Activation mechanisms of clinically distinct B-Raf V600E and V600K mutants
Abbreviations
-
- MAPK
-
- mitogen-activated protein kinase
-
- PDB
-
- Protein Data Bank
-
- NtA
-
- N-terminal acidic motif
-
- A-loop
-
- activation loop
Dear Editor,
B-Raf, the main effector of Ras in the mitogen-activated protein kinase (MAPK) pathway, is among the most highly mutated kinases in human cancer [1]. About 40%-60% of melanoma patients harbor B-Raf mutations, of which ∼90% involve V600E and V600K. B-RafV600E is more frequent (60%-80%) than B-RafV600K (10%-30%). Substitution of a Val codon by Glu requires a single nucleotide change, whereas Val to Lys requires two [2]. This is in line with melanoma patients harboring the V600K mutation, who usually suffer from higher sun exposure that may induce increased DNA damage [3]. Since both mutations occur at the same position of the kinase domain and are mutated to charged residues, it was believed that the B-Raf V600E and V600K mutants would share a similar behavior, and in clinical trials, patients with V600E and V600K mutations have been recruited into the same cohort. However, emerging data suggest that B-Raf V600E and V600K mutants are not identical [4]. V600K tumors are more aggressive than V600E. Patients harboring V600K have a higher risk for relapse and shorter survival than those with V600E [5]. V600K tumors are less responsive to some kinase inhibitors but benefit from immunotherapy treatments [6]. These results imply that the two mutants may have intrinsic molecular differences. However, little is known about the underlying mechanism and the structural basis mediating these differences.
B-Raf V600E and V600K mutants can be constitutively activated as monomers [7]. On the other hand, it has been shown that the dimerization of B-RafV600 mutants does take place and is important for MAPK signal transduction in cancer [8]. Thus, our premise was that while B-RafV600 mutants can be active as monomers, their dimerization can enhance their signaling. To explore both scenarios, we studied B-Raf V600E and V600K mutants in the monomeric, homo-dimeric, and hetero-dimeric states at atomic resolution, aiming to identify structural features that may differentiate between B-Raf V600E and V600K mutants and help clarify their clinical outcomes. The study methods are described in the Supplementary Materials.
The V600 residue plays essential roles in maintaining the inactive conformation of B-Raf's kinase domain [9, 10]. Its mutations to E600 or K600 result in two structural changes involving (i) longer side-chain and (ii) loss of hydrophobicity. Our modeling shows that the long side-chains in the B-Raf V600E and V600K mutants caused structural collapse of the N-lobe hydrophobic surface in the inactive OFF-state conformation. The charged side-chains of E600 and K600 in the mutants disfavor hydrophobic residues in this region (Supplementary Figure S1). These structural effects may synergistically destabilize the inactive OFF-state conformation, shifting the populations towards the active state. In contrast, in the active ON-state conformation, the mutations to E600 and K600 led to neither structural collapse nor disfavored residue contacts. The E600 and K600 in the B-Raf mutants approached the bottom region of the αC-helix, pointing away from the ATP pocket. This can be confirmed by the crystal structures of the B-RafV600E mutants in the Protein Data Bank (PDB). Except for structures with large inhibitors that prevent the “IN” αC-helix, the B-RafV600E mutants adopt the active ON-state conformation (Supplementary Figure S2). These observations indicate that both B-Raf V600E and V600K mutants favor the active ON-state conformation.
In the active ON-state conformation, the monomeric B-Raf V600E and V600K mutants exhibited notable structural differences in local residue contacts (Figure 1A). In B-RafV600E, the negatively charged E600 residue formed a salt bridge with the positively charged K507 residue in the αC-helix's basic box, 506RKTR509, resembling the pS602 in the active wild-type B-Raf [10] (Figure 1B). It also frequently interacted with another basic residue, R603, in the activation loop (A-loop), which may help stabilize the nearby E600-K507 interactions (Figure 1A). In contrast, the monomeric B-RafV600K mutant is much less compact at the αC/N-terminal acidic motif (NtA) region. The positively charged K600 residue failed to establish favorable interactions with either the αC-helix basic box or R603 in the A-loop and was more exposed to the solvent.
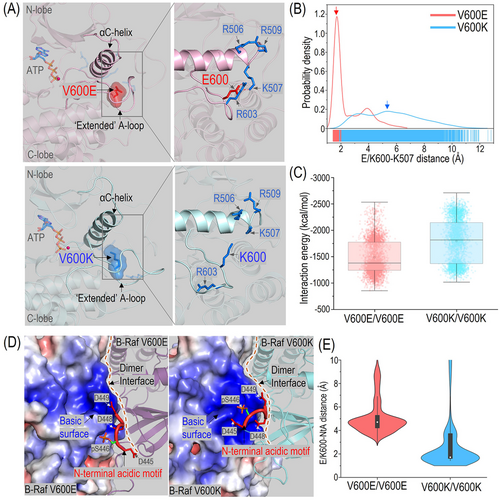
Structural features that differentiate between the B-Raf V600E and V600K mutants. (A) Snapshots of monomeric B-Raf V600E and V600K mutants in the active ON-state conformation. E600 in B-Raf V600E mutant forms salt bridges with K507 in the αC-helix and R603 in the A-loop. K600 in B-Raf V600K lacks these interactions and is more exposed. (B) Residue distances between E600 or K600 in B-Raf mutants and K507 in the αC-helix. (C) Interaction energies between protomers for B-Raf V600E and V600K homodimers. (D) Snapshots of residue contacts at the homodimer interfaces of B-Raf V600E and V600K mutants. In the homodimers, one protomer (left side) is represented by the electrostatic potential surface calculated by the ABPS and another protomer is shown in a cartoon drawing. The phosphorylated residue pS446 and acidic residues, D445, D448 and D449, in the NtA motif are shown as sticks. (E) Residue distances between E600 or K600 in B-Raf mutants and the charged residues in the NtA motif. Abbreviations: A-loop: activation loop; ABPS: Advanced Possion-Boltzmann Solver; NtA: N-terminal acidic;
Raf dimerizes in a “side-to-side” way for activation [10]. Interestingly, from the simulations, we observed that the distinct local contacts observed in monomeric B-Raf V600E and V600K mutants were located at the “side-to-side” dimer interface and affected their homodimerization. The interaction energies between the two protomers suggest that the dimer interfaces in the B-RafV600K homodimers were stronger than in the B-RafV600E homodimers (Figure 1C). Further analysis indicated this was largely because the K600 residue in the B-RafV600K mutant made the basic surface of the αC-helix broader and more extensive (Figure 1D). In this region, the basic residues, K600, R603, R506, K507, and R509, may accommodate the acidic NtA motif 445DSSDD449 from another protomer.
In most cases, K600 was directly involved in the NtA interactions and formed salt bridges (Figure 1E). Differently, the electrostatic potential of the basic surface in B-RafV600E mutants was weaker than in B-RafV600K (Figure 1D). The E600-K507 and E600-R603 salt bridges in the B-RafV600E mutants neutralized the basic surface and changed the local morphology at the dimer interface where NtA binds. E600 barely established direct contact with the charged residues in the NtA motif at the dimer interface (Figure 1E).
We also modeled and simulated the heterodimers of B-RafV600E and B-RafV600K with Raf-1 (C-Raf). The results suggested that the effects of the V600E and V600K mutations on the heterodimer interfaces were less significant (Supplementary Figure S3). It has been suggested that B-Raf's NtA phosphorylation is constitutive, while Raf-1's is not. Compared to B-Raf, Raf-1's NtA motif is less charged, with one acidic residue, D337, in 337DSSYY341. At the heterodimer interfaces, Raf-1's NtA motifs were less attracted by the charged E600/V600 residues in the A-loop and the αC-helix basic box, frequently moving away from the dimer interface.
Both V600E and V600K mutations can populate the ON-state kinase domain conformation, but the B-RafV600K homodimer has a stronger dimer interface, implying that B-RafV600K mutants may favor dimerization more than B-RafV600E. This appears to align with the observed more aggressive B-RafV600K tumors over B-RafV600E[4, 5]. While expression profiles, cell types, and underlying patient mutation loads may play essential roles in the clinics, we expect that the stronger homodimer interfaces in B-RafV600K mutants may contribute to the clinical profiles of the B-Raf tumors. Stronger interfaces, with reduced degradation, could lead to higher populations of the active dimer. We expect that the higher the number of constitutively active Raf molecules in the dimeric state, the higher the number of phosphorylated MEKs and the stronger the oncogenic signal propagating down the MAPK pathway to phosphorylate ERK could increase the level of cell proliferation.
In conclusion, we observed that the structural features that distinguish B-Raf V600E from V600K mutants were located at the dimer interface. This provides a novel structural opportunity for targeting the distinct dimer interfaces in the B-Raf V600E and V600K mutants. The dimer interface has been the target of the first and next-generation Raf inhibitors. Inhibitors, such as PLX8396 and PHI1, have been shown to be selective to Raf dimers. These distinct structural features at the dimer interface may help develop more potent and selective inhibitors against B-Raf cancers. Allosteric drugs targeting these features, or a combination of allosteric and drugs such as encorafenib (targeting B-Raf) plus binimetinib (MEK1), or mutant-selective PROTACs for degradation, may be an alternative strategy. Allosteric interface-targeting small molecules have shown some success.
DECLARATIONS
AUTHOR CONTRIBUTIONS
Mingzhen Zhang: conceptualization, investigation, analysis and writing the original draft. Ryan Maloney: writing, review and editing. Yonglan Liu: writing, review and editing. Hyunbum Jang: conceptualization, writing, review and editing. Ruth Nussinov: conceptualization, supervision, writing, review and editing.
CONFLICT OF INTEREST DISCLOSURES
There are no conflicts of interest to declare.
ACKNOWLEDGMENTS
The calculations were performed using the high-performance computational facilities of the Biowulf PC/Linux cluster at the National Institutes of Health, Bethesda, MD (https://hpc.nih.gov/).
AVAILABILITY OF DATA AND MATERIALS
Not appliable.
CONSENT FOR PUBLICATION
Not appliable.
FUNDING
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261201500003I. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not appliable.




