predictive performance than the other two biomarkers (Figure 4D). Taken together, residual ctDNA after ACT could effectively predict recurrence of disease with higher performance metrics, suggesting its potential significance in prognosis evaluation.
3.6 Correlations of ERBB4 mutational status in primary tumors with RFS
Although both residual postoperative and post-ACT ctDNA could screen out patients with high risk of recurrence, some patients (22.2%, 16/72) who were negative for ctDNA after surgery or adjuvant chemotherapy still experienced disease relapse. To investigate the biological explanations of this phenomenon, we performed an exploratory analysis of molecular alterations of primary tumors. For patients with undetectable ctDNA during surveillance, the ERBB4 mutation rate of primary tumors was significantly higher in patients who relapsed than those who were disease-free (25.0% vs. 3.6%, P = 0.020). Besides, ERBB4 mutational status could also discriminate patients with and without disease recurrence in the whole cohort (P = 0.038; Supplementary Figure S3A). Patients who harbored ERBB4 mutation in tumor tissues had significantly reduced RFS compared to those who were ERBB4 wildtype (Supplementary Figure S3B). The external cohort of STAD from TCGA similarly indicated that GC patients who harbored ERBB4 mutation had shorter DFS than patients without ERBB4 mutation (Supplementary Figure S3C-D).
3.7 Incorporation of ctDNA after definitive treatment improved model's performance for RFS prediction
To assess the overall probability of recurrence, we combined all the risk factors, including pathological stage, lymphovascular invasion, nerve invasion, ERBB4 mutational status presented in primary tumors and ctDNA status (postoperative or post-ACT if available), and constructed a nomogram for RFS prediction (Figure 5A). In particular, the prediction of RFS generated by the model with ctDNA status (C-index = 0.78, 95% CI = 0.71–0.84) was more accurate than that of the model without ctDNA status (C-index = 0.71, 95% CI = 0.64–0.79, P = 0.009; Figure 5B). Additionally, we separately plotted time-dependent ROC curves using the recurrence probabilities computed by the predictive model with and without ctDNA status. The AUC of the model with ctDNA status versus the model without were 0.83 versus 0.74 at 2-year RFS (P = 0.006), and 0.84 versus 0.75 at 3-year RFS (P = 0.002), respectively (Figure 5C). Altogether, these findings indicated that the intergration of tissue-based and circulating tumor features could achieve better risk prediction.

4 DISCUSSION
This prospective cohort study evaluated the power of ctDNA detection to predict risk of recurrence in patients with stage II/III GC. The results underlined that residual ctDNA after definitive treatment could identify patients with higher likelihood of relapse. Compared to recent studies addressing the same issues within relatively small cohorts [18-20], we confirmed that both postoperative and post-ACT ctDNA positivity were still independent predictors of RFS after adjusting for all risk factors in multivariate analyses.
In the present study, we used a 425-gene panel-based NGS approach to capture tumor-specific somatic mutations in primary tumors and applied them to detect ctDNA and we regarded a variant detected in plasma as cancer-derived only when it also presents in at least 2 primary gastric tumors. The population-based rather than highly personalized tumor-informed strategy was applied due to strong genomic heterogeneity within GCs, the mutation feature of a single tumor tissue cannot represent the whole mutational spectrum [36, 37]. Besides, the European Society for Medical Oncology (ESMO) Precision Medicine Working Group recently recommends to sample at least 1 week after surgery to analyze ctDNA MRD [38]. Here, liquid biopsy was performed within 1 week after surgery considering the short half-life (∼2 h) of ctDNA [39]. Undoubtedly, the population-based strategy for defining ctDNA positivity and sampling within 1 week after surgery might lead to decrease in specificity and produce false-positives, in addition to the lack of an independent validation cohort, which became the limitation of the present study. However, ctDNA evaluation performed as early as 1-7 days postoperatively in this study still showed comparable performance of sensitivity with many reported tumor-informed ctDNA analyses between 4 and 10 weeks postoperatively [13, 18, 19, 40]. In clinical practice, high sensitivity of MRD detection will be critical if MRD assays are applied for guiding a patient with ctDNA MRD to receive adjuvant therapy.
Preoperatively, ctDNA positive rate was 38.0% in our cohort, which was in the low end of preoperative detection rates ranging from 37% to 57% in localized gastroesophageal cancer [18, 19]. Besides, preoperative ctDNA did not demonstrate a strong correlation with baseline clinicopathological features and showed limited value for predicting recurrence. These results showed discrepancies with reports [19, 41], which indicated that ctDNA shedding rates were affected by factors such as T stage and tumor volume. Further, similar to our findings, other studies also illustrated consistency in the clinical features between preoperative positive and negative ctDNA subgroups [14, 42]. These data portray that our understanding of the mechanisms of ctDNA shedding in GC actually remains unclear [43].
Through serial ctDNA monitoring, we highlighted the importance of post-ACT ctDNA as a prognostic biomarker for it reflected much higher risk of relapse than postoperative ctDNA (HR = 14.99 vs. 2.74), which was not emphasized in previous researches assessing the association between ctDNA detection and recurrence risk in resectable GC. Yang et al. [19] reported similar results with quite a small sample size (n = 23), and the other two studies lacked evaluation of residual ctDNA after ACT for predicting recurrence [18, 20]. Actually, postoperative ctDNA was not invariable during the whole clinical course as we showed. After accomplishing ACT with curative intent, dynamic shifting of ctDNA status was observed and in good concordance with treatment response. More importantly, post-ACT ctDNA demonstrated a predictive advantage over postoperative ctDNA (sensitivity = 77.8% vs. 42.4%; specificity = 90.6% vs. 83.6%). Therefore, ctDNA detection after adjuvant therapy deserves more attention, as their existence indicated an extremely high risk of recurrence and the reminder of resistance to adjuvant therapy.
Of note, a subset of patients who were ctDNA-negative after definitive treatment still experienced relapse (22.2% in the present study and 32.0% in a study by Yang et al. [19]). Exploratory analysis of primary tumors revealed that ERBB4-mutant GC was probably a more malignant subtype with poor prognosis in both our and external cohorts, as evidenced by the finding that ERBB4 mutation played an oncogenic role in GC from our group [44]. Besides, a comprehensive model established with all risk factors including ctDNA status showed a high accuracy in recurrence risk prediction. Consequently, the utility of ctDNA detection for predicting recurrence should be combined with the clinicopathological and biological features (e.g., molecular alterations, pathological stage) presented in tumors to achieve better predictive accuracy.
For a long time, it is debatable whether we should perform ctDNA testing for postoperative patients. In 2022, the DYNAMIC study (ACTRN12615000381583) provided prospective evidence that ctDNA-based adjuvant therapies for colon cancer reduced the use of adjuvant chemotherapy without comprising RFS [45]. Other ctDNA MRD-based adjuvant therapies were explored in the IMvigor011 study (NCT04660344) [46] for muscle-invasive urothelial cancer, the PEGASUS trial (NCT04259944) [47] for stage II-III colon cancer and the MERMAID-1 study (NCT04385368) [48] for stage II-III non-small cell lung cancer. Moreover, for patients with residual ctDNA after completion of adjuvant therapies, whether they should extend treatment period or receive other treatment regimens are still worthy of further exploration. Reassuringly, we find that several prospective studies are exploring “molecular metastatic” treatment after completing standard adjuvant therapy with molecular relapse monitoring in breast cancer [TRAK-ER (NCT04985266), DARE (NCT04567420) and LEADER (NCT03285412) trials].
5 CONCLUSIONS
Taken together, this prospective, large-scale cohort study provides evidence supporting that the existence of ctDNA after definitive treatment, especially after ACT, reflects high risk of relapse in patients with stage II/III GC, and the combination of tissue-based and circulating cancer features could better evaluate the outcomes of patients. In the future, ctDNA may serve as an effective tool to guide GC management, including early intervention and response evaluation in the post-surgical setting, which needs validation through randomized clinical trials.
DECLARATIONS
AUTHOR CONTRIBUTIONS
SQY: Formal analysis; visualization; data curation and writing. RCN: Formal analysis; visualization; data curation and writing. YSH: Formal analysis; visualization; data curation and writing. YBC: Formal analysis; visualization; data curation and writing. SYW: Formal analysis and methodology. XWS: Supervision of the ctDNA protocol. YFL: Data curation. ZKL: Data curation and methodology. YXC: Formal analysis and methodology. YCY: Data curation. YX: Methodology. HBQ: Data curation. YL: Data curation. WW: Data curation. ZXL: Methodology. QZ: Methodology. RHX: Supervision of the study design and writing. ZWZ: Supervision of the study design; writing and funding acquisition. FW: Supervision of the data procession; writing and funding acquisition.
ACKNOWLEDGMENTS
We thank all patients and their families for participating in this study.
CONFLICT INTEREST
The authors disclose no conflicts.
FUNDING
This research was support by the Science and Technology Program of Guangdong (2019B020227002, to RHX), the CAMS Innovation Fund for Medical Sciences (CIFMS) (2019-I2M-5-036, to RHX), the International Cooperation and Exchanges National Natural Science Foundation of China (82061160373, to FW), the National Natural Science Foundation of China (General Program, 81872011, to FW); the Sun Yat-sen University Clinical Research 5010 Program (2018014, to FW); the Young Physician Scientist Program of Sun Yat-sen University Cancer Center (16zxqk03, to FW); the Guangdong Esophageal Cancer Institute Science and Technology Program (M202210, to SQY). The funders played no role in the study's design, conduct, or reporting.
AVAILABILITY OF DATA AND MATERIALS
The key raw data have been deposited into the Research Data Deposit (www.researchdata.org.cn), with the approval number of RDDA2023290160. The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study was conducted in accordance with the Declaration of Helsinki and Good Clinical Practice Guidelines after approval by the institutional independent ethics committee of Sun Yat-sen University Cancer Center (Ethics approval ID: B2016-050-01). All participants signed the informed consent form before participating in the study.
CONSENT FOR PUBLICATION
Not applicable.




