Multiplexed imaging of tumor immune microenvironmental markers in locally advanced or metastatic non-small-cell lung cancer characterizes the features of response to PD-1 blockade plus chemotherapy
Trial registration: ClinicalTrials.gov identifier: NCT03134872.
Abstract
Background
Although programmed cell death 1 (PD-1) blockade plus chemotherapy can significantly prolong the progression-free survival (PFS) and overall survival (OS) in first-line settings in patients with driver-negative advanced non-small-cell lung cancer (NSCLC), the predictive biomarkers remain undetermined. Here, we investigated the predictive value of tumor immune microenvironmental marker expression to characterize the response features to PD-1 blockade plus chemotherapy.
Methods
Tumor tissue samples at baseline were prospectively collected from 144 locally advanced or metastatic NSCLC patients without driver gene alterations who received camrelizumab plus chemotherapy or chemotherapy alone. Tumor immune microenvironmental markers, including PD-1 ligand (PD-L1), CD8, CD68, CD4 and forkhead box P3, were assessed using multiplex immunofluorescence (mIF) assays. Kaplan-Meier curves were used to determine treatment outcome differences according to their expression status. Mutational profiles were compared between tumors with distinct expression levels of these markers and their combinations.
Results
Responders had significantly higher CD8/PD-L1 (P = 0.015) or CD68/PD-L1 co-expression levels (P = 0.021) than non-responders in the camrelizumab plus chemotherapy group, while no difference was observed in the chemotherapy group. Patients with high CD8/PD-L1 or CD68/PD-L1 co-expression level was associated with significantly longer PFS (P = 0.002, P = 0.024; respectively) and OS (P = 0.006, P = 0.026; respectively) than those with low co-expression in camrelizumab plus chemotherapy group. When comparing survival in the camrelizumab plus chemotherapy with chemotherapy by CD8/PD-L1 co-expression stratification, significantly better PFS (P = 0.003) and OS (P = 0.032) were observed in high co-expression subgroups. The predictive value of CD8/PD-L1 and CD68/PD-L1 co-expression remained statistically significant for PFS and OS when adjusting clinicopathological features. Although the prevalence of TP53 or KRAS mutations was similar between patients with and without CD8/PD-L1 or CD68/PD-L1 co-expression, the positive groups had a significantly higher proportion of TP53/KRAS co-mutations than the negative groups (both 13.0% vs. 0.0%, P = 0.023). Notably, enriched PI3K (P = 0.012) and cell cycle pathway (P = 0.021) were found in the CD8/PD-L1 co-expression group.
Conclusion
Tumor immune microenvironmental marker expression, especially CD8/PD-L1 or CD68/PD-L1 co-expression, was associated with the efficacy of PD-1 blockade plus chemotherapy as first-line treatment in patients with advanced NSCLC.
Abbreviations
-
- ALK
-
- anaplastic lymphoma kinase
-
- CNV
-
- copy number variation
-
- CR
-
- complete response
-
- DCR
-
- disease control rate
-
- ECOG PS
-
- Eastern Cooperative Oncology Group performance status
-
- EGFR
-
- epidermal growth factor receptor
-
- FFPE
-
- formalin-fixed paraffin-embedded
-
- FOXP3
-
- forkhead box P3
-
- HLA
-
- human leukocyte antigen
-
- HR
-
- hazard ratio
-
- ICIs
-
- immune checkpoint inhibitors
-
- IFN-γ
-
- interferon-gamma
-
- InDels
-
- insertions/deletions
-
- ITT
-
- intention-to-treat
-
- LUSC
-
- lung squamous cell carcinoma
-
- MHC-II
-
- major histocompatibility complex class II
-
- mIF
-
- multiplex immunofluorescence
-
- mIHC
-
- multiplex immunohistochemistry
-
- NGS
-
- next generation sequencing
-
- NSCLC
-
- non-small-cell lung cancer
-
- ORR
-
- objective response rate
-
- OS
-
- overall survival
-
- PD
-
- disease progression
-
- PD-1
-
- programmed cell death 1
-
- PD-L1
-
- programmed death-ligand 1
-
- PFS
-
- progression-free survival
-
- PR
-
- partial response
-
- SD
-
- stable disease
-
- SNV
-
- single nucleotide variation
-
- TILs
-
- tumor-infiltrating lymphocytes
-
- TMB
-
- tumor mutation burden
-
- TME
-
- tumor microenvironment
-
- TNB
-
- tumor neoantigen burden
-
- VAF
-
- variant allele frequency
-
- WES
-
- whole-exome sequencing
1 BACKGROUND
Lung cancers remain the leading cause of cancer-related death worldwide. Approximately 85% of them are non-small-cell lung cancer (NSCLC). Without druggable genetic aberrations, advanced-stage patients often suffer from poor survival due to limited treatment strategies [1]. Recently, immune checkpoint inhibitors (ICIs) targeting programmed cell death 1 (PD-1) or its ligand (PD-L1) have revolutionized the treatment landscape of driver-negative locally advanced or metastatic NSCLC [1-3]. A series of global or regional phase III trials have consistently demonstrated that anti-PD-1/PD-L1 antibody plus chemotherapy could significantly improve the progression-free survival (PFS) and/or overall survival (OS) compared with chemotherapy alone in the first-line setting in locally advanced or metastatic NSCLC, irrespective of the PD-L1 expression status [4-9]. Nevertheless, the reported objective response rate (ORR) ranged from 48% to 64% in the intention-to-treat (ITT) population, suggesting that nearly half of them could not benefit from this regimen [1]. Hence, there is an urgent need to identify reliable predictive biomarkers for this therapeutic strategy.
To date, there are three officially approved predictive biomarkers to guide the clinical application of anti-PD-1/PD-L1 monotherapy, including PD-L1 expression, tumor mutational burden (TMB) and microsatellite instability status [10], with moderate predictive performance. Besides that, CD8+ tumor-infiltrating lymphocytes (TILs) [11, 12], interferon-gamma (IFN-γ) gene signature [13], tertiary lymphoid structures [14, 15] and specific genetic aberrations [16, 17] also showed potential in predicting the efficacy of ICI monotherapy in various solid tumors. Considering the significant role of the tumor immune microenvironment in successful antitumor immunity, there is increasing recognition that incorporating distinct tumor immune microenvironmental features could be essential for developing more reliable predictive biomarkers [18]. For example, the combination of TMB and PD-L1 expression or T cell-inflamed gene expression profile showed an improved predictive utility compared with single biomarkers for anti-PD-1/PD-L1 monotherapy [19]. Similarly, previous studies have reported that tumor microenvironment (TME) classification based on PD-L1 expression and CD8+ TILs or CD68+ macrophages were associated with response and survival in advanced NSCLC patients treated with anti-PD-1/PD-L1 monotherapy [20-22], suggesting the robustly predictive value of the co-expression of distinct tumor immune microenvironmental markers. More importantly, evaluating the combination of the tumor immune microenvironmental features with multiplex immunohistochemistry (IHC) or multiplex immunofluorescence (mIF) showed improved performance over PD-L1 expression and TMB [21]. However, as for PD-1 blockade plus chemotherapy, both the PD-L1 expression and TMB were ineffective in predicting response. Only major histocompatibility complex class II (MHC-II) antigen presentation pathway expression was recently reported to be useful for predicting the benefit of PD-1 blockade plus chemotherapy in non-squamous NSCLC [23]. Thus, whether incorporating distinct tumor immune microenvironmental features could serve as a robust biomarker for PD-1 blockade plus chemotherapy in locally advanced or metastatic NSCLC remains undetermined.
In our previously reported phase III trial (CameL, NCT03134872), the results showed that camrelizumab plus chemotherapy could significantly improve PFS (median: 11.3 vs. 8.3 months; hazard ratio [HR] = 0.60, 95% confidence interval [CI], 0.45-0.79; P = 0.0001) compared with chemotherapy alone as first-line treatment for patients with locally advanced or metastatic, non-squamous NSCLC patients without epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK) alterations [7]. Initial biomarker analysis revealed neither PD-L1 expression nor TMB could predict response to camrelizumab plus chemotherapy. Here, we aimed to investigate the predictive value of tumor immune microenvironmental markers expression detected by mIF assays in pretreatment tumor tissue samples, including PD-L1, CD8, CD68, CD4, forkhead box P3 (FOXP3) and their combinations to characterize the features of response to PD-1 blockade plus chemotherapy. The reasons for choosing CD68, CD4 and FOXP3 were because CD68 is the classic surface marker of macrophages, CD4 is another important marker of T cells to antitumor immunity, and FOXP3 is the classic surface marker of regulatory T cells (Tregs). Besides PD-L1 and CD8, these markers’ expression levels could partly represent the relative abundance of CD68+ macrophages, CD4+ T cells and FOXP3+ Tregs. These three types of immune cells play a significant role in successful antitumor immunity. Thus, detecting CD68, CD4 and FOXP3, together with PD-L1 and CD8, could be more useful to characterize the features of response to PD-1 blockade plus chemotherapy or chemotherapy alone in locally advanced or metastatic NSCLC. Given the close relationship between tumor genotype and immunophenotype, we also compared the mutational landscape of tumors with the distinct expression levels of these markers.
2 PATIENTS AND METHODS
2.1 Study design
CameL is a randomized, open-label, multicenter, phase III trial conducted in 52 medical centers in China. The clinical protocol was approved by the respective institutional review boards and ethics committees. All participants provided written informed consent. This study was the pre-defined biomarker analysis detailed in our previous publication and its appendix [7]. Briefly, patients with the following criteria were eligible: aged 18-70 years, histologically or cytologically confirmed stage IIIB-IV non-squamous NSCLC (as per the International Association for the Study of Lung Cancer Staging Handbook in Thoracic Oncology, 8th Edition) without EGFR and ALK alteration, Eastern Cooperative Oncology Group performance status (ECOG PS) of 0 or 1, no previous systemic chemotherapy, at least one measurable lesion per Response Evaluation Criteria in Solid Tumors version 1.1 (RECIST v1.1), and a life expectancy of ≥3 months. Patients were excluded if they had untreated central nervous system metastases and corticosteroid use within 2 weeks before study treatment was recorded. Details of the study design, including full inclusion and exclusion criteria, are shown in our previous article [7].
2.2 Sample collection
According to the previous protocol, we collected the fresh samples at baseline or archival tumor tissues within 6 months before the protocol-defined treatments. Pretreatment blood samples (8-10 mL) were also collected in ethylene diamine tetraacetic acid (EDTA)-coated tubes (BD Biosciences, Franklin Lakes, NJ, USA) and centrifuged at 1800g for 10 minutes within 2 hours of collection to separate white blood cells.
2.3 DNA extraction and library preparation
Formalin-fixed paraffin-embedded (FFPE) tumor samples with ≥20% tumor cell content were qualified and included. White blood cell sediments were used for genomic DNA extraction as the germline controls. Genomic DNA from tumor tissues and whole blood control samples were extracted with the QIAamp DNA FFPE Tissue Kit (Qiagen, GmBH, Germany) and DNeasy Blood and Tissue Kit (Qiagen). Library preparations were performed with KAPA Hyper Prep Kit (KAPA Biosystems, Wilmington, MA, USA). Target enrichment was performed using the xGen Exome Research Panel and Hybridization (Integrated DNA Technology, Coralville, IA, USA) and Wash Reagents Kit (Integrated DNA Technology) according to the manufacturer's protocol. Sequencing was performed on the Illumina HiSeq4000 platform using PE150 sequencing chemistry (Illumina, San Diego, CA, USA). For the targeted panel, customized xGen lockdown probes (Integrated DNA Technologies) targeting 425 cancer-relevant genes were used for hybridization enrichment. The capture reaction was performed with Dynabeads M-270 (Life Technologies, Waltham, MA, USA), xGen lockdown hybridization, and wash kit (Integrated DNA Technologies) following the manufacturer's protocols. Captured libraries were obtained using on-bead PCR amplified with Illumina p5 (5′-AATGATACGGCGACCGA-3′) and p7 primers (5′-CAAGCAGAAGACGGCATACGAGAT-3′) in KAPA HiFi HotStart ReadyMix (KAPA Biosystems), followed by purification using Agencourt AMPure XP beads (Beckman Coulter, Pasadena, CA, US). Libraries were quantified with quantitative PCR using the KAPA Library Quantification kit (KAPA Biosystems), and the size was determined using the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). The target enriched library was then sequenced on the HiSeq4000 NGS platform (Illumina), following the manufacturer's instructions.
2.4 Sequence alignment and data processing
Base calling was performed on bcl2fastq V.2.16.0.10 [24] to generate sequence reads in the FASTQ format. Quality control was performed using the Trimmomatic software [25]. High-quality reads were mapped to the human genome (hg19, GRCh37 Genome Reference Consortium Human Reference 37) using the Burrows-Wheeler Aligner (BWA) V.0.7.12 with BurrowsWheeler Aligner's maximal exact matches (BWA-MEM) [26, 27] algorithm and default parameters to create SAM files. Picard V.1.119 (https://broadinstitute.github.io/picard/) was used to convert SAM files to compressed BAM files, which were then sorted according to chromosome coordinates. The Genome Analysis Toolkit (GATK) [26] was used to locally realign the BAM files at intervals with insertions/deletion (indels) mismatches and recalibrate base quality scores of reads in BAM files.
2.5 Data filtering and variants calling
Adaptor sequences and low-quality bases of sequenced reads were trimmed using Trimmomatic (v0.36) [25] to obtain clean reads, which were mapped to the human reference genome (hg19) using BWA (v0.7.17). Genome Analysis ToolKit (version 4.0.7.0) [26] was used to sort, mask duplication and recalibrate the base quality score for the mapping result recorded in the BAM files produced by the above step. Single nucleotide variants (SNVs) and small InDels were called via MuTect2 [28] with tumor-normal mode. To avoid false-positive results, SNVs and InDels that appeared on the blacklist (including sequence-specific errors, repeat regions, segmental duplications and lowly mappable regions recorded in ENCODE [https://www.encodeproject.org/]) were removed. Only SNVs and InDels with coverage depth not smaller than 40, variant supporting reads not smaller than 4, and variant allele frequency not smaller than 5% were retained as filtered somatic mutations. After annotation by ANNOVAR [29], we filtered out variants either in introns or synonymous mutations. Furthermore, variants with minor allele frequency ≥1% in the Exome Aggregation Consortium (ExAC) [30] and Genome Aggregation Database (gnomAD) (http://gnomad.broadinstitute.org/) were removed. Lastly, the mutations that were not recorded in the COSMIC database were filtered out.
2.6 Copy number variation (CNV) analysis
In the reference mode, somatic CNVs were identified using the CNVkit (v0.9.5) (https://cnvkit.readthedocs.io/en/v0.9.5/bias.html). The baseline of normalized sequencing depth on targeted regions was constructed based on a panel of normal samples. For each tumor sample, the log2 transformed ratio of normalized sequencing depth on each targeted region between it and the baseline was calculated. If a gene contained at least 5 targeted regions, the median log2 transformed ratio was considered the CNV value of this gene. Gene depletion or amplification was identified if the CNV value of this gene was not larger than 1 or not smaller than 4, respectively.
2.7 TMB calculation
The number of somatic nonsynonymous SNVs (allele frequency ≥0.005) was quantified, and the value was extrapolated to the whole exome using a validated algorithm [31]. TMB was measured in mutations per Mb. The TMB in this study was defined as the number of somatic, coding, base substitutions, and indel mutations per megabases of the genome examined. Only the regions with sequencing depth larger than 100× after deduplication were used for TMB calculation. Germline alterations in the Single Nucleotide Polymorphism database (dbSNP) or occurring with two or more counts in the ExAC database were not counted. To calculate the TMB per megabases, the total number of mutations counted was divided by the size of the coding region of the targeted territory.
2.8 Neoantigen identification
Human leukocyte antigen (HLA) class I alleles were called from the matched normal exome sequencing data for each patient using HLA-HD (v1.2.0.1) [32]. Neoepitope presentation was then predicted for tumor-specific peptides of length 9-11 using the eluted-ligand mode of NetMHCpan-4.0 [33]. The main criteria for neoantigens included (i) derived from tumor-specific alterations (including missense, frameshift, inframe indels, and fusions); (ii) high predicted affinity to HLA class I alleles [half maximal inhibitory concentration (IC50) < 500 nmol/L] with k-mer of 9-11 length; (iii) fold change >10 comparing to wild-type binding affinity. HLA binding affinity was predicted via the IEDB-recommended model using all variant-containing 9-11 mer for HLA-A/B/C binding estimations. HLA typing for patients was performed in silico using HLA-ATHLATES [34] according to the recommended algorithm. Tumor neoantigen burden (TNB) was used to calculate the number of all identified neoantigens.
2.9 mIF staining
Manual mIF staining was performed in 4-μm sequential histologic tumor sections obtained from FFPE tumor blocks using the Opal 7-Color fIHC Kit (Akoya Biosciences, Marlborough, MA, USA) based on the Tyramide Signal Amplification (TSA) kit (Biotium, Fremont, CA, USA). The stained slides were scanned using a Vectra multispectral microscope (Akoya Biosciences). The IF markers were grouped into the panel consisting of FOXP3 (dilution 1:100; Abcam, Shanghai, China), CD8 (dilution 1:400; Zsbio, Beijing, China), PD-L1 (clone SP142, dilution 1:25, Zsbio), CD4 (dilution 1:100; Zsbio), and CD68 (dilution 1:500; Zsbio). Briefly, the slides were deparaffinized, rehydrated, and subjected to epitope retrieval by boiling in Tris-EDTA buffer (pH 9.0; Zsbio) for 20 minutes at 97°C. Endogenous peroxidases were then blocked by incubation in Antibody Diluent/Block (Akoya Biosciences) for 10 minutes. Only one antigen was detected in each round, including primary antibody incubation, secondary antibody incubation, and TSA visualization, followed by labeling the next antibody after epitope retrieval and protein blocking as before. In this panel, antigens were detected in the following order: FOXP3, CD8, PD-L1, CD4 and CD68. Finally, these slides were stained with 4’,6-diamidino-2-phenylindole (DAPI; Selleckchem, Shanghai, China) for nuclei and mounted with anti-quenching sealing tablets. Human tonsil FFPE tissues were used with and without primary antibodies as positive and negative controls, respectively.
2.10 mIF image acquisition
The stained slides were scanned with a Vectra 3.0 microscope system (PerkinElmer, Waltham, MA, USA) under fluorescent illumination. From each slide, Vectra automatically captured the fluorescent spectra from 420 nm to 720 nm at 20-nm intervals with the same exposure time and then combined the captured images to create a single stack image that retained the particular spectral signature of all markers. After the specimens were scanned at low magnification (×10), the specific fields in the tumor area were scanned at high resolution (×20) to capture various elements of tumor heterogeneity.
2.11 Multispectral analysis
Tumor multispectral images were analyzed by two experienced pathologists using the tissue segmentation tool of the InForm 3.0 software (Akoya Biosciences). Representative multispectral images were selected as training samples to build an algorithm (tissue segmentation, cell segmentation, phenotyping tool and positivity score) using the InForm software. All markers in the panel were analyzed one by one at a time in the same project, and all cases were analyzed with the same algorithm. After batch analysis, the performance of the algorithm was evaluated visually for all cases. The expression level was recorded as the positive cell density score and percent of stained cells. For PD-L1 expression, we extracted its tumor proportion score (defined as the percentage of viable tumor cells showing partial or complete membrane staining at any intensity) from our previous report [7]. The definition of PD-L1 tumor proportion score differed from the definition of the percent of positively stained cells. PD-L1 tumor proportion score, measured by the 22C3 assay using immunohistochemical staining, was defined as the percentage of viable tumor cells showing partial or complete membrane staining at any intensity. The percent of positively stained cells in this study, measured by the SP142 assay using multiplex immunofluorescence staining, was defined as the positive marker expression group as that with a percent of positively stained cells ≥ median level of the study cohort.
2.12 Statistical analysis
Responders were defined as patients who achieved complete response (CR) or partial response (PR), and non-responders as patients who achieved stable disease (SD) or disease progression (PD) according to the most recent high-quality literature [19, 35]. Categorical variables were analyzed using the chi-squared or Fisher's exact test, and continuous variables were analyzed by analysis of variance (ANOVA) and Tukey's multiple comparison tests. Mann-Whitney U tests or Kruskal-Wallis rank-sum tests were used to compare continuous variables across multiple groups. Associations between treatment response and different expression levels of tumor immune microenvironmental markers were conducted using the Mann-Whitney U tests. The correlation between positive cell density score and percent of stained cells of a specific marker was conducted using Spearman's correlation analysis. ORR and disease control rate (DCR) were analyzed, and the corresponding 95% CI was estimated using the Clopper-Pearson method. Between-group comparisons were assessed using the stratified Cochran-Mantel-Haenszel method. The Kaplan-Meier curves were used to estimate the median PFS and OS, with the 95% CIs estimated using the Brookmeyer and Crowley method. Between-group comparisons in PFS and OS were assessed using a stratified log-rank test. HR and associated 95% CI was calculated based on a stratified Cox proportional hazards model. All statistical analyses were conducted using GraphPad PRISM 6.0 (GraphPad Software, San Diego, CA, USA) and the SPSS statistical software version 22.0 (SPSS Inc., Chicago, IL, USA). Two-sided P < 0.05 was considered statistically significant.
3 RESULTS
3.1 Patient characteristics
Pretreatment tissue samples from 144 patients with previously untreated stage IIIB-IV non-squamous NSCLC (77 from camrelizumab plus chemotherapy group and 67 from chemotherapy only group) were collected for this biomarker analysis (Supplementary Figure S1 and Table 1). Baseline clinical features, including age, sex, smoking history, ECOG PS, disease stage, brain metastasis, PD-L1 expression level and clinical outcomes, were balanced between the biomarker evaluable cohort and ITT population (Supplementary Table S1 and Table 1). Survival outcomes of the biomarker evaluable cohort were consistent with those of the ITT population (data not shown). Although age distribution was statistically different, other clinical parameters, including sex, smoking history, ECOG PS, disease stage, brain metastasis, histological type and PD-L1 expression level, were balanced between camrelizumab plus chemotherapy and chemotherapy only groups in this analysis (Table 1). Patients treated with camrelizumab plus chemotherapy had significantly higher ORR than those treated with chemotherapy (59.7% vs. 37.3%, P = 0.007).
| Camrelizumb plus chemotherapy | Placebo plus chemotherapy | ||
|---|---|---|---|
| Items | (n = 77 [53.5%]) | (n = 67 [46.5%]) | P value |
| Age | |||
| ≥65 years | 12 (15.6%) | 24 (35.8%) | 0.005 |
| <65 years | 65 (84.4%) | 43 (64.2%) | |
| Sex | |||
| Male | 54 (70.1%) | 49 (73.1%) | 0.690 |
| Female | 23 (29.9%) | 18 (26.9%) | |
| Smoking history | |||
| ≥400 cigarette-years | 54 (70.1%) | 42 (62.7%) | 0.345 |
| <400 cigarette-years | 23 (29.9%) | 25 (37.3%) | |
| ECOG performance status | |||
| 0 | 23 (29.9%) | 15 (22.3%) | 0.310 |
| 1 | 54 (70.1%) | 52 (77.7%) | |
| Disease stage* | |||
| IIIB/IIIC | 15 (19.5%) | 10 (14.9%) | 0.472 |
| IV | 62 (80.5%) | 57 (85.1%) | |
| Histological type | |||
| Adenocarcinoma | 75 (97.4%) | 65 (97.0%) | 0.714 |
| Non-adenocarcinoma | 2 (2.6%) | 2 (3.0%) | |
| Brain metastases at baseline | |||
| Yes | 1 (1.3%) | 2 (3.0%) | 0.903 |
| No | 76 (98.7%) | 65 (97.0%) | |
| PD-L1 tumor proportion score | |||
| <1% | 17 (22.1%) | 20 (29.9%) | 0.287 |
| ≥1% | 59 (76.6%) | 42 (62.7%) | |
| 1%-49% | 49 (63.6%) | 33 (49.3%) | |
| ≥50% | 10 (13.0%) | 9 (13.4%) | |
| NE | 1 (1.3%) | 5 (7.5%) | |
| Best Response | |||
| CR | 0 (0.0%) | 0 (0.0%) | |
| PR | 46 (59.7%) | 25 (37.3%) | 0.007 |
| SD | 21 (27.3%) | 27 (40.3%) | |
| PD | 5 (6.5%) | 11 (16.4%) | |
| NE | 5 (6.5%) | 4 (6.0%) | |
- ECOG, Eastern Cooperative Oncology Group. CR, complete response. PR, partial response. SD, stable disease. PD, disease progression. NE, not evaluable.
- Non-adenocarcinoma: two patients with large cell carcinoma in the camrelizumab plus chemotherapy group; two patients with adenosquamous carcinoma in the chemotherapy group.
- * Stage was recorded as per the International Association for the Study of Lung Cancer Staging Handbook in Thoracic Oncology, 8th Edition.
3.2 Correlations between the expression level of tumor immune microenvironmental markers and treatment response
Among the biomarker evaluable cohort, all of the included pretreatment tissue samples were qualified and could be used for mIF assays. Tumor immune microenvironmental markers, including PD-L1, CD8, CD68, CD4 and FOXP3, were investigated (Figure 1A). We first evaluated the correlations between the expression level of each marker and objective response in the camrelizumab plus chemotherapy group. As shown in Figure 1B, responders had both significantly higher PD-L1 positive cell density score (P = 0.013) and percent of stained cells (P = 0.020) than non-responders. Markedly higher CD8 positive cell density score (P = 0.046) and percent of CD68-stained cells (P = 0.048) were also observed in responders, while CD4 and FOXP3 expression levels showed no correlation with objective response (Figure 1B).
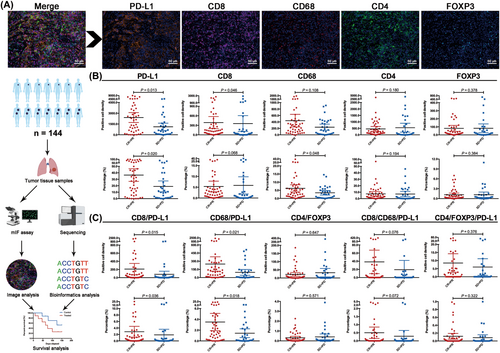
Next, we investigated the correlations between two/three of their biologically rational combinations and objective response. We found that responders had significantly higher CD8/PD-L1 and CD68/PD-L1 positive cell density scores and percent of stained cells than non-responders, whereas CD4/FOXP3, CD8/CD68/PD-L1 and CD4/FOXP3/PD-L1 showed similar co-expression level between responders and non-responders (Figure 1C). Intriguingly, patients with objective response (CR + PR) had similar expression levels of these tumor immune microenvironmental markers to those with SD + PD in the chemotherapy-only group (Supplementary Figure S2).
3.3 Predictive value of each tumor immune microenvironmental marker expression
To investigate the predictive value of each marker expression, we defined the positive marker expression group as that with a positive cell density score or percent of stained cells ≥ median level in the analyzed population. As shown in Supplementary Figure S3A, PD-L1 positive cell density score was only correlated with significantly longer PFS (P = 0.012), while OS was comparable between positive and negative cell density score groups (P = 0.111) in patients who received camrelizumab plus chemotherapy; CD8 and FOXP3 positive cell density scores correlated with longer OS, but not PFS, in the camrelizumab plus chemotherapy group, respectively; both CD68 and CD4 positive cell density scores did not correlate with PFS and OS in the camrelizumab plus chemotherapy group. A high percentage of CD8 or CD4 stained cells was associated with longer OS (P = 0.018, P = 0.042; respectively), while a high percent of PD-L1, CD68 and FOXP3 stained cells did not correlate with PFS and OS in this group (Supplementary Figure S3B). Comparatively, positive cell density scores and percent of stained cells for all of these tumor immune microenvironmental markers showed no correlation with PFS and OS in the chemotherapy-only group (Supplementary Figure S4).
3.4 Predictive value of their biologically rational combinations
Whether biologically rational combinations of these markers from mIF assays were predictive for PD-1 blockade plus chemotherapy remains unknown. Herein, we analyzed the associations between different markers’ combinations and clinical outcomes in the camrelizumab plus chemotherapy group. The results showed that CD8/PD-L1, CD68/PD-L1, CD4/FOXP3/PD-L1 and CD8/CD68/PD-L1 (but not CD4/FOXP3) co-expression levels by calculating cell density score and percent of stained cell could predict survival benefit from camrelizumab plus chemotherapy (Supplementary Figure S5). According to the P values, high CD8/PD-L1 and CD68/PD-L1 co-expression levels showed a superior predictive value (Supplementary Figure S5). Considering the biological and statistical significance and convenient clinical application, we used the percent of stained cells to define the high versus low co-expression groups since the percent of stained cells was significantly associated with cell density score (Supplementary Figure S6). Similarly, we defined the high co-expression group as that with a percent of stained cells ≥ median level and the low co-expression group as that with a percent of stained cells < median level in the analyzed population. The representative images are shown in Figure 2A. The ORR of camrelizumab plus chemotherapy was significantly higher in patients with high CD8/PD-L1 co-expression level (n = 39) than those with low co-expression level (n = 38) (ORR: 71.8% vs. 47.4%, P = 0.029; Figure 2B), while ORR was similar between high (n = 34) and low (n = 33) co-expression groups in the chemotherapy-only group (ORR: 38.3% vs. 36.4%, P > 0.05; Figure 2B). Patients with high CD8/PD-L1 co-expression levels were associated with significantly longer PFS (P = 0.002; Figure 2C) and OS (P = 0.006; Figure 2D) than those with low CD8/PD-L1 co-expression level in camrelizumab plus chemotherapy group. Moreover, when comparing survival between the camrelizumab plus chemotherapy group and chemotherapy only group by CD8/PD-L1 co-expression stratification, markedly longer PFS (P = 0.003) and OS (P = 0.032) were observed in patients with high CD8/PD-L1 co-expression level, whereas no differences for PFS and OS were found in those with low CD8/PD-L1 co-expression level (Figure 2C-D).
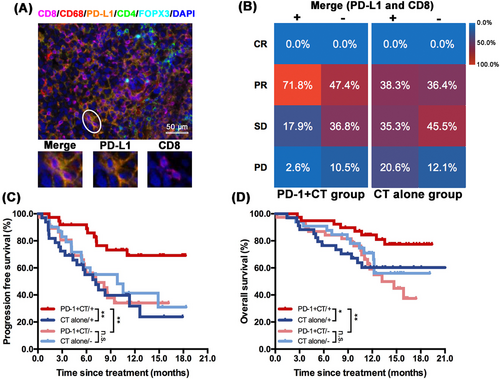
Similarly, high CD68/PD-L1 co-expression (n = 39; representative images showed in Figure 3A) level was also associated with numerically better ORR (69.2% vs. 50.0%, P = 0.085; Figure 3B) in camrelizumab plus chemotherapy group, though it did not reach the statistical significance mainly due to the limited sample size. Notably, patients with high CD68/PD-L1 co-expression level had significantly prolonged PFS (P = 0.024; Figure 3C) and OS (P = 0.026; Figure 3D) than those with low CD68/PD-L1 co-expression level (n = 38) in camrelizumab plus chemotherapy group. Camrelizumab plus chemotherapy also showed significantly better PFS (P = 0.004; Figure 3C) and OS (P = 0.022; Figure 3D) than chemotherapy in patients with high CD68/PD-L1 co-expression levels, whereas PFS and OS were analogous between camrelizumab plus chemotherapy and chemotherapy alone in patients with low CD68/PD-L1 co-expression level. The ORR was similar between high (n = 34) and low (n = 33) CD68/PD-L1 co-expression subgroups in chemotherapy group (38.2% vs. 36.4%, P > 0.05; Figure 3B). When we adjusted the clinicopathological features, the predictive value of CD8/PD-L1 and CD68/PD-L1 co-expression remained statistically significant for both PFS and OS in the camrelizumab plus chemotherapy group (Supplementary Table S2). Neither CD8/PD-L1 nor CD68/PD-L1 positive cell density score and percent of stained cells showed a predictive value for treatment response and survival outcomes in patients treated with chemotherapy (Supplementary Figure S7). We have also explored the significance of CD8/CD68/PD-L1 co-expression as a predictive biomarker in patients who received camrelizumab plus chemotherapy. The results showed that positive CD8/CD68/PD-L1 co-expression was also associated with significantly longer PFS (P = 0.049) and OS (P = 0.043) than negative groups (Supplementary Figure S5). However, it did not correlate with better response (P = 0.076 for positive cell density; P = 0.072 for positive percent of stained cell; Figure 1C). Hence, positive CD8/CD68/PD-L1 co-expression did not show a superior predictive value than positive CD8/PD-L1 or CD68/PD-L1 co-expression in the camrelizumab plus chemotherapy group.

3.5 Mutational profiles of tumors with distinct co-expression levels of CD8/PD-L1 or CD68/PD-L1
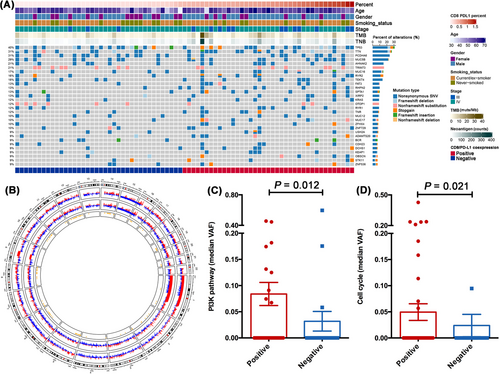
Next, we surveyed whether different co-expression levels of CD8/PD-L1 or CD68/PD-L1 had distinct mutational profiles. All pretreatment tissue samples from the 144 patients were eligible for targeted-panel sequencing (Supplementary Figure S1). Tumors with high CD8/PD-L1 or CD68/PD-L1 co-expression levels had similar TMB and TNB levels to those with low CD8/PD-L1 or CD68/PD-L1 co-expression levels (Figure 4A and Supplementary Figure S8A). The most frequently altered genes were TP53 (40%), TTN (31%), PCDHA6 (31%), MUC5B (29%), AHNAK2 (21%), TRIM73 (21%), TEKT4 (18%), MUC16 (17%), RYR2 (16%), FAT3 (13%), RHPN2 (13%), SPTA1 (13%), XIRP2 (13%), and KRAS (12%) in all included patients (n = 144). Among them, tumors with high CD8/PD-L1 co-expression levels had a significantly higher proportion of RHPN2 mutations (23.2% vs. 3.1%, P = 0.020) than those with low CD8/PD-L1 co-expression levels, while none of them showed differences between high versus low CD68/PD-L1 co-expression groups (Supplementary Figure S8A). Although the prevalence of TP53 or KRAS mutations was similar between high and low CD8/PD-L1 or CD68/PD-L1 co-expression groups, tumors with high CD8/PD-L1 or CD68/PD-L1 co-expression levels had a significantly higher proportion of TP53/KRAS co-mutations than those with low co-expression level (both 13.0% vs. 0.0%, P = 0.023; Figure 4A and Supplementary Figure S8A). CNV analysis showed significant deletion of chromosomes 5p and 8q in the CD8/PD-L1 positive group and focal amplification of chromosomes 5p, 7p, and 14q in the CD8/PD-L1 negative group (Figure 4B), while CNV was obvious in chromosomes 8q, 10q, 12q and 14q between CD68/PD-L1 positive and negative group (Supplementary Figure S8B). Notably, tumors from CD8/PD-L1 positive group compared with those from the negative group were enriched in the PI3K (P = 0.012; Figure 4C) and cell cycle pathway (P = 0.021; Figure 4D), whereas these two pathways were not different between CD68/PD-L1 positive and negative groups (P = 0.171, P = 0.235; Supplementary Figure S8C-D).
4 DISCUSSION
PD-1 blockade plus chemotherapy has become the new standard of care as first-line treatment for patients with locally advanced or metastatic NSCLC [1-8], but the predictive biomarkers remain largely unknown. In this study, we found that responders had significantly higher CD8/PD-L1 or CD68/PD-L1 co-expression levels than non-responders in patients treated with camrelizumab plus chemotherapy, while it showed no difference in patients treated with chemotherapy alone. Moreover, high CD8/PD-L1 or CD68/PD-L1 co-expression levels were also associated with significantly longer PFS and OS in the camrelizumab plus chemotherapy group. Furthermore, we observed that tumors with high CD8/PD-L1 or CD68/PD-L1 co-expression levels had a significantly higher proportion of TP53/KRAS co-mutations. Meanwhile, tumors with high CD8/PD-L1 co-expression levels were enriched in the cell cycle and PI3K pathway compared with those with low co-expression levels. Taken together, these findings demonstrate that biologically rational combinations of tumor immune microenvironmental markers, especially CD8/PD-L1 and CD68/PD-L1 co-expression, could serve as predictive biomarkers for PD-1 blockade plus chemotherapy in patients with locally advanced or metastatic NSCLC.
Given the complexity of a successful antitumor immune response, accumulating evidence showed that incorporating distinct tumor immune microenvironmental features is key to developing robust predictors of response. Since the first proposal that TME could be classified into four different subtypes according to the PD-L1 expression and presence or absence of TILs [36], several studies have attempted to investigate the association of this classification with distinct clinical outcomes from anti-PD-1/PD-L1 monotherapy. Zhang et al.[22] retrospectively collected the data of 378 patients with NSCLC and analyzed PD-L1 expression and CD8-positive TILs by mIHC assay. They reported that a higher level of CD8 and PD-L1 double-positive TILs was correlated with a better outcome to anti-PD-1/PD-L1 monotherapy, while it was associated with significantly worse PFS in the non-immunotherapy cohort. A recent study assessed the efficacy of anti-PD-1/PD-L1 monotherapy based on the four different TME subtypes in advanced NSCLC and revealed distinct treatment responses and PFS according to the different subtypes [21]. Importantly, in this present study, we further extended the predictive potent of CD8 and PD-L1. We observed that patients with positive CD8/PD-L1 co-expression detected by mIF assay had significantly better ORR, PFS and OS than those with negative CD8/PD-L1 co-expression when receiving PD-1 blockade plus chemotherapy group. In contrast, pretreatment PD-L1 expression and tissue TMB failed to show their predictive values for PD-1 blockade plus chemotherapy in locally advanced or metastatic NSCLC [7, 37].
Additionally, our results also demonstrated that CD68/PD-L1 co-expression was also associated with longer PFS and OS in the PD-1 blockade plus chemotherapy group. CD68 is the classic tumor-associated macrophage marker, a double-edged sword in antitumor immunity. Previous studies revealed CD68+ macrophages were the predominant immune cell type expressing PD-L1 [38, 39]. A high level of PD-L1 in macrophages was significantly correlated with a high level of PD-L1 expression in tumor cells and CD8+ T cells infiltration, indicating an inflamed TME phenotype [38, 39]. Positive PD-L1 expression in CD68+ macrophages was associated with significantly better treatment outcomes in patients treated with anti-PD-1/PD-L1 monotherapy [39, 40]. Moreover, a recent study reported that colocalization of PD-L1 expression in tumor cells and CD68+ macrophages was associated with higher rates of pathological complete response to durvalumab and chemotherapy in triple-negative breast cancer [38]. Collectively, these findings suggest that TME classification based on PD-L1 expression and CD68+ macrophages could also serve as potential predictive biomarkers for PD-1 blockade plus chemotherapy in locally advanced or metastatic NSCLC.
Accumulating evidence revealed that tumor genotypes might impact TME phenotypes [41, 42]. Thus, we further explored their mutational profiles and found that tumors from the positive CD8/PD-L1 or CD68/PD-L1 co-expression group had a significantly higher proportion of TP53/KRAS co-mutations, together with enriched cell cycle and PI3K pathway compared with those from the negative group. Similarly, a previous study reported that TP53/KRAS co-mutant NSCLC had the highest proportion of PD-L1+/CD8A+ than those with single mutation or wild-type tumors, together with superior clinical benefit to PD-1 inhibitors [43]. Shirasawa et al.[21] reported that advanced NSCLC with TP53/KRAS co-mutations were more likely to have Type I (PD-L1+/TIL+) tumors, and four of five patients with TP53/KRAS co-mutations responded to PD-1 inhibitors. Nevertheless, the reason why tumors from positive CD8/PD-L1 or CD68/PD-L1 co-expression group had enriched cell cycle and PI3K pathway remain largely unknown. Future investigations are warranted to clarify the potential mechanism of TP53/KRAS co-mutations, cell cycle, and PI3K pathway on TME phenotype and response to anti-PD-1/PD-L1 based treatments.
Traditional IHC cannot capture the accurate spatial information of tumor immune microenvironmental markers expression. The organization of TME is predictive and prognostic in multiple solid tumors. mIHC/mIF assays have many advantages, including simultaneous visualization of multiple protein markers, clear spatial information, and co-expression localization of different markers, thus accurately providing the details of cellular co-expression and geography within the TME [44-46]. In a recent meta-analysis, Lu et al. [44] investigated the indirect comparison of biomarker modalities for predicting the response to anti-PD-1/PD-L1 monotherapy and found that mIHC/mIF, in evaluating TME features, was associated with improved performance over PD-L1 expression, TMB or gene expression profiling. More recently, Berry et al. [47] analyzed only six markers (PD-1, PD-L1, CD8, FOXP3, CD163 and Sox10/S100) expression patterns using mIF assays to generate the multispectral imaging with the “AstroPath” platform and found that it could accurately stratify long-term survival after anti-PD-1/PD-L1 monotherapy. These results support our strategy of incorporating spatial and co-expression information of distinct tumor immune microenvironmental features to develop biologically rational biomarkers for immunotherapy and chemotherapy. Moreover, as workflows are optimized and technical advances allow for more detection channels, we envision its clinical application in the near future.
Several limitations should be acknowledged. First, approximately 40% of the ITT population were included in this biomarker evaluable cohort. Although the baseline characteristics and clinical outcomes were similar between these two cohorts, selection bias could be inevitable. Second, considering the limited sample size, we did not further divide the biomarker evaluable cohort into a training and validation set. The lack of validation, either orthogonally or in a separate dataset, and sensitivity and specificity testing could limit the immediately clinical applicability of the current findings. Hence, the present results should be cautiously interpreted and still need further independent validation with large sample size. Third, we leveraged the median level as the cutoff to define the positive versus negative marker expression groups. Although the cutoff is popular in the research setting and helpful for us to clarify the relevant investigations due to its briefness, it may not be optimal. Fourth, this study only included Chinese patients with advanced NSCLC. Considering the distinct genotype and immunophenotypes of patients of different ethnicities, the current findings should be cautiously applied to patients of other races. Lastly, all available tumor samples were biopsy specimens, and we could only evaluate the tumor immune microenvironmental markers expression in the tumor area but not in the stromal area.
5 CONCLUSIONS
Our data demonstrated that biologically rational combinations of different tumor immune microenvironmental markers by mIF assays could provide a method to seek the predictive biomarker of PD-1 plus chemotherapy in locally advanced or metastatic NSCLC. The predictive value of CD8/PD-L1 or CD68/PD-L1 co-expression for camrelizumab plus chemotherapy as first-line treatment in patients with locally advanced or metastatic NSCLC is worthy of further investigation.
DECLARATIONS
AUTHOR CONTRIBUTIONS
Caicun Zhou, Tao Jiang, Shengxiang Ren, and Fengying Wu conceived and designed this study. Fengying Wu, Gongyan Chen, Yunchao Huang, Jianying Zhou, Lizhu Lin, Jifeng Feng, Zhehai Wang, Yongqian Shu, Jianhua Shi, Yi Hu, Qiming Wang, Ying Cheng, Jianhua Chen, Xiaoyan Lin, Yongsheng Wang, Jianan Huang, Jiuwei Cui, Lejie Cao, Yunpeng Liu, Yiping Zhang, Yueyin Pan, Jun Zhao, LiPing Wang, Jianhua Chang, Qun Chen, Xiubao Ren, Wei Zhang, Yun Fan, Zhiyong He, Jian Fang, Kangsheng Gu, Xiaorong Dong, Shengxiang Ren, and Caicun Zhou enrolled patients and collected the data. Tao Jiang, Shengxiang Ren, and Fengying Wu were responsible for statistical analysis, and all authors participated in data interpretation. The manuscript was drafted by Caicun Zhou, Tao Jiang, Shengxiang Ren, and Fengying Wu and was reviewed and/or revised by all authors. The final version was approved to be submitted by all authors. Nanjing Geneseeq Technology Inc. conducted the next-generation sequencing of the included samples. Genecast Biotechnology Co. conducted the multiplex immunofluorescence assays of the included samples and the biomarker analysis. The authors and corresponding authors are responsible for all aspects of this work, including data presentation and accuracy.
ACKNOWLEDGMENTS
We are grateful to all patients, their families, and staff at the study centers.
COMPETING INTERESTS
Caicun Zhou reported honoraria as a speaker from Roche, Lily China, Boehringer Ingelheim, Merck, Hengrui, Qilu, Sanofi, Merck Sharp & Dohme, Innovent Biologics, C-Stone, Luye Pharma, TopAlliance Biosciences, and Amoy Diagnostics; and advisor fees for Innovent Biologics, Hengrui, Qilu, and TopAlliance Biosciences. Shengxiang Ren reported honoraria as a speaker from Boehringer Ingelheim, Lilly, Merck Sharp & Dohme, Roche, Hengrui, and Junshi, advisor fees for Roche, Merck Sharp & Dohme, and Boehringer Ingelheim and research funding from Hengrui. Tao Zhang, Wei Shi and Jianjun Zou were employees of Hengrui. No other disclosures were reported.
FUNDING
This study was also supported in part by grants from the National Natural Science Foundation of China (No. 81871865, 81874036, 81972167 and 82102859), the Backbone Program of Shanghai Pulmonary Hospital (No. FKGG1802), Shanghai Pujiang Talent Plan (No. 2019PJD048), Shanghai Science and Technology Committee Foundation (NO. 19411950300), Shanghai Key disciplines of Respiratory (No. 2017ZZ02012), Oncology development incentive program of Shanghai Pulmonary Hospital, Shanghai Multidisciplinary Cooperative Project for Diagnosis and Treatment of Major Diseases, and Key Clinical Project Development Program of Shanghai.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This manuscript was the biomarker analysis of the previous trials, registered with ClinicalTrials.gov, NCT03134872. The respective institutional review boards approved the clinical protocol and ethics committees, and all participants provided written informed consent.
CONSENT FOR PUBLICATION
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding authors upon reasonable request. All requests for raw data will be reviewed by the Shanghai Pulmonary Hospital, Tongji University School of Medicine, and Jiangsu Hengrui Pharmaceuticals.




