miR-99a-5p modulates doxorubicin resistance via the COX-2/ABCG2 axis in triple-negative breast cancer: from the discovery to in vivo studies
Abbreviations
-
- ABCG2
-
- ATP-binding cassette subfamily G member 2
-
- ALT
-
- alanine transaminase
-
- AST
-
- aspartate aminotransferase
-
- ATCC
-
- American Type Culture Collection
-
- BC
-
- breast cancer
-
- BET
-
- Brunauer−Emmett−Teller
-
- BJH
-
- Barret-Joyner-Halenda
-
- CI
-
- confidence interval
-
- COX-2
-
- cyclooxygenase-2
-
- CRE
-
- creatinine
-
- CTAB
-
- Cetyltrimethylammonium bromide
-
- DAPI
-
- 4',6-diamidino-2-phenylindole
-
- DLS
-
- dynamic light scattering
-
- DRFS
-
- distant-relapse free survival
-
- FDR
-
- false discovery rate
-
- FTIR
-
- Fourier-transform infrared
-
- GEO
-
- Gene Expression Omnibus
-
- H&E
-
- hematoxylin and eosin
-
- H&E
-
- hematoxylin and eosin
-
- HA
-
- hyaluronic acid
-
- HR
-
- hazard ratio
-
- IC50
-
- half-maximal inhibitory concentration
-
- ICP-MS
-
- inductively coupled plasma mass spectrometry
-
- METABRIC
-
- Molecular Taxonomy of Breast Cancer International Consortium
-
- MFI
-
- mean fluorescence intensity
-
- MSNs
-
- mesoporous silica nanoparticles
-
- myh7
-
- Myosin Heavy Chain 7
-
- NCBI
-
- National Center for Biotechnology Information
-
- OS
-
- overall survival
-
- PEI
-
- polyethyleneimine
-
- PTFE
-
- polytetrafluoroethylene
-
- PXRD
-
- Powder X-ray diffraction
-
- qRT-PCR
-
- quantitative real-time PCR
-
- RIPA
-
- radioimmunoprecipitation
-
- RNU43
-
- U43 Small Nuclear RNA
-
- SAED
-
- selected area electron diffraction
-
- SD
-
- standard deviation
-
- SDS-PAGE
-
- sodium dodecyl sulfate–polyacrylamide gel electrophoresis
-
- SEM
-
- standard error of the mean
-
- siRNAs
-
- short interfering RNAs
-
- TCGA
-
- The Cancer Genome Atlas
-
- TEM
-
- transmission electron microscopy
-
- TEOS
-
- tetraethylorthosilicate
-
- TGA
-
- thermogravimetric analysis
-
- TMAH
-
- tetramethylammonium hydroxide solution
-
- TNBC
-
- triple-negative breast cancer
-
- URE
-
- urea
-
- UTR
-
- untranslated region
Breast cancer (BC) is the most commonly diagnosed cancer and the fifth cause of cancer-related death worldwide [1]. Despite the advances in BC targeted therapies, cytotoxic chemotherapy is still widely used [2]. However, around 20%-30% of BC patients develop metastasis after treatment as a consequence of drug resistance [3]. In this context, microRNAs have emerged as potential therapeutic targets to overcome therapy resistance [4]. Therefore, we aimed to elucidate the molecular mechanisms underlying resistance to doxorubicin, one of the most effective chemotherapeutic agents used in BC. Methods are detailed in Supplementary Materials.
To identify miRNAs involved in doxorubicin resistance, the miRNA expression profiles of MDA-MB-231R cells with acquired doxorubicin resistance and their parental cell line MDA-MB-231 were compared (Supplementary Figure S1A). miR-99a-5p was found as the most differentially expressed miRNA, being downregulated in resistant cells (fold change = -17.83, P = 0.027) (Figure 1A).
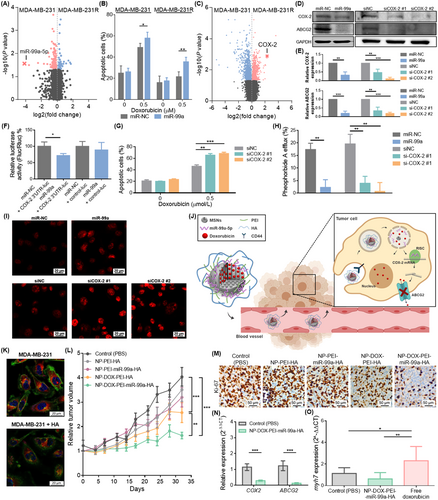
We previously described miR-99a-5p as a good diagnostic biomarker downregulated in BC tissues when compared to healthy breast tissues [5]. To determine its clinical relevance in BC, we examined the association between miR-99a-5p expression and distant relapse-free survival (DRFS) in a cohort of 210 BC patients (GSE22216). miR-99a-5p was overexpressed in patients free of progression (n = 131) compared to patients with distant relapse (n = 79) (fold change = 1.72; P = 0.019), and its low expression was associated with shorter DRFS (P = 0.004; Supplementary Figure S1B-C). Additionally, its association with overall survival (OS) was assessed in the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC, n = 1262) and The Cancer Genome Atlas (TCGA) cohorts (n = 1062), which confirmed that low miR-99a-5p expression was associated with poor prognosis (Supplementary Figure S1D-E).
Therefore, we hypothesized that miR-99a-5p could increase doxorubicin sensitivity. miR-99a-5p was described to increase sensitivity to radiation and cisplatin [6, 7], but its role in doxorubicin resistance was still unexplored. We demonstrated that miR-99a-5p overexpression decreased the half-maximal inhibitory concentration (IC50) of doxorubicin (Figure 1B) and increased doxorubicin-induced apoptosis in MDA-MB-231 and MDA-MB-231R cells (Supplementary Figure S1F-G).
To elucidate the molecular mechanism by which miR-99a-5p sensitizes cells to doxorubicin, we compared the gene expression profiles of MDA-MB-231R and MDA-MB-231 cells by microarray analysis. To identify potential miR-99a-5p targets, mRNAs significantly upregulated in MDA-MB-231R cells were selected (Supplementary Figure S2A) and compared with predicted miR-99a-5p targets from miRWalk 2.0 (Supplementary Table S1). We found cyclooxygenase-2 (COX-2) as a potential target with the highest differential expression (fold change = 4.94; P = 0.001) (Figure 1C), and proved its upregulation in MDA-MB-231R cells (Supplementary Figure S2B-C). COX-2 is implicated not only in inflammation but also in tumorigenesis, treatment resistance, and poor prognosis in cancer patients [8].
To clarify whether miR-99a-5p modulates doxorubicin sensitivity through COX-2, we confirmed that miR-99a-5p overexpression inhibited COX-2 expression and that it is a direct target of miR-99a-5p (Figure 1D-F, Supplementary Figure S2D). Then, we corroborated that COX-2 downregulation sensitized BC cells to doxorubicin, as it reduced IC50 values (Supplementary Figure S2E-F) and increased doxorubicin-induced apoptosis (Figure 1G).
The ATP-binding cassette subfamily G member 2 (ABCG2), a transporter involved in doxorubicin efflux, was suggested to be regulated by COX-2 [9]. Hence, we hypothesized that ABCG2 could be a downstream target of miR-99a-5p through COX-2. We found that miR-99a-5p downregulated the expression of ABCG2 (Figure 1D-E, Supplementary Figure S3A) and decreased the efflux of the ABCG2-specific substrate pheophorbide A (Figure 1H), thus confirming an inhibitory effect on ABCG2 activity. Besides, we proved that overexpression of miR-99a-5p increased the intracellular accumulation of doxorubicin (Figure 1I, Supplementary Figure S3B). Despite ABCG2 being listed as a predicted target of miR-99a-5p by miRWalk 2.0, luciferase reporter gene assay showed no direct interaction (Supplementary Figure S3C). Therefore, we proposed COX-2 as a link between miR-99a-5p and ABCG2. Our results evidenced that silencing of COX-2 downregulated ABCG2 (Figure 1D,E, Supplementary Figure S3D), reduced the pheophorbide A efflux (Figure 1H), and increased the intracellular accumulation of doxorubicin (Figure 1I, Supplementary Figure S3B). Besides, ABCG2 was upregulated in MDA-MB-231R cells (Supplementary Figure S3E-F). Collectively, these data suggest that miR-99a-5p increased sensitivity to doxorubicin in part by modulation of ABCG2 through COX-2.
To translate our findings, we designed mesoporous silica nanoparticles (MSNs) to deliver a combination of miR-99a-5p and doxorubicin to tumor cells. MSNs were loaded with doxorubicin and capped with consecutive layers of polyethyleneimine (PEI), miR-99a-5p, and hyaluronic acid (HA) (NP-DOX-PEI-miR-99a-HA) (Supplementary Figure S4A). PEI favors lysosomal escape, which is necessary to ensure the miRNA delivery into the cytoplasm, and HA is a biocompatible molecule that acts as a cap, inhibiting the release of the nanoparticle's content before internalization by cells, and targets CD44, which plays an important role in BC drug resistance (Figure 1J). Empty nanoparticles (NP-PEI-HA), nanoparticles containing only miR-99a-5p (NP-PEI-miR-99a-HA), or only doxorubicin (NP-DOX-PEI-HA) were evaluated as controls.
Nanoparticles were characterized by standard techniques that confirmed their successful preparation (Supplementary Figure S4B-H, Supplementary Table S2). The amount of encapsulated doxorubicin in NP-DOX-PEI-miR-99a-HA (70 mg/g SiO2) was determined by thermogravimetric analyses and high-performance liquid chromatography. Besides, we verified that doxorubicin was released in lysosomal extract, whereas there was a very low delivery in phosphate-buffered saline (Supplementary Figure S4I), thus indicating that nanoparticles would deliver their content in lysosomes after being uptaken by cells. Moreover, the capacity of NP-DOX-PEI-miR-99a-HA to specifically target CD44 was confirmed by a competition assay. Pretreatment of MDA-MB-231 cells with free HA inhibited the uptake of nanoparticles (60.5%, P < 0.001) (Figure 1K, Supplementary Figure S4J), thus evidencing the NP-DOX-PEI-miR-99a-HA targeting to CD44.
We next examined the in vivo efficacy of the designed nanoparticles in a murine orthotopic xenograft model of triple-negative BC (TNBC). NP-DOX-PEI-miR-99a-HA induced the highest inhibition of tumor growth and cell proliferation; NP-PEI-HA and NP-PEI-mir-99a-HA displayed no significant differences with control, and NP-DOX-PEI-HA showed a moderate capacity to inhibit tumor growth (Figure 1L-M, Supplementary Figure S5A). Two days after last treatment, the tumor volume inhibition rates by NP-DOX-PEI-HA and NP-DOX-PEI-miR-99a-HA were 36.3% and 59.5%, respectively. Furthermore, the functional effect of miR-99a-5p was confirmed as COX-2 and ABCG2 were downregulated in tumors treated with NP-DOX-PEI-miR-99a-HA (Figure 1N). Biodistribution studies evidenced the ability of nanoparticles to target tumors, yet were also found in the kidneys, lungs, spleen, and liver (Supplementary Figure S5B). Moreover, nanoparticles had no toxicity in vivo (Supplementary Figure S5C-D).
Cardiotoxicity is the major adverse effect of doxorubicin [10]. To assess the potential benefit of doxorubicin encapsulation in combination with miR-99a-5p, doses of free doxorubicin and NP-DOX-PEI-miR-99a-HA with similar therapeutic efficacy were selected. Mice were treated with 1.6 mg/kg of free doxorubicin, whereas the equivalent dose of doxorubicin in nanoparticles was approximately 0.7 mg/kg, (44.0% of free drug) (Supplementary Figure S5E). Besides, higher levels of the marker of cardiac atrophy myh7 (myosin heavy chain 7) (Figure 1O) and the presence of cytoplasmic vacuoles in cardiac tissue (Supplementary Figure S5F) evidenced that free doxorubicin induced cardiotoxicity, while NP-DOX-PEI-miR-99a-HA showed no differences with the control. These experiments proved the nanoparticles’ ability to diminish the required dose of doxorubicin and the consequent cardiotoxicity.
In conclusion, we have shown that miR-99a-5p modulated doxorubicin resistance in TNBC. To translate our findings, we designed a nanoparticle for specific co-delivery of miR-99a-5p and doxorubicin that demonstrated its therapeutic potential in vivo, being more effective and less toxic than free doxorubicin alone.
DECLARATIONS
ACKNOWLEDGMENTS
We are especially grateful to the associations Amunt Contra el Cáncer, Asociación de la Lucha Contra el Cáncer de Montesa, and Corazón Solidario Contra el Cáncer. Funding for open access charge: Universitat Politècnica de València.
CONFLICT OF INTERESTS
The authors declare no conflict of interests.
FUNDING INFORMATION
This work was supported by Spanish Government and co-financed by FEDER Funds (PI18/01219, PI21/01351, PI18/00382, and RTI2018-100910-B-C41), CIBER-BBN (CB07/01/2012), CIBERONC (CB16/12/00481) and the Generalitat Valenciana (project PROMETEO 2018/024). I.G.-C. was funded by Generalitat Valenciana (ACIF/2016/030), A.A. and A.L. were funded by Asociación Española Contra el Cancer. J.F.B., was funded by Instituto de Salud Carlos III and the European Social Fund for the financial support “Sara Borrell” (CD19/00038), V.C.-N., was funded by Ministerio de Ciencia e Innovación (FPU grant), and J.M.C. was funded by Sociedad Española de Oncología Médica (Río Hortega-SEOM).
AUTHOR CONTRIBUTIONS
IGC: Conceptualization, methodology, data analysis, writing—original draft preparation. AAA, AL (Ana Lameirinhas), JFB, VCN, FR, SZ, JMG: Methodology, data analysis. AL (Ana Lluch), BB: Conceptualization, funding acquisition. FS: data acquisition and analysis. JMC, RMM, PE: Supervision, conceptualization, writing—reviewing and editing, funding acquisition.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
All animal experiments were approved by the Institutional Review Board of Biomedical Research Institute INCLIVA (Valencia, Spain) (2020/VSC/PEA/0131).
CONSENT FOR PUBLICATION
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
The raw data required to reproduce these findings are available from the authors upon request.




