The E3 ubiquitin ligase HUWE1 acts through the N-Myc-DLL1-NOTCH1 signaling axis to suppress glioblastoma progression
Abstract
Background
Elucidation of the post-transcriptional modification has led to novel strategies to treat intractable tumors, especially glioblastoma (GBM). The ubiquitin-proteasome system (UPS) mediates a reversible, stringent and stepwise post-translational modification which is closely associated with malignant processes of GBM. To this end, developing novel therapeutic approaches to target the UPS may contribute to the treatment of this disease. This study aimed to screen the vital and aberrantly regulated component of the UPS in GBM. Based on the molecular identification, functional characterization, and mechanism investigation, we sought to elaborate a novel therapeutic strategy to target this vital factor to combat GBM.
Methods
We combined glioma datasets and human patient samples to screen and identify aberrantly regulated E3 ubiquitin ligase. Multidimensional database analysis and molecular and functional experiments in vivo and in vitro were used to evaluate the roles of HECT, UBA and WWE domain-containing E3 ubiquitin ligase 1 (HUWE1) in GBM. dCas9 synergistic activation mediator system and recombinant adeno-associated virus (rAAV) were used to endogenously overexpress full-length HUWE1 in vitro and in glioma orthotopic xenografts.
Results
Low expression of HUWE1 was closely associated with worse prognosis of GBM patients. The ubiquitination and subsequent degradation of N-Myc mediated by HUWE1, leading to the inactivation of downstream Delta-like 1 (DLL1)-NOTCH1 signaling pathways, inhibited the proliferation, invasion, and migration of GBM cells in vitro and in vivo. A rAAV dual-vector system for packaging and delivery of dCas9-VP64 was used to augment endogenous HUWE1 expression in vivo and showed an antitumor activity in glioma orthotopic xenografts.
Conclusions
The E3 ubiquitin ligase HUWE1 acts through the N-Myc-DLL1-NOTCH1 signaling axis to suppress GBM progression. Antitumor activity of rAAV dual-vector delivering dCas9-HUWE1 system uncovers a promising therapeutic strategy for GBM.
List of abbreviations
-
- ANOVA
-
- Analysis of Variance
-
- ATM
-
- Ataxia Telangiectasia Mutated
-
- Atoh1
-
- Atonal BHLH Transcription Factor 1
-
- bFGF
-
- Basic fibrobast growth factor
-
- CDKN2A
-
- Cyclin Dependent Kinase Inhibitor 2A
-
- ChIP
-
- Chromatin immunoprecipitation
-
- c-Myc
-
- Myc Proto-Oncogene Protein
-
- CNS
-
- central nervous system
-
- DCX
-
- Doublecortin
-
- DLL1
-
- Delta Like Canonical Notch Ligand 1
-
- DMEM
-
- Dulbecco's Modified Eagle's Medium
-
- Dvl
-
- Dishevelled Segment Polarity Protein
-
- EGF
-
- Epidermal growth factor
-
- FBXW7
-
- F-Box and WD repeat domain-containing 7
-
- GAPDH
-
- Glyceraldehyde-3-Phosphate Dehydrogenase
-
- GAPDH
-
- Glyceraldehyde-3-Phosphate Dehydrogenase
-
- GBM
-
- glioblastoma
-
- GO
-
- Gene Ontology
-
- GSEA
-
- Gene set enrichment analysis
-
- GSEA
-
- gene set enrichment analysis
-
- HES5
-
- Hes Family BHLH Transcription Factor 5
-
- HEY1
-
- Hes Related Family BHLH Transcription Factor With YRPW Motif 1
-
- hGFAP
-
- human glial fibrillary acidic protein
-
- HUWE1
-
- HECT, UBA And WWE Domain Containing E3 Ubiquitin Protein Ligase 1
-
- KD
-
- knockdown
-
- KEGG
-
- Kyoto Encyclopedia of Genes and Genomes
-
- KLHL23
-
- Kelch Like Family Member 23
-
- Mcl-1
-
- MCL1 Apoptosis Regulator
-
- Miz1
-
- Myc-Interacting Zinc Finger Protein 1
-
- MYCN
-
- MYCN Proto-Oncogene
-
- N1ICD
-
- NOTCH1 intracellular domain
-
- NC
-
- negative control
-
- OD
-
- Optical Density
-
- PLA
-
- Proximity ligation assay
-
- rAAV
-
- recombinant adeno-associated virus
-
- SAM
-
- synergistic activation mediator
-
- SD
-
- Standard Deviation
-
- SEM
-
- Standard Error of Mean
-
- sgRNA
-
- single guide RNA
-
- shRNA
-
- short hairpin RNA
-
- siRNA
-
- small interferring RNA
-
- SOX8
-
- SRY-Box Transcription Factor 8
-
- TCGA
-
- The Cancer Genome Atlas
-
- UBE2C
-
- ubiquitin-conjugating enzyme E2 C
-
- UBE2QL1
-
- ubiquitin-conjugating enzyme E2 Q family like 1
-
- UPS
-
- ubiquitin-proteasome system
-
- WHO
-
- World Health Organization
1 BACKGROUND
Glioma is the most common primary intracranial tumor in adults, with an incidence of approximately 5-8/100,000, accounting for approximately 80% of intracranial malignancies, among which glioblastoma (GBM) has the highest degree of malignancy and the poorest prognosis [1]. At present, gliomas are mainly treated by surgical resection, radiotherapy and chemotherapy. Tumor-treating fields is recommended as an adjuvant treatment for high-grade gliomas [2]. The simultaneous addition of temozolomide as a chemotherapeutic adjuvant in radiotherapy can moderately improve the survival rate of young patients with good performance and has become the standard treatment [3, 4]. Despite recent advances in combination therapy, the extensive genetic diversity of this disease leads to resistance to standardized treatments [5, 6]. Therefore, a deeper understanding of the occurrence and progression of GBM will benefit the development of personalized treatment.
GBM manifests as a complex heterogeneous disease at the genetic and epigenetic levels. In addition to simple genetic mutation and amplification, post-transcriptional modification of proteins is now being revealed by technology on a larger scale, which has led to novel strategies to treat tumors. Ubiquitylation is a reversible, stringent and stepwise post-translational modification involving a series of enzymatic reactions. Aberrant ubiquitylation is closely related to various malignant tumor processes [7]. Within this complex milieu, E3 ubiquitin ligases target a broad spectrum of substrates and confer specificity to ubiquitination; thus, they are involved in many tumor-critical functions, such as cell proliferation, energy metabolism, and DNA repair, and are thus a determinant underlying cancer initiation and evolution and an important class of drug targets [8, 9].
The HECT domain-containing E3 ubiquitin ligase (HUWE1, also known as ARF-BP1/Mule) was first identified as a testis E3 ligase, which can ubiquitinate almost all core histones in vitro [10]. HUWE1 regulates the complex interplay among cell proliferation, differentiation and DNA damage response in a physiologically, pathophysiologically and cytologically dependent pattern [11-14]. Previously published data showed that HUWE1 was mainly implicated in nervous system development, function and tumor initiation and progression, including prostate, breast, colon, lung, gastric, and laryngeal cancers [15]. Based on diverse cytological contexts, the substrates of HUWE1 are cell-specific: tumor suppressors [p53, Ataxia Telangiectasia Mutated (ATM), Cyclin Dependent Kinase Inhibitor 2A (CDKN2A), Dishevelled Segment Polarity Protein (Dvl)] [16, 17] and oncoproteins [Myc Proto-Oncogene Protein (c-Myc), MCL1 Apoptosis Regulator (Mcl-1), Myc-Interacting Zinc Finger Protein 1 (Miz1), Atonal BHLH Transcription Factor 1 (Atoh1)] [18-20]. Consequently, the roles of HUWE1 in tumorigenesis remain controversial. Therefore, signatures specific to the cell context must be considered to identify the functions of HUWE1. Recently, published studies have revealed that HUWE1 was under expressed in some brain tumors, indicating its potential tumor suppressor role [21]. Nevertheless, the functions and molecular basis of HUWE1 in GBM, the most challenging central nervous system tumor, are partly unknown and warrant further investigation.
In the present study, we evaluated the expression of HUWE1 in glioma tissues and paired adjacent normal tissues and the prognostic value of HUWE1 in GBM. In vivo and in vitro models were used to analyze the functions of HUWE1 in GBM progression and its underlying mechanisms of action. We also evaluated the antitumor activity of rAAV dual-vector delivering dCas9-HUWE1 system and anticipated that targeting HUWE1 may be a potential therapeutic strategy for GBM.
2 METHODS AND MATERIALS
2.1 Patients and tumor specimens
This study was authorized by the ethics committee of the Third Military Medical University (TMMU, Chongqing, China). Surgical specimens were obtained from GBM patients who were hospitalized in the Department of Neurosurgery, Southwest Hospital between 2014 and 2017 and subjected to neuro-navigation and fluorescein-guided surgery after a clear evaluation. Written informed consents were obtained from patients or their guardians. The histopathological examinations of all samples were independently performed by at least two neuropathologists according to the World Health Organization's (WHO) classification (2016). The clinicopathological information of these patients is summarized in Supplementary Table S1.
2.2 Cell culture
The human GBM cell lines LN229 (ATCC-CRL-2611) and T98G (ATCC-CRL-1690) have been authenticated in a previous study [22]. The human normal glial cell line HEB was bestowed by Professor Guang-Mei Yan from Department of Pharmacology, Zhongshan School of Medicine, Sun Yat-sen University (Guangzhou, Guangdong, China). The primary GBM cell lines GBM1, GBM2 and GBM3 were established using tumor specimens from GBM patients (TMMU) [23]. To isolate glioma cells from GBM tumors, tumor resection tissues were collected and cut into small pieces and then were isolated using the Papain Dissociation System (Worthington Biochemical, Lakewood, NJ, USA) according to the manufacturer's instructions. The tumor tissues were further mechanically dissociated, and the suspension was filtered with a 70 μm cell strainer (BD Biosciences, San Jose, CA, USA) to remove tissue pieces. All GBM cells and HEB cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM, Gibco, Grand Island, NY, USA) with 10% fetal bovine serum (FBS, Gibco), 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C in humidified air with 5% CO2. GBM3 cells were cultured in Neurobasal medium (Invitrogen, Carlsbad, CA, USA) with 20 ng/mL epidermal growth factor (EGF, PeproTech, Rocky Hill, NJ, USA), 20 ng/mL basic fibrobast growth factor (bFGF, PeproTech) and 20 μL/mL B27 Supplement (Life Technologies, Carlsbad, CA, USA).
2.3 Gene expression analyses
An independent cohort of 667 GBM specimens was obtained from The Cancer Genome Atlas (TCGA) database (https://tcgadata.nci.nih.gov/tcga). A total of 472 GBM specimens from the Rembrandt database (https://gdoc.georgetown.edu/gdoc/workflows) and 284 from the Gravendeel database (http://gliovis.bioinfo.cnio.es/) were used to validate the gene expression, glioma WHO grade, and prognostic values of HUWE1 and MYCN Proto-Oncogene (MYCN) in glioma patients. We mined the gene expression data of an independent cohort of 472 GBM specimens from the Rembrandt database and 284 from the Gravendeel database, genes with a fold change > 2 or < -2 (compared with normal brain tissues) and P value < 0.05 were selected; 1298 genes in the Rembrandt database and 2125 genes in Gravendeel were screened. The intersections of the two gene sets were further compared with the ubiquitin-mediated proteolysis gene set in the KEGG. Cancer cell invasion and glioma invasion gene sets were defined in published literature [24, 25] and listed in Supplementary Table S2.
2.4 RNA-sequencing (RNA-seq) analyses
Total cellular RNA was extracted from HUWE1-knockdown (KD) and shCtrl-transfected LN229 cells and purified using a Qiagen RNeasy kit (Qiagen, Hilden, NRW, Germany) according to the manufacturer's protocol. Gene set enrichment analysis (GSEA) was performed using GSEA v2.2.0 (Broad Institute, Cambridge, MA, USA) to enrich genes that showed significant differences in expression between the HUWE1-KD and shCtrl-transfected groups. Gene Ontology (GO) with DAVID software (https://david.ncifcrf.gov/) was used for gene set annotation. The statistical significance for the nominal P value was set to 0.05.
2.5 qRT-PCR
Total RNA was extracted from GBM cells with an RNA Extraction Kit (Fastagen, Shanghai, China) according to the manufacturer's instructions. qRT-PCR was performed using a Bio-Rad CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) as previously described [26]. The cycling conditions were 5 min at 95°C, 40 cycles of 10 s at 95°C, 15 s at 60°C, 20 s at 72°C, and 5 s at 85°C. Assays were performed in triplicate. Primer sequences are listed in Supplementary Table S3.
2.6 Western blotting
Western blotting was performed as previously described [27]. In brief, GBM cells were lysed on ice for 30 min with non-ionic detergent-containing buffers (RIPA lysis buffers) lysis buffer (Thermo Scientific, Waltham, MA, USA) supplemented with protease inhibitor cocktail (1:100, Thermo Scientific). Proteins were separated via sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes. Blots were then probed using primary antibodies as follow: anti-HUWE1 (#5695, 1:1000, Cell Signaling, Danvers, MA, USA), anti-NOTCH1 intracellular domain (N1ICD) (#3608, 1:1000, Cell Signaling), anti-N2ICD (#5732, 1:1000, Cell Signaling), anti-N3ICD (#5276, 1:1000, Cell Signaling), anti-N4ICD (#2423, 1:1000, Cell Signaling), anti-delta-like canonical Notch ligand 1(DLL1) (#2588, 1:1000, Cell Signaling), anti-DLL3 (#78110, 1:1000,CellSignaling), anti-P53 (#2527, 1:1000, Cell Signaling), anti-c-Myc (#5605, 1:1000, Cell Signaling), anti-N-Myc (#9405, 1:1000, Cell Signaling), anti-Miz1 (#14300, 1:1000, Cell Signaling), anti-Mcl1 (#94296, 1:1000, Cell Signaling), anti-HA-Tag (#3724, 1:1000, Cell Signaling), anti-His-Tag (#12698, 1:1000, Cell Signaling), anti-Flag-Tag (#14793, 1:1000, Cell Signaling), anti-LaminB1 (#13435, 1:1000, Cell Signaling), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (#2118, 1:5000, Cell Signaling), and anti-β-Tubulin (#2146, 1:10000, Cell Signaling).
2.7 Transwell chamber migration and invasion assays
An 8.0-μm-pore Transwell membrane (Millipore, Billerica, MA, USA) was first coated with 1 mg/mL Matrigel (BD Biosciences). Then, 500 μL of DMEM containing 10% FBS was added to the lower chambers, and GBM cells were seeded in the upper chambers at a density of 3 × 104 cells in 200 μL of serum-free DMEM. Then, the cells were incubated at 37°C with 5% CO2 for 24 h, and the filter membranes were fixed with 4% formaldehyde for 15 min. Afterward, the cells in the upper chamber were stained with crystal violet for 15 min before the noninvasive cells were scraped with a cotton tip. The invasive cell number was manually counted in five randomly selected fields under a light microscope at 200×. The migration of cells was examined using a Transwell membrane without coating Matrigel. After 12 h of incubation, the upper chambers were removed, and the next steps were performed as described for the invasion assay. As to Transwell chamber migration and invasion assays of DAPT (a γ-secretase inhibitor, to block NOTCH cleavage and NICD production) treated GBM cells, GBM cells were cultured with DAPT (25 μmol/L) throughout the experiment.
2.8 Plasmid transfection and dual-luciferase reporter assay
GBM cells were seeded in 24-well plates at a density of 1 × 105 cells/well and co-transfected with 600 ng MYCN promoter reporter vector and 600 ng Renilla control vector (GeneChem, Shanghai, China) using Lipofectamine 2000 (Invitrogen). After 24 h of incubation, cells were transfected with DLL1 and DLL3 expression vectors or corresponding empty vector (GeneChem). Luciferase activities were measured 48 h after transfection using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). The results are presented after normalization to the measured values of firefly luciferase.
2.9 Co-immunoprecipitation
GBM cells were lysed on ice for 40 min with RIPA lysis buffer supplemented with protease inhibitor cocktail. The supernatant proteins were collected, incubated with indicated primary antibodies or corresponding isotype IgG (#3900, Cell Signaling), and analyzed by using a Pierce™ Co-Immunoprecipitation Kit (Thermo Scientific) according to the manufacturer's protocol.
2.10 In vivo ubiquitination assay
The ubiquitination assay was conducted as previously described [28]. GBM cells were transfected with HA-UB, K6R, K11R, K48R, K63R, K6R/K48R, and K48-only ubiquitin and K6-only ubiquitin plasmids (GeneChem) before treatment with the proteasome inhibitor MG132 (100 μmol/L, Selleck, Houston, TX, USA) for 6 h. Then, the cells were collected for co-immunoprecipitation with anti-N-Myc (ab16898, Abcam, Cambridge, MA, UK) followed by Western blotting of ubiquitin. Precipitation with isotype IgG was used as a negative control.
2.11 In vitro limiting dilution assay
GBM3 cells were implanted into a 96-well plate at a density of 0, 5, 10, 20, 40 cells/well with 10 replicates for each concentration. Seven days after implantation, the number of tumor spheres in each well was determined, and the sphere formation efficiency was calculated using online extreme limiting dilution analysis (http://bioinf.wehi.edu.au/software/elda/).
2.12 Chromatin immunoprecipitation (ChIP)
ChIP was performed using the SimpleChIP® Enzymatic Chromatin IP Kit (Cell Signaling) according to the manufacturer's protocol. Isotype IgG (#2985, Cell Signaling) and anti-N-Myc were used to immunoprecipitate chromatin fragments. The promoter regions of DLL1 and DLL3 were separated into 9 parts for subsequent qRT-PCR, and the primer pairs are listed in Supplementary Table S4.
2.13 Cell Counting Kit-8 (CCK-8) assay and IC50 evaluation
GBM cells were seeded in 96-well plates with 100 μL of DMEM containing 10% FBS at a density of 1500 cells/well with six replicate wells per group. CCK-8 (Dojindo, Kumamoto, Japan) assays were employed for quantitation of cell viability using a fluoroanalyzer (Floskan Ascent, Waltham, MA, USA) with an optical density (OD) value of 450 nm for 8 continuous days.
2.14 CRISPR synergistic activation mediator (SAM) system
Plasmids encoding NLS-dCas9-VP64 and MS2-P65-HSF1 expression plasmids were obtained from GeneChem. The corresponding single guide RNAs (sgRNAs) were designed and ligated into the sgRNA-MS2-P65-HSF1-Neo vector. The lentivirus was used to deliver dCAS9-VP64-Puro and sgRNA-MS2-P65-HSF1-Neo vectors into GBM cells. Then, the infected cells were selected in 5 μg/mL puromycin (InvivoGen, Cayla, France) or 130 μg/mL G418 (A100859, Sangon Biotech, Shanghai, China). The sequences of the relative sgRNAs are listed in Supplementary Table S5. All the expression plasmids were confirmed by sequencing.
2.15 Generation of recombinant AAV vectors
A rAAV dual-vector delivering dCas9-HUWE1 system was obtained from GeneChem. In brief, the SadCas9-VP64 gene and three sgRNAs of HUWE1 were packaged into two separate rAAV vectors. sgRNA NC was packaged into an empty vector and served as control. The sequences of the sgRNAs are listed in Supplementary Table S5. All the expression plasmids were confirmed by sequencing.
2.16 Lentiviral vector construction and RNA interference
HUWE1 short hairpin RNA (shRNA) lentiviral vectors and a non-targeting control shRNA vector were purchased from HANBIO (Shanghai, China). Three siRNAs (RiboBio, Shanghai, China) were used to disturb MYCN expression in GBM cells, and scrambled RNAs were used as negative control. All RNAs described above were transfected into cells using Lipofectamine 2000 (Thermo Fisher Scientific) following the manufacturer's instructions. All sequences are listed in Supplementary Tables S6.
2.17 Orthotopic implantation of GBM cells and bioluminescence imaging
The animal study was performed in accordance with the protocol approved by the Institutional Animal Care and Use Committee of Southwest Hospital, TMMU. Six-week-old immunodeficient NOD/SCID female mice were purchased from the Laboratory Animal Center of TMMU. The mice were anaesthetized, and GBM2 and LN229 cells were intracranially injected at a density of 2 × 105 cells in 5 μL of PBS. Tumor growth was monitored by bioluminescence imaging using In Vivo Imaging System (IVIS) Spectrum (Perkin-Elmer, Waltham, MA, USA) and Living Image Software for IVIS (Perkin-Elmer).
Mice were killed by means of transcardiac perfusion with 4% paraformaldehyde upon presentation of neurological deficits with ataxia, cachexia, lethargy, or seizure. Mice brains were collected and then sampled for subsequent Harris hematoxylin, H&E and immunohistochemical (IHC) staining.
2.18 IHC and immunoreactivity scores
IHC staining of HUWE1 in GBM xenografts and GBM specimens was performed using a Dako REAL EnVision Detection System (DAKO, Carpinteria, CA, USA) according to the manufacturer's protocol. Anti-HUWE1 (HPA002548, Sigma) was used as the primary antibody at a concentration of 5 μg/mL. All images were obtained under a Leica DM4000B microscope equipped with a QImaging EXiAqua camera (QImaging, Surrey, BC, Canada). The staining signal of HUWE1 in GBM specimens was scored according to the intensity of positive cells as described previously [29]. In brief, we randomly captured six regions of IHC slides using a DM4000B microscope (Leica, Wetzlar, Germany). The integrated optical density (IOD) of the positive site (brown) was measured with Image-Pro Plus software (version 5.0, Rockville, MD, USA). The same parameter settings were applied for all images. The mean IOD value was identified as the cutoff value.
2.19 Immunofluorescence staining
Immunofluorescence staining was performed as previously described [30]. Primary antibodies, including anti-HUWE1 and anti-N-Myc, were applied in the study. Cell nuclei were counterstained with Hoechst 33342 (#4082, Cell Signaling).
2.20 Proximity ligation assay (PLA)
PLAs were used to detect protein-protein associations in situ. LN229 and GBM2 cells were seeded in 24-well chamber slides at a density of 10,000 cells/well and then blocked and incubated with primary antibodies: anti-HUWE1 (ab70161, Abcam), anti-N-Myc (#84406, Cell Signaling), anti-MDM2 (#86934, Cell Signaling), anti-Flag (#14793, Cell Signaling), anti-HA (#3724, Cell Signaling), and anti-P53 (#2524, Cell Signaling). PLAs were performed using a Duolink® In Situ Red Starter Kit (DUO92101, Sigma-Aldrich, Saint Louis, MO, USA) in accordance with the protocol. Fluorescence images were acquired using a Leica TCS SP5 confocal microscope (Leica, Abberior Instruments).
2.21 Statistical analyses
All experiments were performed at least three times with triplicate samples. Statistical analysis was performed using SPSS statistical software (SPSS 16.0, Chicago, CA, USA) and GraphPad Prism 6 software (GraphPad, La Jolla, CA, USA). Statistical significance was assessed by one-way analysis of variance (ANOVA) and Student-Newman-Keuls multiple comparison tests, and P < 0.05 was considered significant. The in vitro and in vivo experiments included a standard sample size based on prior studies [31]. Survival analyses were performed using the Kaplan-Meier method, with the log-rank test used for comparison. Overall survival (OS) was defined as the interval from the date of diagnosis to death of any cause. All quantitative data are presented as the mean ± standard deviation (SD) or mean ± standard error of mean (SEM).
3 RESULTS
3.1 Low expression of the E3 ubiquitin ligase HUWE1 was identified in glioma and indicated a poor prognosis
To characterize the dysregulated ubiquitin-proteasome system (UPS) in gliomas, we mined the gene expression data of an independent cohort of 472 GBM specimens from the Rembrandt database and 284 from the Gravendeel database [32]. 1298 genes in the Rembrandt database and 2125 genes in Gravendeel were screened and only four candidate genes, HUWE1, F-Box and WD repeat domain-containing 7 (FBXW7), ubiquitin-conjugating enzyme E2 C (UBE2C) and ubiquitin-conjugating enzyme E2 Q family like 1 (UBE2QL1), fit the selection criteria (Figure 1A). Among these genes, UBE2C and UBE2QL1 are E2 ubiquitin-conjugating enzymes, and FBXW7 has been intensively studied in GBM. To this end, we further investigated HUWE1.
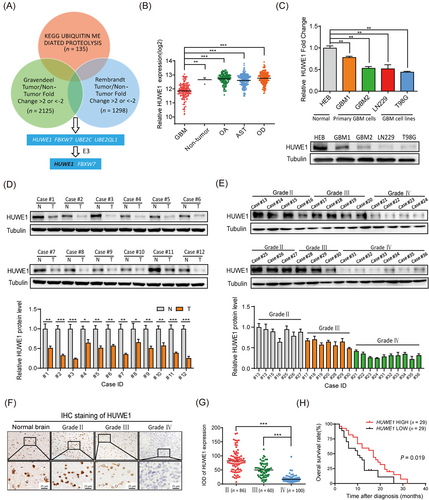
We first evaluated the association of HUWE1 with glioma classification (Supplementary Figure S1A). We found that HUWE1 expression was significantly declined in GBM, compared with other histological subtypes (Figure 1B). In support of this finding, we tested the expression of HUWE1 in GBM cell lines and primary GBM cells. The results showed that HUWE1 expression was significantly decreased in GBM, consistent with the data obtained from patient tissue samples (Figure 1C-E). In addition, IHC staining of HUWE1 in 246 glioma specimens [median age, 46 years (range, 28-71 years); obtained from Southwest Hospital 2014-2017] showed a steadily declining trend from samples at WHO grade II to IV (Figure 1F-G). We analyzed the association between HUWE1 expression and clinicopathological factors of 246 patients with glioma (Supplementary Table S7). Furthermore, we analyzed the clinical prognostic information of our own GBM patient cohorts (n = 58). These data revealed that low HUWE1 expression was associated with poor prognosis of GBM patients (Figure 1H), which was concordant with its prognostic value calculated using data from the TCGA GBM, Gravendeel and Rembrandt GBM databases (Supplementary Figure S1B). Taken together, these results show that the expression of the E3 ubiquitin ligase HUWE1 is reduced in GBM, and this reduction is associated with a poor prognosis, indicating that this molecule might be an important tumor-suppressing factor.
3.2 HUWE1 suppressed GBM progression in vitro and in a mouse xenograft model
To decipher the functional importance of HUWE1 in GBM, we established HUWE1-KD and NC LN229, GBM1, GBM2 and GBM3 cells (Supplementary Figure S2A) and examined the global changes in gene expression in HUWE1-KD and NC LN229 cells, a widely used GBM cell line, using microarray analysis. A volcano plot showed that 1722 genes had upregulated expression and 1092 had downregulated expression in HUWE1-KD LN229 cells (Figure 2A). GO and GSEA showed that these upregulated genes were closely associated with the migratory and invasive capacities of LN229 cells. Additionally, epithelial cell proliferation (GO:0050673) was highly ranked in the gene annotation, indicating that HUWE1 might regulate the proliferation of LN229 cells (Figure 2B). To further validate the function of HUWE1, we first separated glioma patient data obtained from TCGA GBM, Rembrandt, and Gravendeel databases into HUWE1-low and HUWE1-high groups according to the median mRNA expression of HUWE1. GSEA revealed that genes from the HUWE1-low group were significantly enriched in cell migration and invasion in the context of the cancer cell invasion and glioma invasion gene sets, which was consistent with the above bioinformatics results (Figure 2C).
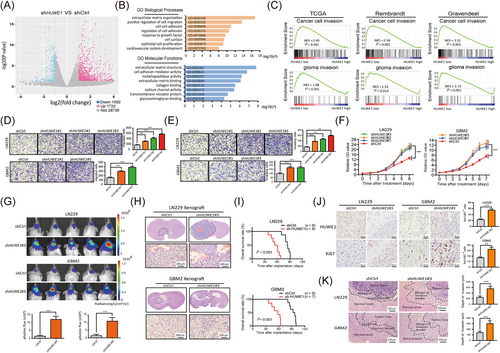
To functionally elucidate the tumor-suppressing roles of HUWE1 in GBM cells, we performed Transwell chamber migration and invasion assays, showing that HUWE1 KD significantly increased tumor cell motility (Figure 2D-E, Supplementary Figure S2B). As confirmed by CCK-8 assays, the proliferative capacity of these cells was strongly improved after disruption of HUWE1 (Figure 2F, Supplementary Figure S2C). Similarly, we found that disrupting HUWE1 expression attenuated the sphere-forming and self-renewal capacity of GBM3 cells (Supplementary Figure S2D).
We further established a mouse xenograft model through intracranial implantation of HUWE1-KD and NC LN229 and GBM2 cells, both have been confirmed with a good intracranial tumorigenicity [33], in NOD/SCID mice. Bioluminescence imaging showed successful tumor growth after ablation of HUWE1, as confirmed by H&E staining of whole brain slices of these xenograft tumors (Figure 2G-H). In addition, the survival rate of the tumor-bearing mice implanted with the HUWE1-KD cells was dramatically shortened compared with that of NC cells (Figure 2I). Next, higher intensities of Ki67 were observed in the xenograft tumors formed by HUWE1-KD cells than those derived from NC cells (Figure 2J). Interestingly, we found a broader range and more jagged edges in the margin of the invasive border of the HUWE1-KD xenograft tumors (Figure 2K), suggesting that disruption of HUWE1 endowed GBM cells with an enhanced invasive ability. Taken together, these results indicate that HUWE1 suppresses GBM cell proliferation and invasion.
3.3 HUWE1 was essential for the degradation of N-Myc in GBM cells
Due to the complex interplay and intrinsically weak interaction between E3 ligases and substrates, traditional immunoprecipitation followed by mass spectrometry may not comprehensively identify the substrates [34]. To this end, we performed conjoint analysis to refine the downstream targets of HUWE1. We first utilized UbiBrowser, an integrated bioinformatics platform [33], to predict the candidate substrates of HUWE1. Then, we combined these 700 candidates with our mass spectrometry data obtained from the quantification proteomics analysis of HUWE1-KD and NC cells and immunoprecipitation proteomics of HUWE1 (Figure 3A). We identified MYCN and UBXN1. Given the higher ranking position and tumor-promoting action of MYCN [35], we provided detailed insight into MYCN.
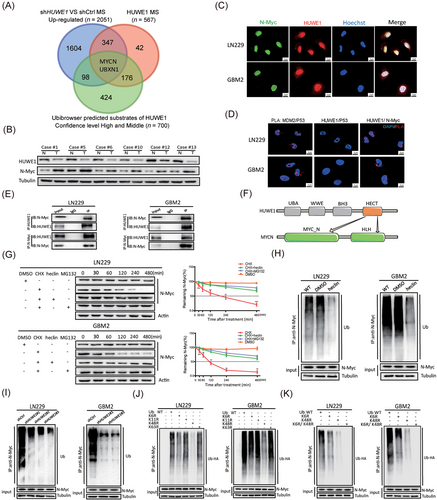
We first tested the expression of N-Myc in GBM patient specimens. N-Myc was negatively correlated with HUWE1 (Figure 3B, Supplementary Figure S3A-3B), implying a possible interaction. To further analyze the interaction of HUWE1 and N-Myc in situ, we carried out confocal microscopy and PLAs. Fluorescence images showed that HUWE1 and N-Myc were mostly co-localized in the nuclei of GBM cells, but fluorescence signals were not observed between HUWE1 and P53 (Figure 3C-D). Furthermore, a physical interaction between endogenous and exogenous HUWE1 and N-Myc was confirmed by coimmunoprecipitation (Figure 3E, Supplementary Figure S3C). This observation was consistent with the finding that disrupting HUWE1 significantly increased the expression of N-Myc; however, other reported HUWE1 substrates, including C-Myc, P53, Mcl1 and Miz1, showed no detectable changes (Supplementary Figure S3D), indicating that this protein interaction is specific and cellular context-dependent. Interestingly, we found a discrepancy in the mRNA and protein expression levels of MYCN between GBM and HEB cells, and disrupting HUWE1 barely changed MYCN expression (Supplementary Figure S3E-3F), revealing a possible strong post-transcriptional modification of N-Myc. To this end, we tested the half-life of N-Myc in HEB cells and found that it was obviously shorter than that in GBM cells (Supplementary Figure S3G).
Combined with the above results, we hypothesized that the abnormal accumulation of N-Myc in GBM cells was caused by the deficiency in HUWE1. Emerging literature indicates that four functional domains of HUWE1 participate in the ubiquitination progress [36]. We found that HUWE1 recognizes the N-terminus and HLH motif of N-Myc through the HECT domain using the UbiBrowser data platform (Figure 3F). As validated by the cycloheximide (CHX) pulse-chase assay, the half-life of N-Myc was significantly delayed after treatment with the synthetic inhibitor CHX, HECT ligase inhibitor heclin [37] and proteasome inhibitor MG-132 for the indicated time (Figure 3G). In addition, disrupting HUWE1 expression could significantly prolong the half-life of N-Myc (Supplementary Figure S3H). Further, in vivo ubiquitination assays showed that N-Myc-linked polyubiquitin chains were observably abolished by treatment with heclin or disruption of HUWE1 (Figure 3H-I), which was consistent with the attenuated fluorescence signals of ubiquitin/N-Myc complexes in HUWE1-KD GBM cells (Supplementary Figure S3I).
A growing body of literature has shown that HECT E3 ligases assemble atypical Ub chains [28]. In particular, HUWE1 assembles K6, K11, K48 and a small amount of K63-linked ubiquitin [38]. In validation studies, we constructed ubiquitin molecules carrying point mutations in Lys6, 11, 48, and 63 (K6R, K11R, K48R and K63R). In vivo ubiquitination assays showed that K48R decreased the polyubiquitin chains of N-Myc in some but not all GBM cells, while K6R had a slight effect on the formation of polyubiquitin chains (Figure 3J). To this end, we constructed ubiquitin molecules simultaneously carrying point mutations in Lys6 and Lys48 (K6R/K48R) and transfected into GBM cells. As expected, the polyubiquitin chains attached to N-Myc were significantly diminished (Figure 3K). In addition, we constructed K48-only ubiquitin and K6-only ubiquitin (Supplementary Figure S3J). PLA fluorescence signals showed the interaction between N-Myc and such mutant ubiquitins (Supplementary Figure S3K).
Collectively, these results establish that polyubiquitin chains attached to N-Myc are mainly through K48. However, K6-linked ubiquitination might functionally regulate N-Myc. Based on the regulatory mechanism of HUWE1 on N-Myc, we asked whether disrupting MYCN could rescue the phenotypes caused by HUWE1 KD. With combined silencing of MYCN and HUWE1, the motility, invasion and proliferation of GBM cells were partially rescued (Supplementary Figure S4). Taken together, these results indicate that HUWE1 promotes K48- and K6-linked ubiquitination of N-Myc, and K48-linked polyubiquitin chains target it for degradation.
3.4 N-Myc activated NOTCH1 signaling pathway by binding to DLL1 promoter in GBM cells
We revealed the interaction between N-Myc and HUWE1, thereby highlighting the roles of N-Myc in GBM. However, the mechanisms underlying how N-Myc regulates the progression of GBM cells have not been fully elucidated. To unbiasedly identify the downstream targets of N-Myc, we evaluated gene expression associated with MYCN by analyzing TCGA GBM databases. KEGG pathway enrichment using DAVID software showed that MYCN-associated genes (adjusted P < 0.05 and r > 0) were significantly enriched in the NOTCH signaling pathway (P = 0.031). Interestingly, among the top 20 genes, 8 genes [DLL3, DLL1, Hes Family BHLH Transcription Factor 5 (HES5), SRY-Box Transcription Factor 8 (SOX8), Doublecortin (DCX), SOX11, HES6, and Kelch Like Family Member 23 (KLHL23)] had a physical or a functional relationship with the NOTCH signaling pathway, suggesting that NOTCH was activated by N-Myc in GBM. More interestingly, DLL1 and DLL3, two canonical ligands of the NOTCH signaling pathway, were concurrently expressed with MYCN, ranking in the top 5 (Figure 4A).
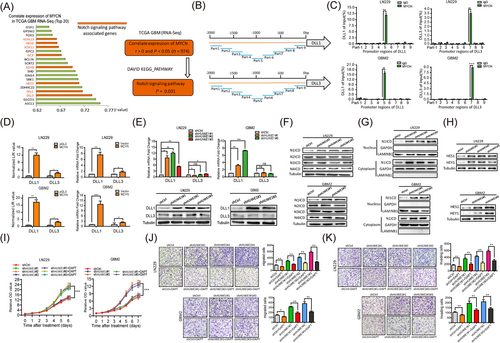
We thus assumed that N-Myc transcriptionally regulated DLL1 and DLL3 to activate the NOTCH signaling pathway. To test this hypothesis, we first divided the promoter regions of DLL1 and DLL3 into 9 continuous segments and performed ChIP assays. We observed that N-Myc was immunoprecipitated along with the corresponding promoter regions of DLL1 and DLL3 by an anti-N-Myc antibody (Figure 4B-C).
Next, we generated a dual-luciferase reporter assay system by co-transfecting reporter plasmids containing DLL1 and DLL3 promoter sequences and MYCN expression plasmids into GBM cells. We found that N-Myc transcriptionally activated DLL1, whereas the activation of DLL3 was barely detectable (Figure 4D). In addition, we found that disrupting HUWE1 increased luciferase activity of DLL1 in HUWE1-KD cells (Supplementary Figure S5A). Similarly, disruption of HUWE1 observably increased DLL1 but not DLL3 at the protein and mRNA levels (Figure 4E), indicating that the downstream targets of the HUWE1-N-Myc axis are DLL3-independent. To further evaluate whether the NOTCH signaling pathway was activated, we first tested four active intracellular domains of NOTCH: N1ICD, N2ICD, N3ICD and N4ICD. Among these, N1ICD expression was significantly increased in the HUWE1-KD cells, while the changes were barely detectable in other intracellular domains (Figure 4F).
To rule out the possibility that HUWE1 affected the mRNA expression of NOTCH instead of functional regulation, we performed qRT-PCR assays. As expected, there was no significant difference in the mRNA levels of NOTCH1, -2, -3 and -4 after HUWE1 was disrupted (Supplementary Figure S5B). Notably, the activated NOTCH1 pathway featured a remarkable increase in N1ICD in the cytoplasm and nucleus, as confirmed by nucleocytoplasmic separation experiments (Figure 4G).
Furthermore, Hes family BHLH transcription factor 1 (HES1) and Hes-related family BHLH transcription factor with YRPW motif 1 (HEY1), two NOTCH1 signaling downstream target genes, had observably increased levels (Figure 4H). In addition, we found that N1ICD and HEY1 were markedly increased in these wild-type GBM cells co-cultured with corresponding HUWE1-KD cells (Supplementary Figure S5C), indicating that the NOTCH1 signaling pathway has been activated. In support of this finding, we simultaneously ablated N-Myc and HUWE1 in GBM cells and found that endogenous N1ICD and DLL1 expression could be strongly rescued (Supplementary Figure S5D-5F). Based on the above results, we hypothesized that blocking the NOTCH signaling pathway may compromise the tumor suppressive role of HUWE1 in GBM cells. Treatment with DAPT partly compromised the tumor suppressive effects of HUWE1 on the proliferative, migratory and invasion capacities (Figure 4I-K). Collectively, these results confirm that N-Myc binds to DLL1 promoter to activate the NOTCH1 signaling pathway in GBM cells.
3.5 Endogenous HUWE1 induced by the dCas9 SAM system suppressed GBM progression
Considering the large molecular size of HUWE1, stable exogenous expression of the full-length protein is difficult. To this end, we aimed to apply dCas9 SAM, a simple and efficient RNA-guided CRISPR/dCas9 technology [39], to increase endogenous HUWE1 expression in GBM cells. The schematic diagram illustrates the mechanism of SAM; in brief, dCas9-VP64 and MS2-P65-HSF1 complexes form key components of this system (Figure 5A). In support of this notion, SAM was first delivered to GBM cells with dual lentiviral vectors. Notably, forced expression of endogenous HUWE1 resulted in an observable increase in mRNA and protein levels, thereby leading to a decrease in N-Myc, DLL1, and N1ICD levels and accumulation of polyubiquitin chains of N-Myc (Figure 5B, Supplementary Figure S6).
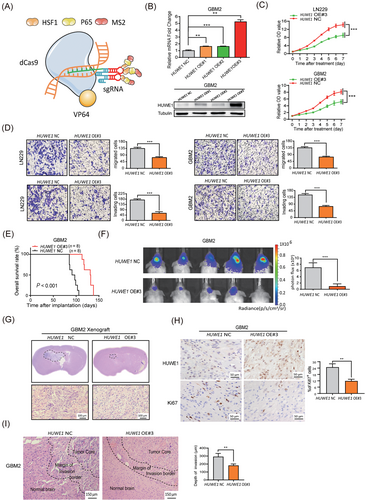
To further validate the tumor suppressive roles of HUWE1, we performed Transwell migration assays and CCK-8 experiments with HUWE1-overexpressing cells. The results showed that forced expression of endogenous HUWE1 significantly suppressed the migratory, invasive, and proliferative capacities of GBM cells in vitro (Figure 5C-D). To further evaluate the tumor suppressive effects of HUWE1 in vivo, we established a mouse xenograft model by implanting HUWE1-overexpressing cells and NC cells into NOD/SCID mice. The tumor-bearing mice transplanted with HUWE1-overexpressing cells survived much longer than the mice that received NC cells (Figure 5E), indicating a prosurvival function of HUWE1. Bioluminescence imaging showed attenuation of tumor growth after forced expression of endogenous HUWE1, which was confirmed by H&E staining of whole brain slices of these xenograft tumors (Figure 5F-G). In contrast to the accumulation of HUWE1, Ki67 was significantly decreased in HUWE1-overexpressing xenografts, as demonstrated by immunohistochemical analysis (Figure 5H). Interestingly, we found that the invasive borders of the HUWE1-overexpressing xenograft tumors shrank, and the boundaries were smoother than those formed by the NC tumors (Figure 5I). These results indicate that HUWE1 induced by the SAM system markedly suppressed GBM cell progression, highlighting this method as a promising therapeutic strategy in treating GBM.
3.6 Antitumor effects of rAAV dual-vector delivering dCas9-HUWE1 system in a GBM xenograft model
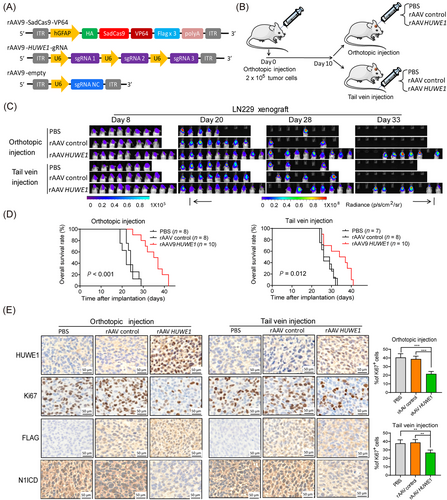
On the basis of the above data, to more carefully evaluate the therapeutic importance of the dCas9-HUWE1 system in GBM, we aimed to transfer it to a GBM xenograft model. To date, rAAV vectors have been reported to directly deliver targeted genes to the central nervous system (CNS) and some solid tumors with superior safety [40]. Moreover, different serotypes of rAAV endowed it with organ and tissue tropism, suggesting that diverse diseases can be precisely targeted [41]. Among different serotypes of rAAV, rAAV9 has the potential to permeate the blood-brain barrier, thereby suggesting the administration of promising CNS therapeutic genes [42].
To this end, we posited that the rAAV9-delivering dCas9-HUWE1 SAM system may be useful for treating GBM. To assess this hypothesis, we constructed a rAAV9 dual-vector system. The dCas9, VP64, and Flag genes and 3 independent sgRNAs were packaged into two separate rAAV9 vectors (Figure 6A). Orthotopic xenografts were established by intracranially injecting LN229 cells (luciferase-expressing) into NOD/SCID mice. Ten days after tumor implantation, rAAV9-HUWE1 vector, rAAV9-control vector, and PBS were administered to the tumor-bearing mice through orthotopic and tail vein injection (Figure 6B). In vivo bioluminescence imaging showed that tumor growth was significantly attenuated after delivering rAAV9 dCas9-HUWE1 relative to PBS and vehicle controls (Figure 6C). Survival curves showed that orthotopic injection of the rAAV9-HUWE1 vector exhibited stronger prosurvival effects than tail vein injection (Figure 6D). Further IHC staining results validated the efficiency of virus infection and showed that HUWE1 expression was substantially elevated after injection of rAAV, consistent with the decline in Ki67 and N1ICD expression (Figure 6E). Notably, this trend was more distinct in the orthotopic injection group than in the vein injection group. Collectively, these data illustrate the antitumor effects of the rAAV-delivering dCas9-HUWE1 system, indicating its promising clinical application.
4 DISCUSSION
In the present study, we provide insight into the UPS in GBM. The UPS is a highly conserved and vital element of post-transcriptional modification in eukaryotes and tightly regulates the turnover of cellular proteins with specificity and precision [43, 44]. To this end, dysregulation of the UPS disrupts the balance between oncogene- and tumor suppressor gene-related proteins, thus facilitating the initiation and progression of cancers. Notably, the UPS extensively regulates prosurvival signaling pathways and maintains glioma stem cells in GBM [45]. Therefore, developing novel therapeutic approaches to target the UPS shows promise in the treatment of GBM [46, 47]. The E3 ligase HUWE1 plays a critical role in UPS, thus making it an attractive therapeutic target in tumors. However, a considerable discrepancy in the roles of HUWE1 in different tumors exists. Recent studies have shown that HUWE1 expression is upregulated in breast, colon, lung, prostate, laryngeal, liver, pancreatic, skin, and thyroid cancers, while in gastric and uterine cancers, HUWE1 expression was significantly downregulated [48]. Given the diverse genetic and epigenetic cellular backgrounds, neither tumor-suppressing nor tumor-promoting roles of HUWE1 have been identified. To obtain unbiased data, we performed conjoint analysis of multi-database to screen and identified HUWE1 as a tumor suppressor gene in GBM. Through validation studies, we showed that HUWE1 could transfer ubiquitin to N-Myc and induce subsequent degradation. In addition, we found that not only K48- but also K6-linked polyubiquitin chains are attached to N-Myc, which is consistent with the latest research showing that HUWE1, in contrast to Nedd4-family E3 ubiquitin ligases, prioritized K6-linked polyubiquitin chains [49]. High expression of MYCN was associated with a good prognosis of GBM patients as shown in the TCGA GBM database. However, these data are not consistent with previous reports that N-Myc promoted GBM progression and predicted unfavorable clinical outcomes [50]. These considerable discrepancies in prognostic value between the gene and protein levels of MYCN indicate an observable post-transcriptional modification of N-Myc. Of note, our present work revealed that HUWE1 ubiquitylated N-Myc and triggered its proteasomal degradation.
Although accumulating evidence links aberrant NOTCH signaling with tumor progression, its oncogenic or tumor-inhibiting effects are controversial. Despite this controversy, the NOTCH1 signaling pathway is considered to be activated in GBM and critical for gliomagenesis [51]. However, the underlying mechanisms concerning the activation of NOTCH1 signaling remain unclear. In the present work, we showed aberrant accumulation of N-Myc caused by a decrease in HUWE1 and transactivated DLL1 through direct binding to promoter regions, thus reinforcing the NOTCH1 signaling pathway in GBM cells. Although both DLL1 and DLL3 were strongly correlated with MYCN, as confirmed by TCGA GBM database and ChIP assays, transcriptional activation mediated by N-Myc was barely detectable in DLL3. A previous research revealed that the canonical ligand DLL1 bound and subsequently activated the NOTCH signaling pathway, while DLL3 was an inhibitory NOTCH ligand [52]. DLL3 prevented the localization of NOTCH and DLL1 to the cell surface in cis and in trans, thus redirecting or retaining NOTCH and DLL1 to late endosomal or lysosomal compartments or Golgi [53]. Our current work is concordant with these data. Interestingly, Zhao et al. [54] showed that HUWE1 operated upstream of the N-Myc-DLL3-Notch1 axis in neural stem cells, which is different from studies showing that DLL3 did not activate NOTCH1 but antagonized its function. The sophisticated regulatory mechanisms of N-Myc require further exploration.
Owing to the large molecular size of HUWE1, it is almost impossible to exogenously express the full-length protein in GBM cells. However, this molecule serves as a tumor suppressor gene in certain cancers, and it is unconvincing to test the antitumor effects of HUWE1 by disturbing it using either siRNA/shRNA or conditional gene knockout. To circumvent this limitation, we used the dCas9-HUWE1 SAM system to force endogenous HUWE1 expression in GBM cells. In subsequent validation studies, we demonstrated HUWE1-induced inhibition of cell proliferation, migration and invasion in vitro and in vivo, which confirmed the tumor-suppressing roles of HUWE1. Based on recent developments concerning the clinical applications of dCas9 in treating infectious diseases and inhibiting tumorigenesis [55], we posited that dCas9-based HUWE1 endogenous expression gene therapy will show potential in treating GBM.
Because rAAV vectors are increasingly popular tools for the biotherapeutic landscape, especially in gene therapy applications, and the rAAV serotype 9 showed highly effective penetration of the blood-brain barrier to transduce astrocytes [56, 57], we constructed rAAV9 for delivery of the dCas9-HUWE1 system. In subsequent validation studies, these rAAV vectors showed a good therapeutic effect in an orthotopic xenograft model. In addition, targeting NOTCH1 in cancers offers an addition to therapeutic strategies recruited for managing cancer. Despite the promising role that γ-secretase inhibitors may hold, their use in the clinical management of cancer faces many challenges [58]. In this present study, rAAV vector-mediated overexpression of HUWE1 attenuated the activation of NOTCH1, thus indicating a potential clinical application in treating GBM. Although the safety and efficacy profiles of rAAV are still controversial, it has been administered in more than 150 clinical trials with good safety and significant clinical benefit in many genetic diseases [59]. However, the application of rAAV vector-mediated gene therapy for cancer is relatively rare.
We recognize some limitations of the present study. First, different from some oncogenes, HUWE1 shows a tumor-inhibiting role in GBM. It was impossible to synthesize or screen small-molecule inhibitors to target this gene. To circumvent this limitation, we utilized rAAV vector-based gene therapy to activate HUWE1. However, the safety and efficacy of this therapeutic model should be more carefully evaluated. Second, although we showed that HUWE1 promoted K48 and K6-linked ubiquitination of N-Myc, and K48-linked polyubiquitin chains mediated turnover of N-Myc, the roles of non-degradative K6-linked polyubiquitination remains poorly understood. Because K6-linked ubiquitin chain did not affect N-Myc protein turnover, we posited that ubiquitylation at this site may regulate its function, thus, further efforts are required to obtain more comprehensive understanding.
5 CONCLUSIONS
Our present work demonstrates that reduced HUWE1 in gliomas is associated with a poor prognosis. The ubiquitination and subsequent degradation of N-Myc mediated by HUWE1, leading to the inactivation of its downstream DLL1-NOTCH1 signaling pathways, promotes the inhibition of the progression of GBM cells. Of note, our study shows that the rAAV-delivering dCas9-HUWE1 system has an antitumor effect in a glioma orthotopic xenograft model, indicating a promising strategy to combat gliomas in future therapies.
DECLARATIONS
ACKNOWLEDGEMENTS
We would like to thank Ze-Yu Yang and Kai-Di Yang for data mining.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study was carried out according to the principles of the Helsinki Declaration and was approved by the Ethics Committee of Third Military Medical University (Army Medical University).
CONSENT FOR PUBLICATION
Not applicable
FUNDING
This research work was supported by grants from National Key R&D Program of China (2016YFA0101200 to XWB), the National Natural Science Foundation of China (81602196 to TL), the Special Grant for Chongqing Postdoctoral Researcher Research Project (xmT2017001 to TL), the Postdoctoral Support Program for Innovative Talent (BX201600022 to TL) and Open Project of Key Laboratory of Tumor Immunopathology of Ministry of Education (2020jsz603 to YY).
AUTHOR CONTRIBUTIONS
YY, LHW and XXZ performed the experiments and collected or analyzed the data; JW, SW, MSZ, ZXY, QHM, YC, HBZ, XQC and JG helped with data collection; YW, XZ, and YHC performed the statistical analysis and helped data interpretation and polished the manuscript; XWB, and TL conceived the study, revised the manuscript and provided the financial support.
Open Research
DATA AVAILABILITY STATEMENT
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.




