Crosstalk between IL-15Rα+ tumor-associated macrophages and breast cancer cells reduces CD8+ T cell recruitment
Wenlong Zhang and Qing Zhang contributed equally to this work.
Abstract
Background
Interleukin-15 (IL-15) is a promising immunotherapeutic agent owing to its powerful immune-activating effects. However, the clinical benefits of these treatments are limited. Crosstalk between tumor cells and immune cells plays an important role in immune escape and immunotherapy drug resistance. Herein, this study aimed to obtain in-depth understanding of crosstalk in the tumor microenvironment for providing potential therapeutic strategies to prevent tumor progression.
Methods
T-cell killing assays and co-culture models were developed to determine the role of crosstalk between macrophages and tumor cells in breast cancer resistant to IL-15. Western blotting, histological analysis, CRISPR-Cas9 knockout, multi-parameter flow cytometry, and tumor cell-macrophage co-injection mouse models were developed to examine the mechanism by which IL-15Rα+ tumor-associated macrophages (TAMs) regulate breast cancer cell resistance to IL-15.
Results
We found that macrophages contributed to the resistance of tumor cells to IL-15, and tumor cells induced macrophages to express high levels of the α subunit of the IL-15 receptor (IL-15Rα). Further investigation showed that IL-15Rα+ TAMs reduced the protein levels of chemokine CX3C chemokine ligand 1 (CX3CL1) in tumor cells to inhibit the recruitment of CD8+ T cells by releasing the IL-15/IL-15Rα complex (IL-15Rc). Administration of an IL-15Rc blocking peptide markedly suppressed breast tumor growth and overcame the resistance of cancer cells to anti- programmed cell death protein 1 (PD-1) antibody immunotherapy. Interestingly, Granulocyte-macrophage colony-stimulating factor (GMCSF) induced γ chain (γc) expression to promote tumor cell-macrophage crosstalk, which facilitated tumor resistance to IL-15. Additionally, we observed that the non-transcriptional regulatory function of hypoxia inducible factor-1alpha (HIF-1α) was essential for IL-15Rc to regulate CX3CL1 expression in tumor cells.
Conclusions
The IL-15Rc-HIF-1α-CX3CL1 signaling pathway serves as a crosstalk between macrophages and tumor cells in the tumor microenvironment of breast cancer. Targeting this pathway may provide a potential therapeutic strategy for enhancing the efficacy of cancer immunotherapy.
Abbreviations
-
- ANOVA
-
- analysis of variance
-
- BSA
-
- bovine albumin
-
- CHX
-
- cycloheximide
-
- CX3CL1
-
- C-X3-C motif chemokine ligand 1
-
- CX3CR1
-
- CX3C chemokine receptor 1
-
- DAPI
-
- 4',6-diamidino-2-phenylindole
-
- DMEM
-
- Dulbecco's modified eagle medium
-
- FDA
-
- Food and Drug Administration
-
- GAPDH
-
- glyceraldehyde-3-phosphate dehydrogenase
-
- GMCSF
-
- granulocyte-macrophage colony-stimulating factor
-
- HIF-1α
-
- hypoxia inducible factor-1alpha
-
- iBMDMs
-
- immortalized bone marrow-derived macrophages
-
- IGF-1
-
- insulin like growth factor-1
-
- IL-15
-
- Interleukin-15
-
- IL-15Rc
-
- IL-15/IL-15Rα complex
-
- IL-15Rα
-
- α subunit of IL-15 receptor
-
- M-CSF
-
- macrophage colony-stimulating factor
-
- MDM2
-
- mouse double minute 2 protein
-
- MDSC
-
- myeloid-derived suppressor cells
-
- MHC II
-
- major histocompatibility complex class II
-
- MMP
-
- matrix metalloproteinase
-
- NK cell
-
- natural killer cell
-
- PD-1
-
- programmed cell death protein 1
-
- PD-L1
-
- programmed death-ligand 1
-
- PI3Kγ
-
- phosphatidylinositol 3-kinase gamma
-
- SDS-PAGE
-
- sodium dodecyl sulfate polyacrylamide gel electrophoresis
-
- STAT3
-
- signal transducer and activator of transcription 3
-
- TAMs
-
- tumor-associated macrophages
-
- TME
-
- tumor microenvironment
-
- γc
-
- common γ chain
1 BACKGROUND
Breast cancer has become the most common cancer among women worldwide, accounting for approximately 2.3 million cases and 685,000 deaths in 2020, and the incidence is increasing dramatically [1]. Cytokines are essential regulators of the immune system and mediate the proliferation and activation signals of immune cells. The activation of antitumor immunity by cytokines has emerged as an important therapeutic strategy for cancer treatment [2]. The cytokines most relevant to immunotherapy are the common gamma receptor (γc) cytokines, especially interleukin-2 (IL-2) and IL-15. IL-15 mainly stimulates the proliferation and cytotoxic function of CD8+ T cells and natural killer (NK) cells and inhibits activation-induced cell death [3, 4]. IL-15 signaling is mediated via heterotrimeric receptors, which contain the β-chain subunits (IL-2R/IL-15Rβ, CD122), a common γ chain (γc, CD132), and a unique α subunit (IL-15Rα) that specifically binds to IL-15 with a high affinity (Kd >10–11 mol/L) [5]. IL-15Rα is widely expressed by macrophages and dendritic cells (DCs), independent of the β and γ subunits [6], and is trans-presented to the β/γ subunit on neighboring CD8+ T cells and NK cells. Administration of IL-15 enhances innate and specific immunity to attack tumors [4]. However, some studies have reported that mice bearing breast carcinoma did not respond to IL-15 treatment [7, 8], even in other tumor models where IL-15 protected tumor cells from apoptosis [9, 10]. In clinical trials, no objective antitumor response has been observed [11, 12], although an increase in circulating NK cells and T cells with a memory phenotype was found in patients receiving IL-15 therapy [13]. Current trials focus on achieving clinical benefits by combining direct targeting and immune treatments, while the mechanisms of tumor resistance to IL-15 have been largely neglected. Understanding these mechanisms will allow the development of better therapeutic strategies.
The target and ultimate effectors of immunotherapy are T cells, which recognize tumor antigens and kill tumor cells in vitro [14, 15]. However, recognition of tumor antigens is not sufficient to eradicate established tumors in vivo [16]. An established tumor is a complex tissue composed of tumor cells and a tumor microenvironment (TME) that promotes growth [17]. Successful tumor-controlling immunotherapy requires the infiltration of activated effector cells into the tumor tissue and destruction of tumor cells, but the TME generally reduces T cell infiltration and suppresses the infiltration of effector cells. Low T-cell infiltration in most tumors results in the poor outcomes of immunotherapy [18]. In fact, previous studies have shown that increased T-cell infiltration is an essential predictive biomarker for a better immunotherapy prognosis [19, 20]. Accumulating evidence has shown that crosstalk between tumor cells and immune cells in the TME plays an important role in the regulation of immune cell infiltration and immune evasion[21]. The identification of crucial immune cell subpopulations, cytokines, and immune checkpoints in this crosstalk has guided the development and US Food and Drug Administration (FDA) approval of immunotherapeutic agents.
Tumor-associated macrophages (TAMs), which account for up to 50% of leukocytes in the TME, have long been considered key regulators of the TME. TAMs are involved in shaping the immunosuppressive microenvironment; regulating the important processes of invasion, metastasis, and drug tolerance; and promoting the progression of various tumor types, such as breast cancer, melanoma, bladder cancer, pancreatic cancer, and non-small cell lung cancer [22]. High-density TAMs are generally associated with poor clinical outcomes in patients with cancer [23, 24]. Additionally, a recent research has shown that TAMs are closely related to immunotherapy resistance, and preclinical experiments have also shown that targeting TAMs may enhance current therapies [25]. The phosphatidylinositol 3-kinase gamma (PI3Kγ) signaling pathway in macrophages mediates the formation of a tumor-immunosuppressive microenvironment. Targeting PI3Kγ in macrophages has been shown to overcome therapeutic resistance to immune checkpoint blockade in a mouse tumor model [26, 27].
Tumor cell-macrophage crosstalk can alter the phenotype and function of both cell types to further promote malignant tumor progression. In this study, we explored the presence and role of this crosstalk in IL-15 therapy for breast cancer. Then, in vitro co-culture and in vivo co-injection models were used to analyze the effect of this crosstalk in macrophages and tumor cells. We anticipated that targeting the intercellular communication mechanism may be a potential therapeutic strategy in combination with IL-15 for breast cancer.
2 MATERIALS AND METHODS
2.1 Reagents
The following primary antibodies and the corresponding isotype controls were purchased from BD Pharmingen, Inc. (San Diego, CA, USA): PE-conjugated anti-mouse antibodies against CD3, CD11b, CD80, CD206 (clone M1), major histocompatibility complex class II (MHC II), NK1.1, programmed death-ligand 1 (PD-L1); FITC-conjugated anti-mouse antibodies against CD4, CD45; APC-conjugated anti-mouse antibodies against CD8, F4/80; APC-Cy7-conjugated anti-mouse CD11c; PE-Cy7-conjugated anti-mouse CD86; and PerCP-conjugated anti-mouse CD40. Anti- programmed cell death protein 1 (PD-1) antibody (clone RPM1-14) was purchased from Bio X Cell (Lebanon, NH, USA). Functional anti-CD3 and anti-CD28 monoclonal antibodies were obtained from Invitrogen (Carlsbad, CA, USA). Penicillin and streptomycin, the DCFH-DA probe, Alexa Fluor® 488-labeled goat anti-rabbit IgG (H+L), Alexa Fluor® 555-labeled donkey anti-mouse IgG (H+L), and HRP-conjugated goat anti-mouse IgG (H+L) were obtained from Beyotime (Haimen, Jiangsu, China). PX-478 and GM6001 were obtained from ApexBio (Houston, TX, USA). Matrigel and macrophage colony-stimulating factor (MCSF) were purchased from Sigma (St. Louis, MO, USA) and GenScript (Nanjing, Jiangsu, China), respectively.
2.2 Cell culture and drug treatment
Murine breast cancer cell line 4T1 and the murine macrophage cell line Raw264.7 were purchased from the Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). Immortalized bone marrow-derived macrophages (iBMDMs) were obtained from Shao Feng's Laboratory (National Institute of Biological Sciences, Beijing, China). 4T1 cells were cultured in RPMI-1640, and iBMDM and Raw264.7 cells were cultured in Dulbecco's modified eagle medium (DMEM). Media was supplemented with 10% fetal bovine serum (FBS), 2 mmol/L L-glutamine, 100 IU/mL penicillin, and 100 IU/mL streptomycin. All cell cultures were maintained at 37°C in 5% CO2. 4T1 cells were treated with the indicated concentrations of PX-478, an inhibitor of hypoxia inducible factor-1alpha (HIF-1α), for 48 h, and then the cells were lysed for the detection of chemokine CX3C chemokine ligand 1 (CX3CL1) and HIF-1α protein levels. To inhibit IL-15Rc release, the cells were cultured alone or cocultured in the presence of the vehicle control or 4 μg/mL GM6001, a broad-spectrum matrix metalloproteinase (MMP) inhibitor. To inhibit CX3CL1 release for further evaluation of the effect of HIF-1α on CX3CL1 release, the cells were cultured in the presence of the vehicle control or 5 μmol/L monensin (Selleck Chemicals, LLC, Houston, TX, USA), a protein transport inhibitor.
2.3 Mice
Nine-week-old female BALB/cJGpt mice and nude mice weighing approximately 20 g were purchased from the Nanjing Biomedical Research Institute of Nanjing University (Nanjing, Jiangsu, China) and bred under specific pathogen-free conditions in our animal facility. All animal experiments were approved by the Institutional Committee for the Protection and Use of Animals of the Nanjing University. The health reports provided by the supplier confirmed that the mice were free of known viral, bacterial, and parasitic pathogens. In the experiment, mice were housed at five per cage, with adequate water and feed, and sterile wood chips were used as bedding. The welfare of animals was assessed daily throughout the animal experiments. Mice that showed signs of disease or stress were either treated (if possible) or euthanized.
2.4 Co-cultures
Raw264.7 cells or iBMDMs were co-cultured with 4T1 tumor cells by using a Transwell culture system with 0.4 μm pore polyester membrane inserts (Corning HTS Transwell, Corning, NY, USA) for 3 days. For the macrophage study, Raw264.7 cells or iBMDMs were seeded in 6-well plates at 1 × 105 cells/well, and 4T1 cells were seeded at 4 × 105 cells/well in Transwell chambers. For the tumor cell study, 4T1 cells were seeded on plates, while Raw264.7 cells or iBMDMs were seeded in Transwell chambers.
2.5 CRISPR-Cas9 knockout of target genes
- gRNA for IL-15Rα: 5′-TGGCCTGGTACATCAAATCA-3′;
- gRNA for CX3CL1: 5′-TTTCGCATTTCGTCATGCCG-3′;
- gRNA for HIF-1α: 5′-AGTGCACCCTAACAAGCCGG-3′.
2.6 Establishment of mouse models
Mice were anesthetized by intraperitoneal injection of pelobarbital sodium (70 mg/kg), and then 200,000 tumor cells were injected into the mammary fat pad of BALB/cJGpt mice at a volume of 20 μL. Cages were randomly selected for specific treatments at the beginning of the experiment. All treatments were started simultaneously, and all groups were evaluated simultaneously. Tumors were established until they were palpable or detectable prior to subsequent treatment. CO2 asphyxiation was used to euthanize the mice at the end of the experiment, and cervical dislocation was performed to ensure death.
For macrophage studies, 4T1 cells were admixed with control iBMDMs or IL-15Rα-knockout (KO) iBMDMs at a 1:1 ratio. Then, 200,000 total cells in a volume of 20 μL were injected into BALB/cJGpt mice. Tumors were measured every 2-3 days, and the volume (1/2 × length × width2) was calculated.
Intraperitoneal administration of diphtheria toxin (25 ng/kg) every 4 days was used to deplete macrophages in CD11b-DTR mice [B6.FVB-Tg(ITGAM-DTR/EGFP)34Lan/J]. Then 1 × 106 EO771 cells, mammary cancer cell line from the C57BL/6 strain, were transplanted one day after the first injection of diphtheria toxin. At 7 days after tumor cell inoculation, 1 × 106 control iBMDMs or IL-15Rα-KO iBMDMs were injected intravenously. The mice were humanely euthanized on day 22, and tumor tissues were harvested and weighed.
Tumor single cells in suspensions were stained for 30 min at 4°C with anti-mouse F4/80 and anti-IL-15Rα antibodies. After rigorous washing with sterile phosphate-buffered saline (PBS), IL-15Rα+ TAMs and IL-15Rα– TAMs were sorted using FACS (FACSAria II SORP, BD Biosciences, Franklin Lakes, NJ, USA). Subsequently, 4T1 cells were mixed with IL-15Rα+ TAMs or IL-15Rα– TAMs at a 1:1 ratio. Then, 200,000 total cells in a volume of 20 μL were injected into the left and right fat pads of BALB/cJGpt mice.
Based on the amino acid sequence of the B helix on IL-15 responsible for binding to IL-15Ra, we designed a peptide (sequence: KVTAMNCFLLELQVILHEYSNM), named B peptide, to block the binding of IL-15 to IL-15Rα. For B peptide treatment experiments, when tumor grew to approximately 3-4 mm in diameter, vehicle, scrambled peptides (0.5 mg peptide/30 μL PBS), B peptides (0.5 mg peptide/30 μL PBS) were injected intratumorally every three days for 2 weeks starting on day 7 after tumor cell injection. The treatments were administered alone or in combination with intravenous injection of anti-PD-1 antibody (100 μg per mouse) every 3 days.
For preparation of tumor single-cell suspensions, breast tumors from mice were cut into small pieces using scissors and then digested with 0.1% collagenase containing 75 μg/mL DNase I pre-warmed at 37°C for 1 h. The digested tissue samples were filtered through 40 μm cell strainers to obtain single-cell suspensions. Red blood cells were subsequently removed with red blood cell lysis buffer and incubated for 2 min at 25°C. Finally, single tumor cell suspensions were analyzed by flow cytometry. To obtain splenocytes for flow cytometry detection of IL-15Rα expression in murine spleen macrophages, the spleen tissue was soaked in PBS, cut into small pieces with scissors, and filtered using a 40 μm cell strainer.
2.7 Construction of lentiviral vectors
We introduced cDNAs encoding HIF-1α, CX3CL1, or IL-15Rα into the pLenti6/v5 lentiviral vector (Invitrogen). Lentiviruses were generated in HEK293T cells using two packaging plasmids (psPAX2 [12260, Addgene] and pMD2G [12259, Addgene]). 4T1 cells were transfected with HIF-1α or CX3CL1 viruses, while Raw264.7 cells were transfected with IL-15Rα virus. The stably expressing cells were selected using blasticidin (Selleck Chemicals).
2.8 Cycloheximide (CHX) chase assay
The half-life of CX3CL1 protein in cells was determined using a CHX chase assay. 4T1 cells were treated with 10 μmol/L CHX (Selleck Chemicals) and lysed at the indicated times after the addition of CHX. CX3CL1 levels were measured using Western blotting.
2.9 Design and synthesis of the B peptide
The B helix of IL-15 primarily interacts with IL-15Rα. The amino acid sequence of the B helix was selected as the B peptide sequence (KVTAMNCFLLELQVILHEYSNM) to block the binding of IL-15 to IL-15Rα. B peptides were synthesized by GenScript. Scrambled peptides were designed and synthesized as control peptides.
2.10 Immunofluorescence
Human breast tumor tissues obtained from breast cancer patients undergoing surgery at Chinese PLA General Hospital (Beijing, China) between 2011 and 2014 were embedded in paraffin blocks. A section from each paraffin block was deparaffinized and rehydrated. Then, sections were boiled 5 min in 10 mmol/L sodium citrate buffer (pH 6.0). After cooling, sections were blocked with 5% bovine serum albumin (BSA) for 45 min. The sections were incubated with anti-CD68 (1:100 dilution), anti-IL-15Rα (1:100 dilution), anti-CX3CL1(1:100 dilution), and anti-HIF-1α (1:100) overnight at 4°C, stained with 4',6-diamidino-2-phenylindole (DAPI) to label nuclei for 15 min. The Alexa Fluor® 488-labeled goat anti-rabbit IgG (H+L) (1:100 dilution) and Alexa Fluor® 555-labeled donkey anti-mouse IgG (H+L) (1:100 dilution) from Beyotime were selected as secondary antibodies, depending on the primary antibody used. CD68, IL-15Rα, CX3CL1, and HIF-1α expression was evaluated using a fluorescence microscope.
All samples were reviewed by experienced pathologists. All patients provided written informed consent before participating in the study. The study protocol was approved by the Institutional Review Board of the Chinese PLA General Hospital.
2.11 Western blotting
Cells were washed twice with PBS, and whole-cell lysis buffer containing protease and phosphatase inhibitors was used to extract the cell protein, which was purchased from Beyotime. Cell lysates were centrifuged at 12,000 rpm at 4°C to remove cell debris, and the Pierce™ BCA assay was used to determine protein concentrations. Protein lysates were electrophoresed on a 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel, followed by immunoblotting on polyvinylidene fluoride membranes (Amersham Biosciences, Piscataway, NJ, USA). Antibodies against CX3CL1 (1:1000; bs-0811R, Bioss, Beijing, China), HIF-1α (1:1000; BS3514, Bioworld Technology, Inc., St. Louis Park, MN, USA), and β-actin (1:5000; AC028, Zen Bio, Chengdu, Sichuan China) were used as primary antibodies and incubated with the membrane overnight at 4°C. After washing, the membranes were incubated with the appropriate secondary antibodies (1:3000; Beyotime)
2.12 Generation of bone marrow-derived macrophages (BMDMs)
Two 8-week-old BALB/cJGpt mice were euthanized using CO2 asphyxiation, and the femurs near the base of the lower limb were cut off, leaving the femurs intact. The muscles around the femur were removed. The femurs were dissected in sterile glass and repeatedly flushed with sterile PBS. A single-cell suspension was prepared and cultured in complete medium containing 20 ng/mL MCSF for 4-6 days.
2.13 Phagocytosis
Fluorescent red latex beads (1 μm diameter, L-2778, Sigma-Aldrich) were used for phagocytosis assays. Prior to the assay, the latex beads were pre-warmed in complete medium (10% FBS in DMEM) for 1 h at 37°C. Next, pre-warmed beads were added to macrophages at a 10:1 ratio and incubated at 37°C for 4 h. Phagocytosis was terminated by the addition of 1 mL of pre-chilled PBS, and macrophages were collected and analyzed using flow cytometry.
2.14 Reactive oxygen species (ROS) measurement
ROS levels in control Raw264.7 and IL-15Rα-OE Raw264.7 cells cultured either alone or in the presence of IL-15 for two days were measured using the Reactive Oxygen Species Assay Kit (Beyotime) according to the manufacturer's instructions.
2.15 Gene expression analysis
- CX3CL1-F: 5’-ACGAAATGCGAAATCATGTGC-3’
- CX3CL1-R: 5’-CTGTGTCGTCTCCAGGACAA-3’
- HIF1A-F: 5’-ACCTTCATCGGAAACTCCAAAG-3’
- HIF1A-R: 5’-ACCTTCATCGGAAACTCCAAAG-3’
- GMCSF-F: 5’-GGCCTTGGAAGCATGTAGAGG-3’
- GMCSF-R: 5’-GGAGAACTCGTTAGAGACGACTT-3’
- MCSF-F: 5’-ATGAGCAGGAGTATTGCCAAGG-3’
- MCSF-R: 5’-TCCATTCCCAATCATGTGGCTA-3’
- GAPDH-F: 5’-AGGTCGGTGTGAACGGATTTG-3’
- GAPDH-R: 5’-TGTAGACCATGTAGTTGAGGTCA-3’
2.16 Formation of IL-15/IL-15Rα complex (IL-15Rc) in vitro
IL-15Rc was formed according to the methods as previously described [29]. Briefly, murine IL-15 (Biolegend, San Diego, CA, USA) and recombinant soluble murine IL-15Rα-Fc (Novoprotein, Shanghai, China) were suspended in PBS at a ratio of 1:6, mixed and incubated for 30 min at 37°C.
2.17 ELISA assay for IL-15Rc level
The concentration of the soluble IL-15Rc was measured using an ELISA kit (eBioscience, San Diego, CA, USA), according to the manufacturer's instructions. Briefly, the cell supernatant was collected and added into a 96-well plate, then incubated with the antibody from the ELISA kit. After washing, the reaction reagent was added, and the absorbance at 450 nm was measured.
2.18 MTT assay
4T1 cells were seeded in 96-well plates (2000 cells/well), and each group was replicated in six wells. Next, 20 μL of MTT solution (Sigma) was added at the indicated times and incubated for 4 h at 37°C. The supernatant was then removed, and 100 μL of dimethyl sulfoxide was added to each well. The optical density was measured at a wavelength of 490 nm.
2.19 Invasion assay
Transwell invasion assays were performed according to standard protocols as previously described [30]. Briefly, 4T1 cells were seed into the upper chamber of Matrigel-coated transwell. After 24 h, the cells on the lower surface of the membrane were stained with 0.1% crystal violet. The number of invaded cells was observed under a microscope. The assay was repeated three times.
2.20 T cell proliferation assay
Splenic CD8+ T cells were isolated by anti-CD8a particles (BD IMag, BD Biosciences, Franklin Lakes, NJ, USA) and were labeled with carboxyfluorescein diacetate succinimidyl ester (CFDA-SE) (Beyotime) and activated by using anti-CD3 and anti-CD28 antibodies supplemented with 200 U/mL IL-2 for 48 h. The activated T cells were then cocultured with IL-15Rα+ TAMs or IL-15Rα– TAMs at ratios of 5:1 for 48 h. T-cell proliferation was analyzed by flow cytometry. The experiments were repeated twice independently.
2.21 T cell cytotoxicity assay
4T1 tumor cells expressing luciferase were used as targets for the T-cell based luciferase killing assay. The ratio of effector to target was titrated continuously from 10:1 to 1:10. The percentage of specific lysis was calculated based on the luciferase signal (total flux) relative to the tumor alone.
2.22 Flow cytometry staining
FcR blocking antibody (1 mg/L, Miltenyi Biotec, Bergisch Gladbach, Germany) was used to pre-incubate cells (106 cells/100 μL) at 4°C for 15 min. Fluorescent antibodies and isotype antibodies were then added, and the cells were further incubated at 4°C for 30 min. PBS was used to wash and suspend cells for flow cytometric analysis.
2.23 Preparation of conditioned medium (CM)
An amount of 1 × 105 BMDMs were seeded in 12-well plates and cultured in DMEM supplemented with 10% FBS in the presence of IL-15 or tumor conditioned medium (TCM) for 48 h. Cell supernatants with cell debris removed were collected for use as conditioned medium. 4T1 cells were cultured in RPMI-1640 medium containing 10% FBS and 30% CM. After 48 h, cells were collected to detect CX3CL1 expression by Western blotting.
2.24 Bioinformatics
Gene expression data from The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/) (Breast Invasive Carcinoma, PanCancer Atlas) were used to analyze the associations of IL-15Rα with CD68 and CD163. Normalized expression data and z-scores for mRNA expression data were downloaded from cBioPortal (www.cbioportal.org).
For the multi-tumor CD8A (encoding CD8 alpha chain, CD8 T-cell signature) and CD4 Pearson correlation analysis, we used the multi-tumor gene-expression microarray dataset from the Expression Project for Oncology (ExpO; http://www.intgen.org/, GEO GSE2109).
Gene expression of CX3CL1 in the TCGA breast cancer cohort was obtained from GEPIA2 (website: gepia2.cancer-pku.cn/#analysis).
We used the PrognoScan platform (http://www.prognoscan.org) to validate the correlation between CD68, CD163, and IL-15Rα expression and relapse-free survival after initiation of monotherapy in tamoxifen-treated patients with breast cancer.
2.25 Statistical analysis
Data are expressed as the mean ± standard error of the mean (SEM). Student's t-test was used to compare data between two groups, while two-way analysis of variance (ANOVA) was used to compare data between three or more groups. Statistical significance was set at P < 0.05.
3 RESULTS
3.1 Macrophages facilitated breast cancer resistance to IL-15 therapy
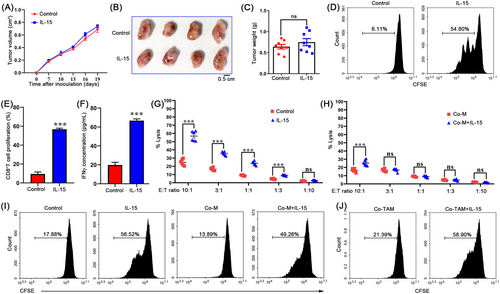
It has been well documented that IL-15 plays a significant antitumor role in tumor immunotherapy by inducing T-cell differentiation and proliferation [31]. Interestingly, we observed resistance to IL-15 therapy in a 4T1 orthotopically grafted breast cancer mouse model (Figure 1A-C). in vitro culture experiments showed that IL-15 effectively enhanced the killing effect of CD8+ T cells sorted from the tumor tissues established using 4T1 tumor cells (Figure 1D-G), suggesting a different mechanism between in vivo and in vitro IL-15 responses. We sought to study the mechanism that mediates breast cancer resistance to IL-15 in the TME. Because immune cells represent the highest proportion of cells in the TME, this suggests that macrophages play an essential role in immune regulation. We evaluated the role of macrophages in 4T1 breast tumor resistance to IL-15. Using a co-culture model of macrophages and tumor cells, we found that macrophages significantly inhibited cytotoxic effect of IL-15 on T cells (Figure 1H). Based on the immunosuppressive effect of TAMs, we believe that macrophages directly inhibit T cell activation, which leads to tumor cell resistance to IL-15. However, macrophages isolated from the co-culture model slightly inhibited T cell proliferation induced by IL-15 (Figure 1I). Similar results were observed for primary TAMs isolated from tumor tissues (Figure 1J). Together, these results suggest that macrophage-tumor cell crosstalk may drive breast cancer resistance to IL-15.
3.2 A TAM subset with high IL-15Rα expression was associated with poor survival in breast cancer patients
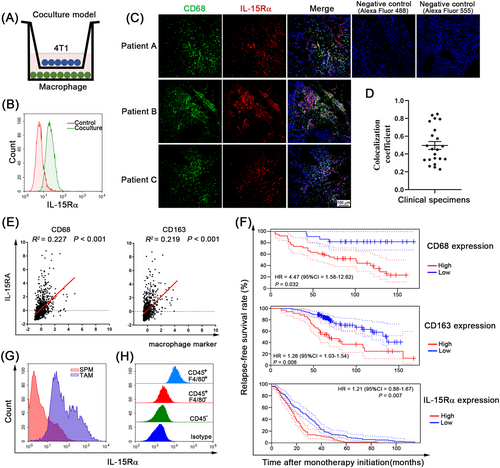
To explore the mechanism by which 4T1 tumors display IL-15 resistance, we introduced a co-culture system where 4T1 tumor cells were cultured with macrophages and found that 4T1 cells induced the expression of IL-15Rα, the α subunit of the IL-15 receptor, in macrophages (Figure 2A-B), as measured by FACS analysis. Similar results were observed using EO771 cells, a murine triple-negative breast cancer cell line (Supplementary Figure S1A). Using confocal microscopy, we further confirmed that IL-15Rα was indeed expressed in macrophages of human breast tumor tissues (Figure 2C). The measured overlap coefficient showed a high degree of colocalization of IL-15Rα and CD68 (a macrophage marker) in the clinical specimens (Figure 2D). Additionally, IL-15Rα was also associated with the macrophage-associated genes CD68 and CD163 (another TAM marker) in The Cancer Genome Atlas (TCGA) dataset (Breast Invasive Carcinoma, PanCancer Atlas) (Figure 2E). The association between IL-15Rα and the clinical outcomes of patients with breast cancer in the PrognoScan database was explored. High levels of macrophage-associated genes (CD68 and CD163) and IL-15Rα were associated with short relapse-free survival (Figure 2F). Next, the IL-15RαHigh TAM population was also identified in the 4T1 orthotopic-grafted breast cancer mouse model and compared with murine spleen macrophages (Figure 2G and Supplementary Figure S1B). Comparing the expression of IL-15Rα in each cell population in the TME, we also found that IL-15Rα+ cells co-expressed the murine macrophage marker F4/80, suggesting that IL-15Rα was mainly expressed in TAMs (Figure 2H and Supplementary Figure S1C). To better characterize the phenotype of IL-15Rα+ TAMs, we performed the flow cytometric analysis to investigate the expression of IL-15Rα on M2-like TAMs (F4/80+CD206+MHC II–) and M1-like TAMs (F4/80+CD206–MHC II+). As shown on Supplementary Figure S1D, IL-15Rα was predominantly enriched on the cell surface of M2-like macrophages. Combined, these results indicate that macrophages highly express IL-15Rα in the TME. It is of great interest to investigate the function of IL-15Rα+ TAMs in tumor tissues and explore their impact on IL-15 therapy.
3.3 IL-15Rα+ TAMs inhibited the infiltration of CD8+ T cells in the TME
To better understand the role of IL-15Rα in the 4T1 breast cancer mouse model (the tumor-immune microenvironment), we measured the proportion of TAMs (IL-15Rα+F4/80+), leukocytes (CD45+), T cells (CD4+, CD8+), NK cells (NK1.1+) and DC cells (MHCII+CD11c+) in the TME on days 7, 14, 21, and 28 after inoculation. Gating strategies for flow cytometry analysis are shown in Supplementary Figure S2. Correlation analysis showed no correlation between IL-15Rα+ TAM content and the content of NK cells, MDSCs, DCs, CD3+ T cells, CD3+CD8+ T cells, or CD3+CD4+ T cells during tumor progression (Supplementary Figure S3A-L). Interestingly, the infiltration of IL-15Rα+ TAMs gradually decreased after peaking at day 14 post-inoculation (Figure 3A), while the ratio of CD8+ to CD4+ T cells decreased dramatically at day 14 (Figure 3B). We found that the ratio of CD8+ to CD4+ T cells was negatively correlated with the frequency of IL-15Rα+ TAMs (Figure 3C). In the TCGA dataset, we also found that patients with high expression of macrophage-associated genes (CD68 and CD163) or IL-15Rα had a lower CD8 to CD4 mRNA level ratio (Supplementary Figure S3M-O), which was consistent with the results of the animal model. These data suggested that IL-15Rα+ TAMs may reduce CD8+ T cells and/or increase CD4+ T cells to decrease the ratio of CD8+ T cells to CD4+ T cells.
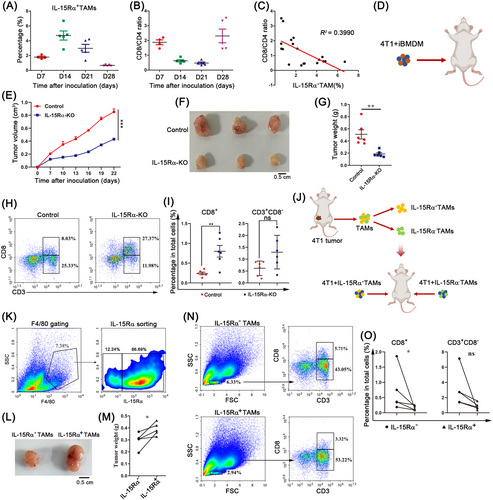
To further confirm the role of macrophage-expressed IL-15Rα in the tumor immune microenvironment, we generated IL-15Rα-KO iBMDMs using the CRISPR-Cas9 system (Supplementary Figure S4). 4T1 tumor cells were implanted in mice in combination with control iBMDMs or IL-15Rα-KO iBMDMs (Figure 3D). Implantation of 4T1 tumor cells mixed with IL-15Rα-KO iBMDMs significantly impaired tumor growth compared to control iBMDMs (Figure 3E-G). Importantly, 4T1 tumor cells co-injected with IL-15Rα-KO iBMDMs showed higher CD8+ T cell content than those co-injected with control iBMDMs (Figure 3H-I). No differences were observed in the number of CD3+CD8– T cells (Figure 3I). To further confirm that the macrophage-expressed IL-15Rα reduces the proportion of CD8+ T cells in the TME, IL-15Rα+ TAMs and IL-15Rα– TAMs were sorted and inoculated with 4T1 tumor cells on the left and right fat pads of other mice (Figure 3J-K). 4T1 tumor cells co-injected with IL-15Rα– TAMs showed smaller tumor size and lower tumor weights (Figure 3L-M) and more CD8+ T cells (Figure 3N-O). We further adopted transgenic CD11b-DTR mice, which have a diphtheria toxin (DT) inducible system that depletes macrophages in various tissues after DT administration. We orthotopically implanted CD11b-DTR mice with EO771 cancer cells. DT administration of depleted existing macrophages was followed by the infusion of control iBMDMs or IL-15Rα-KO iBMDMs. The results showed that the infusion of IL-15Rα-KO iBMDMs significantly inhibited tumor growth (Supplementary Figure S5A-B). Compared to the control iBMDM group, there was no difference in the proportion of TAMs in tumors from the IL-15Rα-KO iBMDM group (Supplementary Figure S5C-D). It is noteworthy that IL-15Rα-KO iBMDM-treated tumors exhibited significantly more CD8+ T cell content (Supplementary Figure S5E). These results demonstrate that IL-15Rα-deficient TAMs increase the number of CD8+ T cells in the TME, thereby enhancing antitumor immunity.
3.4 IL-15Rα+ TAMs released IL-15Rc to decrease chemokine CX3CL1 expression in tumor cells
We examined the mechanism by which macrophage-expressed IL-15Rα reduces the infiltration of CD8+ T cells into the TME. Since the source of T cells depends on self-proliferation and recruitment from the peripheral blood, we first examined the expression of Ki-67 in CD8+ T cells from tumors co-injected with control iBMDMs or IL-15Rα-KO iBMDMs. No differences were observed (Figure 4A-B). Compared to co-cultures with IL-15Rα– TAMs, CD8+ T cell proliferation was unaffected by co-culture with IL-15Rα+ TAMs (Figure 4C), suggesting that macrophage-expressed IL-15Rα did not affect the proliferation of CD8+ T cells. Hence, macrophage-expressed IL-15Rα may affect CD8+ T cell recruitment.
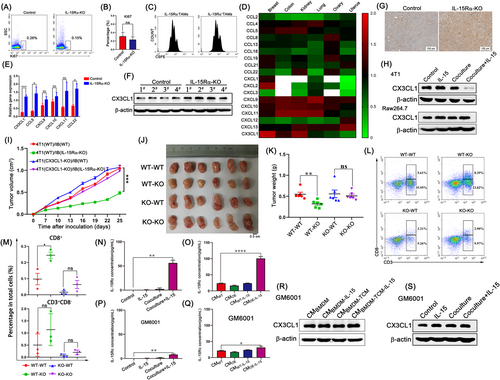
We used an unbiased approach to identify chemokines associated with CD4+ and CD8+ T cell infiltration in cancers using the multi-tumor gene-expression microarray dataset GSE2109 (Supplementary Figure S6). Cross-analysis revealed that among all chemokines, the expression of CX3CL1 was notable correlated with CD8A expression when compared with CD4 expression in breast cancer (Figure 4D). CX3CL1 (fractalkine) is the only member of the CX3C chemokine subfamily [32]. CX3C chemokine receptor 1 (CX3CR1) is the only receptor of CX3CL1 and is mainly expressed on CD8+ T cells, NK cells, DCs, and monocytes [33], indicating that CX3CL1 is more likely to recruit CD8+ T cells than CD4+ T cells. We found that, among all chemokine receptors, the expression of CX3CR1 was significantly higher in CD8+ T cells than in CD4+ T cells (Supplementary Figure S7A), which was further confirmed by flow cytometry and immunofluorescence staining (Supplementary Figure S7B-C). Analysis of the clinical sample database using the GEPIA2 platform showed that CX3CL1 expression in breast cancer tumor tissues was remarkably lower than that in normal tissues (Supplementary Figure S8A). Among patients with breast cancer, those with higher expression of CX3CL1 showed a longer disease-free survival time (Supplementary Figure S8B). The CX3CL1 content gradually decreased with tumor progression (Supplementary Figure S8C-D). Furthermore, we analyzed the expression of T cell-related chemokines in tumor samples co-injected with control iBMDMs or IL-15Rα-KO iBMDMs. The results showed that among all chemokines, CX3CL1 expression was most significantly upregulated in the IL-15Rα-KO group (Figure 4E-G and Supplementary Figure S8E). These data suggest that IL-15Rα+ TAMs may downregulate CX3CL1 expression in tumors to reduce the recruitment of CD8+ T cells.
To further confirm that CX3CL1 promotes CD8+ T-cell mobilization in breast tumors, thereby enhancing antitumor immunity, we generated 4T1 tumor cells that stably overexpressed CX3CL1 (CX3CL1-OE). The in vitro MTT assay and invasion assay showed that overexpression of CX3CL1 promoted 4T1 tumor cell proliferation, although it did not affect invasion ability (Supplementary Figure S9A-B). Overexpression of CX3CL1 in tumor cells significantly inhibited tumor growth in vivo (Supplementary Figure S9C-E) and increased CD8+ T cell content in the TME (Supplementary Figure S9F-G), as well as the percentage of TAMs (Supplementary Figure S9H-I).
To analyze whether CX3CL1 was derived from TAMs or tumor cells, which were the two cell types with the largest proportions in the TME, we performed immunofluorescence staining for IL-15Rα and CX3CL1. The results showed that CX3CL1 and IL-15Rα were not colocalized (Supplementary Figure S10A), suggesting that CX3CL1 was not derived from TAMs. Next, we constructed Raw264.7 macrophage cells that overexpressed IL-15Rα (IL-15Rα-OE) (Supplementary Figure S10B). After IL-15 stimulation, the expression of CX3CL1 remained the same in IL-15Rα-OE Raw264.7 cells (Supplementary Figure S10C). In the presence of IL-15 in the co-culture system, the expression of CX3CL1 in 4T1 tumor cells was significantly downregulated, whereas the expression of CX3CL1 in macrophages was unaltered (Figure 4H), suggesting that IL-15Rα+ TAMs can reduce the expression of CX3CL1 in 4T1 tumor cells. Similarly, CX3CL1 expression was also significantly decreased in EO711 tumor cells in the co-culture system in the presence of IL-15 (Supplementary Figure S10D). Tumor conditioned medium (TCM) can skew BMDM differentiation into TAMs [34] and upregulated IL-15Rα expression (Supplementary Figure S11A). We collected the culture medium from BMDMs cultured alone (CMBMDM), BMDMs stimulated by IL-15 (CMBMDM-IL-15), BMDMs stimulated by TCM (CMBMDM-TCM), or BMDMs stimulated by TCM and IL-15 (CMBMDM-TCM-IL-15), where CMBMDM-TCM-IL-15 decreased the protein level of CX3CL1 in 4T1 tumor cells (Supplementary Figure S11B). To further confirm that IL-15Rα+ TAMs can promote tumor growth by down-regulating the expression of CX3CL1 in tumor cells, we generated CX3CL1-knockout (CX3CL1-KO) 4T1 tumor cells. IL-15Rα-KO iBMDMs showed significant inhibitory effects on tumor growth of control 4T1 tumor cells, whereas CX3CL1-KO 4T1 tumor growth was not impaired (Figure 4I-K). Moreover, CD8+ T cell content did not increase when CX3CL1-KO 4T1 tumor cells were co-injected with IL-15Rα-KO iBMDMs (Figure 4L-M). In conclusion, IL-15Rα+ TAMs reduce the recruitment of CD8+ T cells by downregulating CX3CL1 expression in tumor cells, thus promoting tumor growth.
IL-15Rα functions not only as a membrane-bound receptor but also as a secreted protein [35]. Soluble IL-15Rα proteins generated by proteolytic cleavage can bind IL-15 to form IL-15Rc, which can stimulate neighboring cells. Since IL-15 did not affect the phenotype, phagocytosis, and ROS production of IL-15Rα-OE Raw264.7 macrophages (Supplementary Figure S12A-C), we hypothesized that the complex, formed by IL-15Rα shed while binding IL-15, is the IL-15Rα+ TAM-derived soluble factor that reduces the levels of CX3CL1 in 4T1 tumor cells. To evaluate this hypothesis, we measured the concentration of IL-15Rc in the co-culture system using ELISA. The results showed a higher concentration of IL-15Rc in the culture medium of the co-culture with IL-15 than in the absence of IL-15 (Figure 4N and Supplementary Figure S12D). The IL-15Rc concentration in the IL-15Rα-OE Raw264.7 cell culture supernatant was significantly upregulated after IL-15 stimulation (Figure 4O). GM6001 [36], a broad-spectrum MMP inhibitor, blocked sIL-15Rα release into the culture supernatants (Figure 4P and 4Q). The results showed that GM6001 completely prevented the CMBMDM-TCM-IL-15-induced reduction of CX3CL1 in tumor cells (Figure 4R). Similar results were observed in the co-culture experiments in the presence of GM6001 (Figure 4S). Taken together, these data suggest that IL-15Rc is necessary for the regulation of CX3CL1 levels in 4T1 tumor cells by IL-15Rα+ TAMs.
3.5 GMCSF induced the expression of γc to regulate the response of tumor cells to IL-15Rc
To further explore how IL-15Rc reduces CX3CL1 expression in tumor cells, we constructed IL-15Rc by combining IL-15 and IL-15Rα-Fc in vitro. Unexpectedly, IL-15Rc alone did not reduce CX3CL1 expression in 4T1 tumor cells (Figure 5A). We found that 4T1 tumor cells did not express γc and IL-2R/IL-15Rβ subunits (Supplementary Figure S13), which are responsible for delivering IL-15Rc signaling, while co-cultures with macrophages significantly upregulated the γc subunit but not the IL-2R/IL-15Rβ subunit in 4T1 tumor cells (Figure 5B). Neutralizing antibody blocking γc inhibited the reduction of CX3CL1 expression in tumor cells in the co-culture system, whereas blocking IL-2R/IL-15Rβ had no effect (Figure 5C). This result was further confirmed when IL-15Rc alone reduced CX3CL1 expression in tumor cells overexpressing γc (Figure 5D-E). These data suggest that macrophages promote the tumor cell response to IL-15Rc by upregulating γc expression in tumor cells in an IL-2R/IL-15Rβ-independent manner.
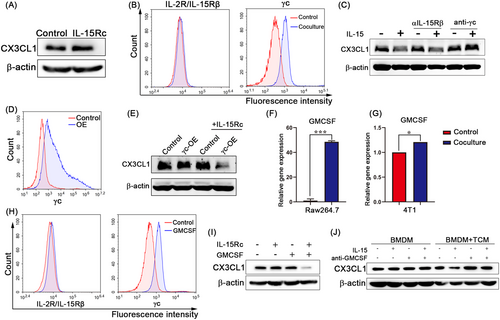
We investigated the soluble factors that upregulate γc expression in tumor cells in a co-culture system. We specifically analyzed the differential genes of secretory proteins in macrophages co-cultured with tumor cells and macrophages alone (GEO: GSE99960). Among these genes, GMCSF was the most upregulated (Supplementary Figure S14). Additionally, in the co-culture system, GMCSF was significantly upregulated in macrophages but only slightly changed in tumor cells (Figure 5F-G). IL-15Rc combined with GMCSF promoted the expression of γc but not IL-2R/IL-15Rβ (Figure 5H) and led to the reduction of CX3CL1 expression in tumor cells (Figure 5I). Neutralizing antibodies against GMCSF treatment potently abrogated the effects of reducing CX3CL1 expression in tumor cells induced by the culture medium of BMDMs stimulated by TCM (Figure 5J). Together, these results suggest that GMCSF acts as a switch to promote the response of tumor cells to IL-15Rc by regulating the expression of γc.
3.6 Non-transcriptional activity of HIF-1α was necessary for IL-15Rc-induced reduction of CX3CL1 expression in tumor cells
Signal transducer and activator of transcription 3 (STAT3) and STAT5 are known downstream targets of IL-15Rc in T cells and NK cells [37]. However, the expression and phosphorylation levels of STAT3 and STAT5 in 4T1 tumor cells were not affected by GMCSF or IL-15Rc (Figure 6A), indicating that STAT3 and STAT5 do not regulate CX3CL1. At least three signaling pathways downregulate CX3CL1 expression: hypoxia, 15d-PGJ2, and soluble IL-6 receptor α (sIL-6Rα) [38]. We observed that after co-culture with macrophages in the presence of IL-15, the CX3CL1 mRNA level in 4T1 tumor cells remained unchanged (Figure 6B). Hypoxia showed no effect on CX3CL1 mRNA degradation (Figure 6C) [39], whereas both 15d-PGJ2 and sIL-6Rα were reported to promote CX3CL1 mRNA degradation [40, 41]. In contrast to previous studies, CX3CL1 protein levels were significantly upregulated in 4T1 tumor cells under hypoxic conditions (Figure 6D). HIF-1α is the master regulator that responds to hypoxic conditions and is correlated with CX3CL1 in 4T1 tumor cells. We explored the possibility that HIF-1α is involved in the regulation of CX3CL1 in IL-15Rα+ TAMs. The protein levels of HIF-1α and CX3CL1 were decreased in 4T1 tumor cells co-cultured with macrophages in the presence of IL-15 under either normoxic or hypoxic conditions (Figure 6E). Besides, IL-15Rc combined with GMCSF also reduced the protein levels of HIF-1α and CX3CL1 in tumor cells (Supplementary Figure S15A). Additionally, the HIF-1α protein level was strongly correlated with the CX3CL1 protein level (R2 = 0.792) in 4T1 tumor progression (Figure 6F-G). To further confirm the relationship between HIF-1α and CX3CL1, the selective HIF-1α inhibitor, PX-478 [42], was used to decrease HIF-1α protein levels. The results showed that CX3CL1 protein levels were significantly decreased in 4T1 tumor cells treated with PX-478, while the mRNA level remained unchanged (Supplementary Figure S15B-C). Additionally, we generated 4T1 tumor cells stably overexpressing HIF-1α (HIF-1α-OE) and HIF-1α-deficient (HIF-1α-KO) 4T1 tumor cells using lentiviral infection and CRISPR-Cas9 system, respectively. CX3CL1 protein levels were increased in HIF-1α-OE 4T1 tumor cells but decreased in HIF-1α-KO 4T1 tumor cells (Figure 6H); however, mRNA levels were unaffected in HIF-1α-OE and HIF-1α-KO cells (Figure 6I). CMBMDM-TCM-IL-15 did not reduce CX3CL1 expression in HIF-1α-KO 4T1 tumor cells (Figure 6J). Taken together, we conclude that HIF-1α is the upstream regulator of CX3CL1 in tumor cells.
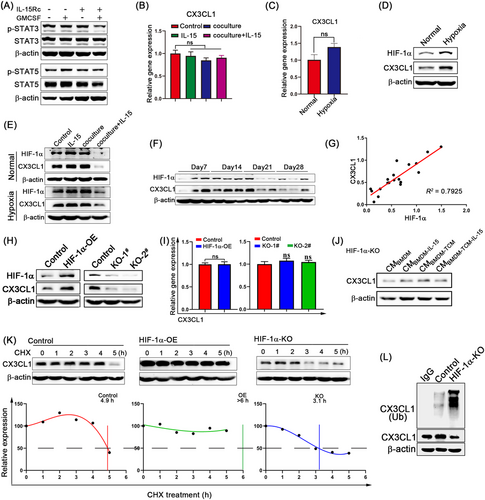
CX3CL1 mRNA levels were not affected by HIF-1α overexpression or knockout, suggesting that HIF-1α regulates CX3CL1 protein levels independent of mRNA transcription. Next, we studied CX3CL1 protein stability by treating 4T1 tumor cells with CHX, a compound that blocks de novo protein synthesis. HIF-1α overexpression significantly stabilized the level of CX3CL1 in cells treated with CHX and prolonged its half-life, while CX3CL1 half-life was shortened in HIF-1α-KO 4T1 tumor cells (Figure 6K). Furthermore, we observed that CX3CL1 ubiquitination was increased in HIF1-1α-KO cells (Figure 6L), which suggested that HIF-1α could suppress the ubiquitination of CX3CL1. Additionally, CX3CL1 expression remained reduced in HIF-1α-KO cells after blocking protein transport using monensin (Supplementary Figure S15D), indicating that HIF-1α did not affect CX3CL1 release. These results indicate that HIF-1α protects CX3CL1 from degradation.
3.7 Blocking the formation of IL-15Rc increased CD8+ T cell infiltration and overcame the resistance of breast cancer to PD-1 blockade
IL-15 has four α-helices, of which the B helix primarily interacts with IL-15Rα and the D helix primarily interacts with the γ-chain (γc) [43]. Based on the amino acid sequence of the B helix, we designed a B peptide to block the binding of IL-15 to IL-15Rα. As expected, B peptide treatment completely abrogated the decrease of CX3CL1 expression in tumor cells in the co-culture experiments (Figure 7A). Similarly, the B peptide totally prevented the CMBMDM-TCM-IL-15-induced decrease in CX3CL1 expression (Figure 7B). Considering the inhibitory effect of the B peptide on IL-15Rα+ TAM functions, we studied the antitumor effect of the B peptide in vivo. Tumor growth was significantly inhibited in mice treated with B peptide compared to vehicle-treated or scramble-treated controls (Figure 7C-D). The body weights of the mice in all experimental groups were unaffected (Figure 7D). Additionally, in vitro MTT and invasion assays showed that the B peptide had no direct effect on the proliferation and invasion ability of tumor cells (Supplementary Figure S16A-C). Next, we analyzed the infiltration of immune cells into the TME. As expected, the CD8+ T cell content was higher in B peptide-treated tumors, while CD3+CD8– T cell content did not change (Figure 7E-F). The B peptide promoted the infiltration of TAMs and NK cells in the TME (Supplementary Figure S16D-G), but did not affect MDSC and DC content (Supplementary Figure S16H-K). We also observed that the B peptide increased the levels of CX3CL1 and HIF-1α in the tumor compared to the control group (Figure 7G-H). Therefore, we conclude that the B peptide, capable of blocking the formation of the IL-15 complex, is a promising candidate for the clinical development of anti-breast cancer drugs.
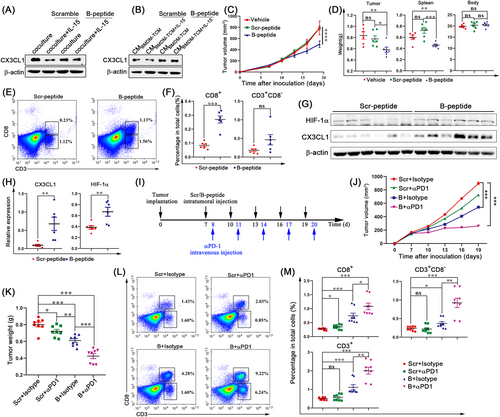
Sufficient T-cell infiltration is essential for tumor responses to PD-1 blockade [44]; for example, 4T1 tumors showed resistance to anti-PD-1 therapy [27]. Since our data showed that the B peptide promoted the infiltration of CD8+ T cells, the possibility of the B peptide overcoming PD-1 resistance in 4T1 tumors could be considered. In fact, the combination of anti-PD-1 and the B peptide significantly delayed tumor growth compared to B peptide or PD-1 blockade alone (Figure 7I-K). More CD8+ T-cell and CD3+CD8– T-cell infiltration was observed in the combined treatment (Figure 7L-M). Additionally, we quantified the expression level of PD-1 in T cells after the treatment with either scramble peptide or B peptide. As shown in Supplementary Figure S16L, B peptide did not influence PD-1 expression in T cells, suggesting that the function of B peptide in enhancing anti-PD-1 efficacy was not through influencing PD-1 expression on T cells. Collectively, these data suggest that the B peptide, which increases CD8+ T-cell infiltration, overcomes the resistance of 4T1 breast cancer to PD-1 blockade.
4 DISCUSSION
The phenotypic and functional properties of macrophages recruited by tumor cells influence their differentiation and the phenotype of tumor cells. This crosstalk plays a central role in cancer development and has emerged as a critical mechanism for drug resistance. Our study showed that the crosstalk between macrophages and tumor cells converted IL-15 signaling to reduce CD8+ T cell recruitment in the breast cancer microenvironment (Supplementary Figure S17).
In this study, we found that tumor cells increased the IL-15Rα expression in macrophages. It is well established that IL-15Rα on the surface of macrophages presents IL-15 in trans to CD8+ T cells and NK cells, which promotes the activation, proliferation, and cytolytic activity of CD8+ T cells and NK cells, thereby exerting antitumor effects [45]. However, how TAM-derived IL-15Rα functions in the TME and whether it contributes to IL-15 resistance in breast cancer remains unknown. Here, we demonstrated the presence of IL-15Rα+ TAMs in breast cancer and characterized their tumor-promoting functions. We characterized the specific expression pattern of IL-15Rα on TAMs and confirmed the positive association between the expression of IL-15Rα and macrophage markers (CD68 and CD163) in patients with breast cancer by analyzing multiple databases (Figure 2E). Moreover, TAM-expressed IL-15Rα reduced the recruitment of CD8+ T cells via IL-15Rc. Thus, our study revealed a novel mechanism of IL-15Rα in cancer cell biology and suggested that IL-15Rα plays key roles in the TME and influences tumor progression.
TAMs contribute to tumor progression at multiple levels, including promoting proliferation and survival, nurturing cancer stem cells, supporting metastasis and an immunosuppressive microenvironment [46]. Numerous studies have revealed extensive heterogeneity among TAM subsets [47]. TAMs can also act as potent effector cells and enhance the activation of cytotoxic killer cells, such as T cells and NK cells, to initiate the antitumor effect [48]. Several clinical studies have shown that certain TAM subsets were associated with good prognosis in patients [49, 50], but more data are needed to further identify the function of specific TAM subsets to help develop more targeted therapies. We specifically investigated the role of IL-15Rα+ TAMs in regulating the tumor immune microenvironment. Correlation analysis showed that the percentage of IL-15Rα+ TAMs was negatively correlated with the ratio of CD8+ to CD4+ T cells (Figure 3C). We demonstrated that IL-15Rα+ TAMs explicitly reduced the recruitment of CD8+ T cells, which is contrary to the macrophage-derived IL-15Rα that can promote the proliferation and activation of CD8+ T cells under physiological conditions. These data indicate that there is a novel mechanism capable of converting IL-15 antitumor signals into tumor-promoting signals. Similarly, a recent study reported that IL-15 promoted the proliferation and insulin like growth factor-1 (IGF-1) production of IL-15Rα+ myeloid cells (CD45+CD11b+GR-1+) in lung cancer in immunodeficient mice [51]. Therefore, whether IL-15Rα+ TAMs exhibit similar functions in tumor types other than breast cancer requires further investigation.
The source of T cells depends on self-proliferation and recruitment from the peripheral blood. After excluding the effect of proliferation, we speculated that CX3CL1 was probably the key factor for IL-15Rα+ TAM function through a comparative analysis of chemokines in breast cancer and their receptors on T cells. The expression of CX3CL1 was reduced in breast cancer tissues compared to normal tissues in the GEPIA2 dataset (Supplementary Figure S8A). Next, we provided evidence that IL-15Rα+ TAMs reduced the expression of CX3CL1 in tumor cells, but not in macrophages (Figure 4H). CX3CL1 has been demonstrated to be involved in the proliferation, survival and invasion of various malignant tumor types [52-54]. MTT assay in vitro showed that CX3CL1 overexpression promoted 4T1 tumor cell proliferation (Supplementary Figure S9A). However, CX3CL1 overexpression in 4T1 tumor cells significantly inhibited tumor growth in vivo and increased CD8+ T cell content in the TME (Supplementary Figure S9C-G), which is consistent with a previous study showing that high expression of CX3CL1 by tumor cells was associated with good prognosis and increased tumor-infiltrating CD8+ T cells, NK cells, and DCs in breast carcinoma patients [55]. Antitumor immune effects are elicited by CX3CL1 by recruiting effector cells into tumor tissues [56-58]. However, the profile and regulatory mechanism of CX3CL1 expression in tumors remains unknown. We provide evidence that IL-15Rα+ TAMs may be an important factor in the regulation of CX3CL1 expression in breast cancer.
The present study confirmed that the IL-15Rc is necessary for reducing CX3CL1 expression in 4T1 tumor cells. In the crosstalk between IL-15Rα+ TAMs and tumor cells, GMCSF acted as a switch to promote tumor cell response to IL-15Rc. Similar results have been reported where GMCSF and IL-15 worked together as immunosuppressors and allowed engraftment of allogeneic B16F10 cells in immunocompetent mice [59]. GMCSF regulated γc expression in tumor cells without affecting IL-2R/IL-15Rβ, suggesting that the response of 4T1 tumor cells to IL-15Rc is not dependent on the conventional γc/β receptor. One study has found the presence of a functional hybrid receptor of the GMCSF receptor β and γc that can bind IL-15Rc in hematopoietic CD34+ cells [60]. Whether the hybrid receptor also exists in 4T1 tumor cells requires further investigation. The source of GMCSF in tumor tissues has not been determined, but we found that the expression of GMCSF in macrophages increased significantly after co-culturing with 4T1 tumor cells in vitro (Figure 5F), suggesting that GMCSF is likely derived from TAMs. Recently, IL-15Rc has become a promising candidate for tumor immunotherapy because it promotes the activation and proliferation of both CD8+ T cells and NK cells [61]. Despite the fact that it did not alter 4T1 primary tumor growth, the IL-15 agonist complex or ALT-803 is currently being evaluated in a phase Ib/II clinical trial in which ALT-803 has been used intravesically in combination with Bacillus Calmette-Guérin (BCG) in adult patients with BCG-naïve non-muscle-invasive bladder cancer (NCT02138734) [8]. The mechanism by which tumor cells respond to IL-15Rc remains unclear. Our data demonstrated that blocking the binding of IL-15 to IL-15Rα by the B peptide inhibited tumor growth and increased the infiltration of CD8+ T cells (Figure 7C-F). Additionally, the B peptide overcame the resistance of 4T1 breast cancer cells to PD-1 blockade (Figure 7J-K). These data suggest that IL-15Rc resistance exists in 4T1 breast tumors. For IL-15Rc to be an effective immunotherapeutic agent, further studies are needed to elucidate the role of IL-15Rc in cancer cell biology.
HIF-1α is a master transcriptional regulator of the hypoxic response pathway and has been widely investigated in various physiological and pathological conditions [62, 63]. A limited number of HIF-1α non-transcriptional pathways have been identified in which HIF-1α functions as a chaperone molecule to facilitate the function of another protein. HIF-1α has been shown to interact with mouse double minute 2 protein (MDM2) to suppress p53 ubiquitination [64]. Additionally, HIF-1α can bind to γ-secretase and directly modulate its enzymatic activity in Alzheimer's disease [65]. We revealed that IL-15Rc downregulated CX3CL1 expression in tumor cells via the non-transcriptional activity of HIF-1α. HIF-1α did not regulate the mRNA levels of CX3CL1; instead, HIF-1α promoted the protein expression levels of CX3CL1 by inhibiting the ubiquitination of CX3CL1. However, the detailed mechanism is still unknown, and further studies are needed to investigate how HIF-1α regulates CX3CL1 ubiquitination.
Our study did have some limitations. First, we used iBMDMs to study the function of IL-15Rα-expressing macrophages. iBMDMs are phenotypically comparable to primary macrophages and are commonly used to examine the mechanisms of macrophage behavior. However, to study the effect of macrophages on tumor growth in vivo, the use of an IL-15Rα genetic ablation mouse model may strengthen the reliability of these results. Second, the results were based on the 4T1 tumor model. Although the 4T1 tumor model is appropriate for breast cancer, it is only representative of triple-negative breast cancer. Therefore, whether these findings can be generalized to other breast cancer subtypes requires further investigation.
5 CONCLUSIONS
Taken together, our current work reveals a crosstalk between macrophages and tumor cells in the breast cancer microenvironment. Our data indicate that the IL-15/IL-15Rα complex formed by IL-15Rα+ TAMs, upon binding to γc on tumor cells upregulated by GMCSF, decreases the stability of the chemokine CX3CL1 protein recruiting CD8+ T cells in tumor cells through the non-transcriptional activity of HIF-1α. Blocking the binding of IL-15 and IL-15Rα increases the infiltration of CD8+ T cells and improves the efficacy of anti-PD-1 immunotherapies.
DECLARATIONS
ACKNOWLEDGEMENT
We are grateful to Prof. Feng Shao (National Institute of Biological Sciences, Beijing, 102206, China) for kindly providing iBMDMs. We also thank Yuxin Wang for the technical support in data mining and the related bioinformatic analysis. This research was financed by grants from the National Key Research and Development Plan (2017YFA0506000), and Guangdong Basic and Applied Basic Research Foundation (2021B1515120016), National Natural Science Foundation of China (82072822), the Key Research and Development Program of Jiangsu Province, China-Social Development Projects (BE2020687).
COMPETING INTERESTS
The authors declare no conflict of interests.
AUTHOR CONTRIBUTIONS
PP.S., HQ.G., and WL.Z. conceived and designed the project. WL.Z., Q.Z., and HF.S. performed the experiments. WL.Z., Q.S., TS.L., and ZL.H. analyzed the data. PP.S., WL.Z., Q.S., and NF.Y. wrote the manuscript. PP.S. financially supported the research. PP.S., WX.W., and NF.Y. supervised the study. All authors read and approved the final manuscript.
ETHICS APPROVAL
Animal welfare and experimental procedures were performed in strict accordance with the Guide for the Care and Use of Laboratory Animals (The Ministry of Science and Technology of China) and the related ethical regulations of our university. The protocol was approved by the State Key Laboratory of Pharmaceutical Biotechnology at Nanjing University, and efforts were made to minimize suffering.
Open Research
DATA AVAILABILITY STATEMENT
All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.




