Transient and DNA-free in vivo CRISPR/Cas9 genome editing for flexible modeling of endometrial carcinogenesis
Abbreviations
-
- CRISPR
-
- Clustered Regularly Interspaced Short Palindromic Repeats
-
- crRNA
-
- CRISPR RNA
-
- PTEN
-
- Phosphatase and Tensin homolog
-
- EC
-
- Endometrial Carcinoma
-
- EIN
-
- Endometrial Intraepithelial Neoplasia
-
- PCR
-
- Polymerase Chain Reaction
-
- CK8
-
- Cytokeratin 8
-
- PI3K/AKT
-
- Phosphoinositide 3-kinase/Akt kinase
-
- Fbxw7
-
- Box And WD Repeat Domain Containing 7
-
- mT
-
- membrane-targeted tdTomato
-
- mG
-
- membrane-targeted GFP
-
- pCA-mT/mG
-
- plasmids encoding membrane-targeted tdTomato and mG membrane-targeted GFP
-
- RNP
-
- Ribonucleoprotein
-
- NGS
-
- Next-Generation-Sequencing
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) (CRISPR/Cas9)-mediated generation of somatically genetically engineered mouse models have emerged as a new approach for in vivo modeling of cancer [1]. Here, we describe a novel DNA-free, easy, rapid, flexible, multiplexable, and robust method to model endometrial neoplasia by CRISPR/Cas9 ribonucleoprotein (RNP) electroporation into the uterus of mice.
First, we tested our CRISPR/Cas9 editing approach with an in vitro CRISPR/Cas9 assay using a plasmid encoding a Locus of X-over P1 (LoxP)-flanked membrane-targeted tandem dimer Tomato (mT) followed by membrane-targeted Green Fluorescent protein (GFP) (pCA-mT/mG). The material and methods used in this study are described in the Supplementary Material file. Upon Cre recombination, LoxP-flanked mT was deleted, and we observed that the cells shifted from red to green fluorescence. Then, we designed a CRISPR RNA (crRNA) targeting the Protospacer Adjacent Motif (PAM) sequence adjacent to LoxP sites flanking mT (Supplementary Figure S1A). The pCA-mT/mG plasmid was incubated with the LoxP-targeting RNP, and the Cas9-mediated cleavage of the target DNA was assessed by agarose gel electrophoresis (Supplementary Figure S1B, arrowhead). Next, we tested the ability of the LoxP-targeting Cas9 RNP to induce gene editing into living cells. Skin fibroblasts isolated from mT/mG reporter mouse [2] were transfected with the LoxP-targeting Cas9 RNP. LoxP-targeting Cas9 RNP caused a switch from red to green in ∼10% of total fibroblasts (Supplementary Figure S1C). These results enabled us to test the ability of the LoxP-targeting Cas9 RNP in vivo. The abdominal cavity of anesthetized mT/mG females was opened longitudinally, and their uterine horns were exposed. Five μL of LoxP-targeting Cas9 RNP mix containing 0.5 μL of 62 mmol/L recombinant Cas9, 1.5 μL of 20 μmol/L crRNA:trans-activating CRISPR RNA (tracrRNA) duplex and 2 μL of Optimized Minimum Essential Medium (Opti-MEM) and 1 μL of Fast Green were injected into one uterine horn. RNP was electroporated using a BTX830 square electroporator by applying 4 pulses of 50 mV during 50 msec spaced by a 950 msec gap (Figure 1A). After 15 days, the uterine horns were dissected, opened longitudinally, and examined for the presence of green cells (Figure 1B). To demonstrate the correct genomic editing of LoxP sites in vivo, DNA extracted from green endometrial cell regions was amplified by polymerase chain reaction (PCR), which amplified a 181 bp product compatible with LoxP cleavage, mT excision and subsequent joining (Supplementary Figures S2A-B). PCR products were submitted to next-generation sequencing (NGS)-amplicon sequencing and analyzed with the Cas9 editing detection software Crispresso2 [3] and Cas-Analyzer [4] with similar results (Supplementary Figure S2B).
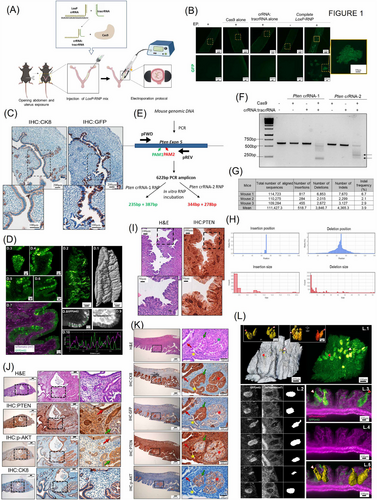
(A) Diagram depicting mouse surgery, intra-uterine RNP delivery and electroporation protocol. (B) Representative images of LoxP-RNP electroporated uteri. Green cells are observed when Cas9 and crRNA:tracrRNA were injected and an electric pulse was applied (EP). In experimental conditions in which Cas9 or crRNA:tracrRNA alone were injected or in the condition in which complete LoxP-RNP was injected, but no EP was applied, no green cells were observed. n = 4 uterine horns per condition. Images at 2X and 10X magnification. (C) Representative images of cytokeratin 8 (CK8) and GFP on consecutive histological sections of mT/mGf/f uteri 15 days after electroporation of LoxP-RNP. Images were acquired at 4X and 10X magnification. (D) Representative lightsheet microscopy images corresponding to mT/mGf/f endometria 15 days after electroporation with LoxP-RNP. (D.1) shows the GFP cell distribution in a 7 mm-long endometrium, (D.2) the surface of the endometrium detected from the tdTomato channel. (D.3-D.6) are subsequent close-up sections in a representative XY location at Z depths, calculated from the tissue surface, of 20 μm (D.3), 108 μm (D.4), 232 μm (D.5), 375 μm (D.6) where a GFP positive mosaic pattern can be seen in the epithelial cells of the endometrium. (D.7) shows the merged fluorescence acquired with the green GFP channel with overlapped tdTomato shown in magenta. (D.8) and (D.9) show the respective distribution of GFP and tdTomato cells along the endometrium epithelium. (D.10) shows the complementary intensity profile (see peaks to valley correspondence), measured on D.8 (green) and D.9 (magenta), along the green dashed line displayed in D.8). Scale bars: (D.3-D.6), (D.8-D.9), 50μm; (D.1-D.2), 500 μm. (D.1-D.2) was reconstructed from a vertical tiled image of 1×5 fields. All images were acquired at a magnification of 7 times (7X) with a lightsheet thickness of approximately 4.5 μm (D.3-D.6) are representative capture fields corresponding to the Supplementary video 1. (E-F) Representative diagram and agarose gel of in vitro CRISPR/Cas9 activity assay and to evaluate Pten RNPs. Arrowheads indicate the DNA fragments produced by Pten crRNA-2 RNP activity. (G-H) Bioinformatic analysis of amplicon sequences from Pten-RNP electroporated mice. Analysis of NGS-Pten amplicons with Cas-analyzer software. The upper table indicates the number of unmodified reads and reads displaying insertions or deletions (indels) in three different mice. Graphs indicated the position and size of indels in a region spanning 70 nt each site of the predicted cutting site. (I) Representative images of H&E staining and immunostaining to detect PTEN on serial histological sections of C57/B6 mice 15 days after electroporation with Pten-RNP. Scale bars: 100μm and 50 μm respectively (J) Representative images of H&E staining, and immunostaining to detect PTEN, phosphorylated AKT (p-AKT), cytokeratin 8 (CK8), on consecutive histological sections of C57/B6 mice uteri 12 weeks post-electroporation with Pten-RNP. Red arrows indicate PTEN expressing wild-type epithelial cells edited in LoxP locus (GFP positive). Green arrows indicate tumoral cells lacking PTEN expression that has been edited in the LoxP locus (GFP positive). Images at 4X and 20X. (K) Representative images of H&E staining, and immunostaining to detect PTEN, phosphorylated AKT (p-AKT), cytokeratin 8 (CK8), and GFP on consecutive histological sections of mT/mG uteri 12 weeks post-electroporation of Pten-RNP and LoxP-RNP. Images show the co-existence of PTEN-deficient endometrial EIN, either positive (green arrow) or negative (red asterisk), for GFP expression with PTEN expressing non-tumoral epithelial cells either positive (red arrow) or negative (yellow asterisk) for GFP expression. Images at 4X and 20X. (L) Lightsheet fluorescence microscopy shows that in vivo CRISPR/Cas9 electroporation of Pten-RNP and LoxP-RNP uteri generates endometrial epithelial lesions throughout the tissue. Representative images and reconstructions of mT/mG endometria 12 weeks after electroporation with Pten-RNP and LoxP-RNP. (L.1) surface rendering (left, from tdTomato channel) and corresponding maximum intensity projection (right, from GFP channel) in a 3×3 mm tissue fragment. Insets show six selected segmented GFP+ lesions by surface rendering from two opposite directions: front (left, yellow surfaces) and back view (right, volumes with different colors). (L.2) shows selected sections of GFP positive lesions (left) and corresponding tdTomato signal (center), with the segmented mask of the lesions. (L.3-L.5) Show an orthogonal cut through three lesions, visualizing tdTomato (L.4), overlapped tdTomato/GFP channels (L.3), and volume rendering of the lesions (L.5). (L.1) and (L.3-L.5) clearly show the pear-shaped lesions with the tip originating from the epithelium surface; see arrows in L.1 and L.5. Red arrowheads with white outline and white arrowheads with red outline point at the same lesions across images L.1, L.3 and L.5. Scales: L.1, 400 μm, insets: left 200 μm, right 100 μm, L.3-L-5, 100 μm. A three-dimensional video of the same sample is shown in Supplementary video 2.
Abbreviations: CRISPR, Clustered Regularly Interspaced Short Palindromic Repeats; Cas9, CRISPR-associated protein 9; RNP; ribonucleoprotein; LoxP, Locus of X-over P1; mT, membrane-targeted tandem dimer Tomato; GFP, Green Fluorescent protein; crRNA, CRISPR RNAs; PAM, Protospacer Adjacent Motif; tracRNA, trans-activating CRISPR RNA; Opti-MEM, Optimized Minimum Essential Medium; PCR, Polymerase Chain Reaction; CK8, cytokeratin-8; Pten, Phosphatase and tensin homolog; Fbxw7, Box And WD Repeat Domain Containing 7; PI3K/AKT, Phosphatidylinositol-4,5-bisphosphate 3-kinase/Alpha serine/threonine-protein kinase; p-Akt; phosphorylated Akt; NGS, next-generation sequencing; indels, insertions or deletions; EIN, endometrial intraepithelial neoplasia; EC, Endometrial Cancer; 3D, three-dimensional; bp; base pairs; IHC; immunohistochemistry; H&E; Eosin and Hematoxylin. pFWD; Forward Primer; pREV; Reverse Primer.
The epithelial nature of edited endometrial cells was demonstrated by cytokeratin-8 (CK8) and GFP immunohistochemistry on consecutive paraffin sections (Figure 1C). We also performed an analysis of GFP expression using lightsheet microscopy. Optically sectioned images and three-dimensional reconstruction of an approximately 7 mm long uterine tissue revealed that the GFP editing was restricted to cells displaying epithelial sheet (Figure 1D, Supplementary Video S1). Lighsheet imaging revealed the tissue-wide mosaic pattern distribution of the GFP-expressing cells.
The above results enabled us to design a protocol for genomic editing of genes involved in endometrial neoplasia. According to the whole genome sequencing [5], we selected three of the most frequently mutated tumor suppressor genes in endometrial cancer (EC): Phosphatase and tensin homolog (Pten), p53 and Box and WD repeat domain containing 7 (Fbxw7). We designed two different crRNAs targeting exon 5 of Pten and one crRNA targeting either p53 or Fbxw7. Nuclease activity of Pten (Figure 1E), p53 and Fbxw7 (Supplementary Figure S3A). RNPs were assessed by in vitro Cas9 cleavage assay using PCR amplicons obtained using primers flanking crRNAs target sites (Figure 1E-F, Supplementary Figure S3A). Pten crRNA-2 produced better cleavage bands and was selected for further experiments (we will refer to it as Pten-RNP, the one containing crRNA-2). Next, we evaluated the ability of these RNPs to produce genomic editing of endometrial cells in vivo. For this purpose, female mice were electroporated with Pten, p53 or Fbxw7 RNPs and sacrificed 15 days after electroporation. DNA from endometrial cells was amplified by PCR using primers flanking crRNAs target sites, and amplicons were submitted to NGS. NGS analysis of p53, Fbxw7 (Supplementary Figure S3B-C) and Pten amplicons (Figure 1G-H, Supplementary Figure S3C) demonstrated the presence of insertions or deletions (indels) on targeted position. To set a proof of principle for the CRISPR/Cas9 in vivo approach as a tool to recapitulate phenotypic effects of EC mutated genes, Pten-targeting Cas9 RNP was electroporated in B6/C57 mice. Previous studies demonstrated that conditional PTEN ablation in endometrial epithelial cells resulted in endometrial intraepithelial neoplasia (EIN)/hyperplasia [6-8]. Fifteen days after electroporation, PTEN immunohistochemistry revealed the presence of cells displaying loss of PTEN staining among others retaining its expression (Figure 1I, Supplementary Figure S3D). To evaluate the pathological effects of CRISPR/Cas9-induced loss of PTEN, uterine horns from electroporated mice were subjected to histopathological evaluation after 12 weeks. Hematoxylin and eosin staining revealed areas of EIN, the precursor lesion of Endometrial Cancer (EC) (CK8 positive but with no PTEN expression) surrounded by normal epithelium (CK8 and PTEN positive) (Figure 1J), suggesting that the CRISPR/Cas9 genomic editing of Pten in epithelial cells caused an increase in Phosphatidylinositol-4,5-bisphosphate 3-kinase/Alpha serine/threonine-protein kinase (PI3K/AKT) signaling pathway activity. Immunohistochemistry on serial endometrial sections with CK8, PTEN and p-AKT antibodies revealed that loss of PTEN expression resulted in subsequent activation of AKT (Figure 1J).
One of the main advantages of the Cas9 system is its ability to simultaneously edit more than one locus at a time [9, 10]. By co-electroporating two or more RNPs targeting different loci, it is feasible to create heterogeneous populations of cells which may be a valuable tool to model tumoral heterogeneity in EC. To evaluate this hypothesis, LoxP-RNP and Pten- RNP were co-electroporated in mT/mG mice, and 12 weeks later, the uteri were submitted to an immunohistochemistry analysis on serial endometrial sections with GFP, CK8, PTEN and p-AKT antibodies (Figure 1K). Immunohistochemistry revealed heterogenous staining with four scenarios. First, a group of cells that only received LoxP-targeting RNP resulted in normal epithelial cells displaying positive staining for GFP and PTEN but negative for p-AKT. Second, a group of cells that only received Pten-RNP resulted in EIN displaying negative staining for GFP and PTEN but positive for p-AKT. Third, a group of cells that received both LoxP- and Pten-targeting RNPs was EIN positive for GFP and p-AKT but negative for PTEN staining. Fourth, a group of cells that did not receive any of the two targeting Cas9-RNPs and displayed negative staining for GFP and p-AKT and positive staining for PTEN.
Lastly, we performed a lightsheet microscopy analysis of mGFP on a whole uterine fragment electroporated with LoxP- and Pten-targeting RNPs. Twelve weeks after electroporation, endometrial tissues were imaged under lightsheet microscopy. Optical section images and three-dimensional reconstructions of uterine tissue revealed GFP expressing EIN (Figure 1L, Supplementary Video S2) with a three-dimensional (3D) “pear” shape, as shown by GFP positive segmentation and 3D rendering.
Based on the observed findings, we believe the in vivo editing of mouse endometrial cells could be a versatile tool for EC modeling that could also be applied to identify candidate genes that regulate other aspects of uterine physiology.
AUTHOR CONTRIBUTION
Conceptualization, Raúl Navaridas and Xavier Dolcet; methodology, Raúl Navaridas, Anna Ruiz-Mitjana, Maria Vidal-Sabanés, Aida Perramon-Güell, Cristina Megino-Luque, Lídia Bardia, Julien Colombelli, David Llobet-Navas and Xavier Dolcet; investigation, Raúl Navaridas, Anna Ruiz-Mitjana, Maria Vidal-Sabanés; results validation, all authors; formal analysis, Joaquim Egea, Mario Encinas, Xavier Matias-Guiu, Xavier Dolcet; resources, Julien Colombelli, Lídia Bardia, Xavier Dolcet; writing, Raúl Navaridas, Julien Colombelli, Xavier Dolcet; supervision, Xavier Dolcet; main project administration, Xavier Dolcet; funding acquisition, Xavier Dolcet.
DECLARATIONS
ACKNOWLEDGMENTS
We thank Sébastien Tosi from the Advanced Digital Microscopy core facility (IRB Barcelona) for support with image analysis tools.
CONFLICT OF INTEREST DISCLOSURES
The authors have no conflicts of interest to declare.
FUNDING
This work was supported by the Spanish Ministry of Science, Innovation and Universities (grant code PID2019-104734RB-I00) and the Spanish Association Against Cancer (grant code LABAE19004LLOB). This work was also funded by the Institute of Health Carlos III (MS17/00063) (co-founded by the European Social Fund [ESF] “investing in your future”).
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
All procedures performed in this study followed the National Institute of Health Guide for the Care and Use of Laboratory Animals and were compliant with the guidelines of Universitat de Lleida (N. 02-02/19)
CONSENT FOR PUBLICATION
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
The datasets analyzed during the current study are available from the corresponding author upon reasonable request.