Phosphatidylserine released from apoptotic cells in tumor induces M2-like macrophage polarization through the PSR-STAT3-JMJD3 axis
Abstract
Background
Understanding how the tumor microenvironment is shaped by various factors is important for the development of new therapeutic strategies. Tumor cells often undergo spontaneous apoptotic cell death in tumor microenvironment, these apoptotic cells are histologically co-localized with immunosuppressive macrophages. However, the mechanism by which tumor cell apoptosis modulates macrophage polarization is not fully understood. In this study, we aimed to explore the tumor promoting effects of apoptotic tumor cells and the signal pathways involved.
Methods
Apoptotic cells and macrophages in tumors were detected by immunohistochemical staining. Morphological analysis was performed with Giemsa staining. Lipids generated from apoptotic cells were detected by liquid chromatography-mass spectrometry. Phosphatidylserine-containing liposomes were prepared to mimic apoptotic cells. The expression of protein was determined by real-time PCR, immunohistochemistry enzyme-linked immunosorbent assay and Western blotting. Mouse malignant ascites and subcutaneous tumor models were designed for in vivo analysis. Transgenic mice with specific genes knocked out and inhibitors specific to certain proteins were used for the mechanistic studies.
Results
The location and the number of apoptotic cells were correlated with that of macrophages in several types of carcinomas. Phosphatidylserine, a lipid molecule generated in apoptotic cells, induced polarization and accumulation of M2-like macrophages in vivo and in vitro. Moreover, sustained administration of phosphoserine promoted tumor growth in the malignant ascites and subcutaneous tumor models. Further analyses suggested that phosphoserine induced a M2-like phenotype in macrophages, which was related to the activation of phosphoserine receptors including T-cell immunoglobin mucin 4 (TIM4) and the FAK-SRC-STAT3 signaling pathway as well as elevated the expression of the histone demethylase Jumonji domain-containing protein 3 (JMJD3). Administration of specific inhibitors of these pathways could reduce tumor progression.
Conclusions
This study suggest that apoptotic cell-generated phosphoserine might be a notable signal for immunosuppressive macrophages in tumors, and the related pathways might be potential therapeutic targets for cancer therapy.
Abbreviations
-
- TAM
-
- tumor-associated macrophage
-
- PS
-
- phosphatidylserine
-
- PSR
-
- phosphatidylserine receptor
-
- FBS
-
- fetal bovine serum
-
- shRNA
-
- short hairpin RNA
-
- FAK
-
- focal adhesion kinase
-
- IL-10
-
- interleukin 10
-
- ELISA
-
- enzyme-linked immunosorbent assay
-
- CM
-
- conditioned medium
-
- LC-MS
-
- liquid chromatography-mass spectrometry
-
- PSL
-
- phosphatidylserine liposome
-
- PC
-
- phosphatidylcholine
-
- PCL
-
- phosphatidylcholine liposome
-
- Chol
-
- cholesterol
-
- RT-PCR
-
- reverse transcription PCR
-
- TGF-β
-
- transforming growth factor beta
-
- SEM
-
- standard error of mean
-
- M-CSF
-
- macrophage colony-stimulating factor
-
- AC
-
- apoptotic tumor cell
-
- ACM
-
- apoptotic cell culture medium
-
- VEGF
-
- vascular endothelial growth factor
-
- Arg1
-
- arginase 1
-
- CCL2
-
- C-C motif chemokine ligand 2
-
- iNOs
-
- inducible nitric oxide synthase
-
- Irf4
-
- interferon-regulatory factor 4
-
- JMJD3
-
- Jumonji domain-containing protein 3
-
- MERTK
-
- C-MER proto-oncogene tyrosine kinase
-
- Mgl1
-
- macrophage galactose-type C-type lectin 1
-
- Mrc1
-
- mannose receptor C type 1
-
- PE
-
- phosphatidylethanolamine
-
- STAT3
-
- signal transducer and activator of transcription 3
-
- TIM4
-
- T-cell immunoglobin mucin 4
-
- TNF-α
-
- tumor necrosis factor alpha
-
- HPF
-
- high power field
-
- DNR
-
- daunorubicin
1 BACKGROUND
The major challenge for immunotherapy of cancer lies in how to turn the “cold” tumor into a “hot” one [1]. Cold tumors are described as an immune desert filled with abundant immunosuppressive cells, such as immunosuppressive tumor-associated macrophages (TAMs), which are known to mostly adopt an alternatively activated (M2-like) phenotype [2, 3] and promote tumor progression by inducing angiogenesis, drug resistance, and immunosuppression [4, 5]. Understanding the origin of the immunosuppressive M2 phenotype and the underlying mechanisms by which tumoral signals affect macrophage polarization is important for finding a strategy to reshape the suppressive microenvironment [6].
In most physiological cases, apoptotic cells are engulfed by macrophages, which results in the histological co-localization of apoptotic cells with macrophages and the release of anti-inflammatory cytokines [7, 8]. Notably, spontaneous apoptotic cell death is also one of the characteristics of the tumor microenvironment, as the typical features of a tumor tissue with ischemia and hypoxia could also lead to the accumulation of apoptotic cells. Interestingly, the macrophages responsible for cell clearance that localize around the apoptotic cells in tumors are recognized as TAMs [9, 10]. Most TAMs have been reported to adopt an M2-like phenotype and promote tumor progression by facilitating angiogenesis and suppressing anti-tumor immunity [11, 12]. To date, the mechanisms underlying the origin of TAMs are not fully understood.
Here, we hypothesized that one or more candidate molecules released from apoptotic tumor cells may modulate macrophage polarization, which in turn yields tumor-promoting effects. To explore this possibility, we determined the composition of crude lipid extracts of apoptotic tumor cells and identified a single molecule, phosphatidylserine (PS), which could induce macrophage polarization towards the M2-like phenotype. We then examined the related receptors and the underlying signaling pathways that are involved in this process.
2 MATERIALS AND METHODS
2.1 Animals
Jumonji domain-containing protein 3 (Jmjd3)-flag knock-in mice were generated by insertion of a Flag-tag downstream of the stop codon of the Jmjd3 gene. Jmjd3 conditional knockout (Jmjd3−/−) mice were generated by targeted deletion of exons 14-20 and were produced by breeding Jmjd3flox/flox mice with LysM-Cre mice. Signal transducer and activator of transcription 3 (Stat3) conditional knockout (Stat3−/−) mice were generated by breeding Stat3flox/flox mice with LysM-Cre mice. Both Jmjd3 knock-in mice and Jmjd3flox/flox mice, were gifts from Prof. Charlie Degui Chen (Chinese Academy of Sciences, Shanghai, China) [13]. Stat3flox/flox mice, C-MER proto-oncogene tyrosine kinase (Mertk)-deficient (Mertk−/−) mice, and LysM-Cre mice were purchased from Jackson Laboratory (Bar Harbor, Maine, USA). Genotyping analysis of genomic DNA from tails was performed before use (representative genotyping analysis gels were shown in Supplementary Figure S1A-C). Female BALB/C and C57/BL6 mice (6 to 8 weeks old, 18-22 g) were purchased from Vital River (Beijing, China). All studies involving mice were approved by the Animal Care and Use Committee of Sichuan University as we did before [14].
2.2 Cell lines, cell culture, and transfection
Tumor cell lines CT26, LL2, and 4T1 were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and tumor cell line ID8 was purchased from Millipore Sigma (St. Louis, MO, USA). These tumor cells were cultured in complete RPMI-1640 (Gibco, Carlsbad, CA, USA) or DMEM medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA), 100 U/mL streptomycin and 100 μg/mL penicillin (Pen Strep; Gibco, Carlsbad, CA, USA), at 37°C in 5% CO2. All cell lines were tested for mycoplasma. Cell lines were not independently authenticated beyond the identity provided from ATCC.
The murine Bcl2 gene (GenBank accession number NM_009741) was cloned by OriGene Technologies Inc (Rockville, MD, USA). The short hairpin RNA plasmids against mouse Bcl2 (shBcl2, TRCN0000226262, target sequence:CCACAAGTGAGTCGACAAAC) and packaging vectors (pMD2.0G and psPAX) were purchased from Sigma-Aldrich (St Louis, MO, USA). Bcl2-overexpressed/shBcl2 CT26 cells were achieved by infected with lentivirus carrying a Bcl2-overexpressing plasmid or a shBcl2 plasmid, respectively, and selected with puromycin (Sigma-Aldrich, St Louis, MO, USA)
2.3 Antibodies and reagents
The following antibodies for flow cytometry were purchased from Biolegend (San Diego, CA, USA CA), including CD45 (PerCP/Cy5.5 130132, APC 103112; CD11b (APC 101212;); F4/80 (PE 123110, APC 123116); CD206 (PE 141706, FITC 141704, APC 141707); TIM4 (PE 130005); CD36 (PE 102605); IL-10 (Pacific Blue, 505019);TGF (TGF-β, Brilliant Violet 421, 141408) The following flow cytometry antibodies were purchased from BD Biosciences (San Jose, CA, USA), including CD45 (PE 553081, FITC 553080); CD11b (PE 553311, FITC 553310).The antibodies including Stabilin-2 (bs-12346R; Bioss, Beijing, China); RAGE (PA5-24787; Thermo Scientific, Carlsbad, CA, USA); MERTK (AF591; R&D Systems, Minneapolis, MN, USA); and BAI 1 (MB39213488; MyBiosource, San Diego, CA, USA) were used in flow cytometry analysis too. Alexa Fluor 488-conjugated donkey-anti-rabbit and Alexa Fluor 594-conjugated donkey-anti-rat antibodies were purchased from Invitrogen (Invitrogen, Carlsbad, CA, USA).
Anti-phospho-FAK (Tyr397, #3283), anti-FAK (#3285), anti-phospho-STAT3 (Tyr705, clone D3A7, #9145), anti-STAT3 (clone 124H6, #9139), anti-Histone H3 (clone D1H2, #4499), Tri-Methyl-Histone H3 (Lys27, clone C36B11, #9733), and anti-β-ACTIN (clone 8H10D10, #3700) antibodies were purchased from Cell Signaling Technology (CST, Beverly, MA, USA); anti-phospho-SRC (Tyr416) and anti-SRC antibodies were obtained from HuaBio (Hangzhou, Zhejiang, China); and an anti-Flag antibody was purchased from Sigma-Aldrich. All these antibodies were used for western blotting.
For the detection of apoptotic cells, the FITC Annexin V-Apoptosis Detection Kit from BD Pharmingen (San Jose, CA, USA) was used. A Multi-Plex Kit (Cat. MCYTOMAG-70K) from Merck Millipore (Billerica, MA, USA) was used for cytokine and chemokine analysis. The interleukin-10 (IL-10) enzyme-linked immunosorbent assay (ELISA) Kit was obtained from eBioscience (San Diego, CA, USA), and the Tumor growth factor-beta (TGF-β) ELISA kit was purchased from DaKeWe Biotech (Shenzhen, Guangdong, China). DNAse I was purchased from Thermo Scientific (Carlsbad, CA, USA), and trypsin was obtained from Gibco (Carlsbad, CA, USA).
The MERTK inhibitor UNC569 [15] was purchased from Merck Millipore, and the JMJD3 inhibitor GSK-J1/4 [16] was purchased from Sigma-Aldrich. The STAT3 inhibitor S3I-201 [17], the SRC inhibitor PP2, and the FAK inhibitor PF-573228 were purchased from Selleck Chemicals (Houston, TX, USA). Daunorubicin hydrochloride (DNR) was purchased from Meilun Biotech (Dalian, Liaoning, China), and latex beads (1 mm, 0.01%) were purchased from Sigma-Aldrich.
2.4 Tumor tissue microarray, tumor specimens, immunohistochemistry, and immunofluorescence
The tumor tissue microarrays were purchased from Shanghai Outdo Biotech Company (Shanghai, China). Apoptotic cells were detected with an anti-cleaved-Caspase3 antibody (CST, Beverly, MA, USA), and M2 macrophages were detected with an anti-CD163 antibody (Abcam,OTI2G12, Cambridge, MA, USA) by immunofluorescence. The antigenic binding sites in the tissue microarray HColA180Su15 (contained 100 colorectal cancer tissues, 8 of which were excluded due to incompleteness) were visualized using the Opal 7-Color Manual IHC Kit (NEL811001KT; PerkinElmer, Waltham, MA, USA) according to the manufacturer's protocol. Multicolour immunohistochemical data were collected with a Vectra Polaris Automated Quantitative Pathology Imaging System (CLS143455; PerkinElmer Waltham, MA, USA) and analyzed with InForm 2.4.2 software (PerkinElmer Waltham, MA, USA). The antigenic binding sites in the tissue microarrays HbreD030 (contained 30 Breast Cancer tissues, 1 of which was excluded due to incompleteness) and HlungA020 (contained 20 Lung Cancer tissues), were visualized by immunohistochemical staining. Briefly, After the microarrays deparaffinization and rehydration, endogenous peroxide was blocked with 3% H2O2. Then the microarrays were soaked 10 mM sodium citrate buffer (pH 6.0) and antigen retrieval was performed with an autoclave. After washed with PBS (phosphate-buffered saline, Servicebio, Wuhan, China), the microarrays were blocked with goat serum and then incubation with indicated primary antibody overnight at 4°C. After PBS washing, the microarrays were incubated with the appropriate secondary antibody with HRP (horseradish peroxidase) conjugated. Then HRP was detected with diaminobenzidine peroxide solution and cell nuclei were gently counterstained with hematoxylin. All the reagents used were purchased from Beyotime Institute of Biotechnology (Shanghai, China). The data were visualized with the Pannoramic MIDI II system (3DHISTECH Ltd., Budapest, Hungary) and analyzed with Case Viewer software (3DHISTECH Ltd., Budapest, Hungary).
Apoptotic cells (cleaved Caspase3+) and macrophages (F4/80+) in CT26, 4T1, and LL2 subcutaneous tumor tissues were detected by immunofluorescence staining and digitally photographed under a fluorescence microscope (Olympus BX53F, Olympus, Tokyo, Japan). Briefly, Tumor frozen sections with O.C.T. compound were fixed with cool acetone, and blocked with goat serum. Then immuno-stained overnight with indicated primary antibodies overnight at 4°C. After PBS washing, the sections were incubated with the appropriate secondary antibodies with specific fluorescein conjugated. Cell nuclei were stained with DAPI (4’,6-diamidino-2-phenylindole, Beyotime, Shanghai, China). The number of macrophages and apoptotic cells per high-power field was counted by two independent pathologists. Ten high-power fields per specimen were analyzed, and the means were used for correlation analysis via Pearson's correlation coefficient I.
2.5 Preparation of apoptotic tumor cells and conditioned medium (CM) and crude lipids of apoptotic tumor cells
Apoptotic tumor cells were harvested by heating CT26 cells in a 40°C water bath for 1 h. The apoptotic rate of heat-induced apoptotic cell was determined by flow cytometry, and the cell proliferative potential was determined with Cell Counting Kit 8 (CCK8, data not shown). Then, apoptotic tumor cells were cultured for another 24 h to obtain CM of apoptotic tumor cells. After centrifugation at 4000 g for 30 min to remove cells and cell debris, CM of apoptotic tumor cells was filtered through a 0.22-μmol/L microporous membrane (Merck Millipore) and stored at -20°C until use. In the apoptotic tumor cell-CM inactivation study, apoptotic tumor cell-CM was heated at 100°C for 20 min or digested with DNAse I (100 U/mL) or trypsin (10 μg/mL) at 37°C for 8 h. The enzyme was inactivated by heating at 85°C for 5 min.
DNR-induced apoptotic tumor cells (DNR-CT26) were harvested by incubating CT26 cells with DNR (5 μg/mL; Meilun Biotech, Dalian, Liaoning, China) for 24 h and washed 3 times with phosphate-buffered saline (PBS). Heat-induced apoptotic tumor cells were cultured in complete RPMI-1640 medium (without FBS) for another 24 h.
Crude lipids were extracted by mixing the apoptotic tumor cells or apoptotic tumor cell-CM with HPLC-grade (high performance liquid chromatography grade) chloroform-methanol (2:1, vol/vol) and dried under a stream of N2 [18]. The crude lipid extract used for macrophage stimulation was re-dissolved in complete RPMI-1640 medium with 10% FBS and sterilized by filtration through a 0.22-μm Millipore microporous membrane. For LC-MS (liquid chromatography-mass spectrometry) analysis, the crude lipid was re-dissolved in methyl alcohol and analyzed with a 3200Q TRAP LC/MS/MS system (AB Sciex, Foster City, CA, USA). In brief, chromatography was carried out on Waters ACQUITY (Waters Technologies Corporation, MA, United States) UHPLC system equipped with a Waters auto sampler and column thermostat. A ACQUITY UPLC CSH C18 (150 × 2.1 mm), 1.7 μm column from Waters Technologies Corporation, maintained at 40 temperature was used for separation of analytes. The mobile phase consisted of 0.1% (v/v) formic acid in water (A) and acetonitrile (B) in binary gradient ratio with a flow rate of 0.2 mL. The gradient proportion was as follows: started with 62% of B, increased to 64% by 10 min; and decreased to 62% by 3 min. The autosampler temperature was maintained at 10 °C and the sample injection volume was 10 μL. The mass spectrometry detector is 3200Q TRAP.
2.6 Liposome preparation
The PS-containing liposome (PSL; PS/PC/Chol = 8/8/5) and phosphatidylcholine (PC)-containing liposome (PCL, PC/Chol = 8/5) were prepared in a film dispersion method. In brief, L-α-PS (sodium salt, Avanti Polar Lipids, Alabaster, AL, USA), PC (Sigma-Aldrich) and cholesterol (Chol; Sigma-Aldrich) were dissolved and mixed in chloroform and evaporated on a rotary evaporator to form the thin film. The thin film was hydrated in sterile 0.9% NaCl (Sichuan Kelun Pharmaceutical Co, Chengdu, Sichuan, China), then sterilized by filtration through a 0.22-μm Millipore microporous membrane, and stored in the dark at 4°C until use (no more than 2 weeks).
2.7 Preparation and polarization of peritoneal macrophages
Mice were anesthetized and intraperitoneally injected with 5-8 mL of RPMI-1640. After abdominal kneading, peritoneal lavage fluids were harvested with a syringe and then centrifuged at 400 g for 3 min to obtain the peritoneal cells for flow cytometry analysis or peritoneal macrophage isolation.
To isolate peritoneal macrophages, peritoneal cells (5 × 106/mL) were incubated in complete RPMI-1640 medium (10% FBS) at 37°C in 5% CO2 for 2 h. Then, the plates were softly washed with medium to remove nonadherent cells, and the adherent cells were collected as peritoneal macrophages [19]. Cell purity was tested by flow cytometry (shown in Supplementary Figure S1D).
Peritoneal macrophages seeded in 6-well plates (approximately 2 × 106 cells/well) were polarized with m-IL-4 (20 ng/mL; Peprotech, Rocky Hill, NJ, USA), LPS (100 ng/mL; Sigma-Aldrich), PSL or PCL (50 μg/mL) in a final volume of 2 mL RPMI-1640 medium or treated with inhibitors (GSK-J1, 30 μmol/L; UNC569, 3 μmol/L; S3I-201, 200 μmol/L; PP2, 10 μmol/L; PF-573228, 30 μmol/L) or a LEAF™ purified anti-mouse Tim-4 antibody (100 μg/mL, clone RMT4-54; Biolegend) for 30 min at 37°C prior to treatment with PSL. After being cultured for the indicated time, the macrophages were harvested for subsequent experiments. The purity of peritoneal macrophages was detected by flow cytometry with the specific macrophage marker CD11b and F4/80.
2.8 Morphological analysis of macrophages
Morphological analysis (Giemsa staining) of peritoneal macrophages seeded on sterile coverslips (WHB-24-CS, diameter: 14 mm; Shanghai, China) was performed with a Liu staining kit (BASO, Zhuhai, Guangdong, China) in accordance with the manufacturer's instructions and photographed with a microscope (Olympus BX53F).
2.9 Tumor models and treatment
To establish a mouse subcutaneous tumor model, 5 × 105 CT26 cells were injected subcutaneously into the right flanks of female BALB/c mice. When the tumor was observable (approximately 7 days after injection), sterile saline or PSL (5 mg/kg/day) were intratumorally injected every day for 14 days. Tumor growth was evaluated by measuring the tumor diameter with calipers every day, and tumor volume was calculated by the following formula: tumor volume = (long diameter) × (short diameter) 2 × 0.5. When the mice were euthanized, the tumors and organs were harvested for further study.
To establish a mouse model of malignant ascites, 2 × 105 CT26 cells were intraperitoneally injected into the peritoneal cavities of female BALB/c mice. Sustained intraperitoneal administration of sterile saline (200 μL), PSL (10 mg/kg), inhibitors (UNC569 20 mg/kg/day, GSK-J4 0.5 mg/kg/day, or S3I-201 5 mg/kg/day) or anti-mouse TIM4 antibody (100 μg/mouse) was initiated 3 days after tumor inoculation. Liposomes were injected every two days for the 80-day survival study. For the short-term study, the administration of liposome and inhibitors was performed every day for 7 days. The antibody was intraperitoneally injected every two days for three times. Then the mice were euthanized, and both peritoneal ascites and tumors were harvested for further study.
2.10 Macrophage peritoneal polarization assay
Macrophage peritoneal polarization was detected by flow cytometry and morphological analysis. After continuous intraperitoneal injection of PSL (10 mg/kg/day) for 3 days, the mice were euthanized, and the peritoneal cells were harvested as described in method 2.7. After being washed with ice-cold PBS, the cells were incubated with the appropriate cocktail of antibodies (CD45, CD11b, F4/80, CD206, 1 μL/test) for 30 min at 4°C or seeded on sterile coverslips (WHB-24-CS, diameter: 14 mm) for further morphological analysis. For intracellular staining (IL-10 or TGF-β staining, 1 μL/test), cells were fixed and permeabilized with the Cytofix/Cytoperm Kit (BD Biosciences) in accordance with the manufacturer's instructions. Then, the cells were sorted with a NovoCyte flow cytometer (ACEA Bioscience, San Diego, CA, USA) after staining and washing. The data were analyzed with FlowJo-V10 CL software (Tree Star, Ashland, OR, USA).
2.11 Reverse transcription PCR (RT-PCR) assay
Total RNA was extracted with the RNA Simple Total RNA Kit (TIANGEN, Beijing, China) and reversely transcribed into cDNA with a reverse transcription kit (TaKaRa Bio, Tokyo, Japan) in accordance with the manufacturer's instructions. RT-PCR was performed with SsoAdvancedTM SYBR Green Supermix (Bio-Rad Laboratories, Milan, Italy) with Bio-Rad CFX96 using a two-step PCR protocol. The relative expression of the target genes to Gapdh was calculated according to the 2−ΔΔCt method.
The program for amplification was one cycle of 95˚C for 3 min, followed by 39 cycles of 95˚C for 10 s and 55˚C for 30 s. The primers used are shown below: the forward primer 5’-GCTGTCTTCCCAAGAGTTGGG-3’ and the reverse primer 5’-ATGGAAGAGACCTTCAGCTAC-3’ for Arg-1; the forward primer 5’-ATGATGTCTGCCAGAGAACC-3’ and the reverse primer 5’-ATCACAGATTTCAGCAACCTTA-3’ for macrophage galactose-type C-type lectin 1 (Mgl1); the forward primer 5’-AAGGCTATCCTGGTGGAAGAA-3’ and the reverse primer 5’-AGGGAAGGGTCAGTCTGTGTT-3’ for mannose receptor C type 1 (Mrc1); the forward primer 5’-CAGGTCCCTGTCATGCTTCT-3’ and the reverse primer 5’-GTCAGCACAGACCTCTCTCT-3’ for Ccl-2; the forward primer 5’-GACAACTTTGGCATTGTGG-3’ and the reverse primer 5’-ATGCAGGGATGATGTTCTG-3’ for IL-6; the forward primer 5’-GGAAGCACGGCAGCAGAATA-3’ and the reverse primer 5’-AACTTGAGGGAGAAGTAGGAATGG-3’ for IL-12; the forward primer 5’-TCTTCTCATTCCTGCTTGTGG-3’ and the reverse primer 5’-GGTCTGGGCCATAGAACTGA-3’ for TNF-α; the forward primer 5’-GGCAGCCTGTGAGACCTTTG-3’ and the reverse primer 5’-CATTGGAAGTGAAGCGTTTCG-3’ for inducible nitric oxide synthase (iNOs); the forward primer 5’-CCCCCATTTCAGCTGACTAA-3’ and the reverse primer 5’-CTGGACCAAGGGGTGTGTT-3’ for Jmjd3; the forward primer 5’-GACCAGTCACACCCAGAAATCCC-3’ and the reverse primer 5’-GTTCCTGTCACCTGGCAACC-3’ for interferon-regulatory factor 4 (Irf4); the forward primer 5’-ACCCAGAAGACTGTGGATGG-3’ and the reverse primer 5’-CACATTGGGGGTAGGAACAC-3’ for Gapdh.
2.12 ELISA and western blotting analysis
Freshly isolated macrophages were stimulated with PCL or PSL (50 μg/mL) for 48 h at 37°C. Then, the supernatant was collected for ELISA analysis. Peritoneal lavage fluids for ELISA analysis were obtained by intraperitoneal injection of 1 mL sterile saline. The concentration of transforming growth factor beta (TGF-β) and IL-10 was measured with a TGF-β ELISA kit (DaKeWe Biotech) and an IL-10 ELISA kit (eBioscience). Peritoneal macrophages stimulated with the indicated reagents (PSL, 50 μg/mL; UNC569, 3 μmol/L; anti-TIM4 antibody, 100 μg/mL; PP2, 10 μmol/L; PF573228, 30 μmol/L; S3I-201, 200 μmol/L; GSK-J1, 30 μmol/L) were lysed on ice with RIPA lysis buffer (Beyotime Institute of Biotechnology) containing proteinase inhibitor cocktail (Sigma-Aldrich) to prepare protein samples. Then, 20-100 μg protein was loaded and separated by SDS-PAGE under reducing conditions and then transferred to PVDF (poly vinylidene fluoride) membranes (Merck Millipore). Membranes were then blocked with 5% skimmed milk for 2 h and incubated with indicated antibody at 4°C overnight. After incubating the bands with HRP-conjugated secondary antibody for 2 h. Bands were detected with Immobilon™ Western Chemiluminescent HRP Substrate (Millipore) and ChemiDoc MP imaging system (Bio-Rad Laboratories). Band intensity was analyzed by Image J 2.1.0 (Java. NIH, USA).
2.13 Statistical analysis
The data were statistically analyzed using the Prism 7.0 (GraphPad Software; San Diego, CA, USA) and presented as the mean ± standard error of mean (SEM). Differences between groups were evaluated by Student's t test. Differences were considered statistically significant if P values < 0.05.
3 RESULTS
3.1 Apoptotic tumor cells correlated with M2-like macrophages in tumors
We observed the co-localization of apoptotic cells and macrophages in human colon cancer specimens stained with cleaved caspase-3 (apoptotic marker) and CD163 (macrophage marker; Figure 1A). The frequencies of TAMs were closely correlated with the presence of apoptotic cells in several types of human tumors, including colon, breast, and lung cancers (Figure 1B-D). Similar results were found in murine models of CT26, 4T1, and LL2 subcutaneous tumors (Figure 1E-H) and orthotopic tumors (Supplementary Figure S1E-G). To further determine the relationship between tumor cell apoptosis and TAM aggregation, CT26 cells were induced to undergo apoptosis either by interfering with Bcl2 expression or by treatment with a chemotherapy drug (DNR) in vitro and in vivo. The expression of Bcl2 and cell proliferation/apoptosis in stable cell lines was detected (Supplementary Figure S2A-C). To understand the effect of apoptotic cells on the tumor microenvironment at an early stage, mice were inoculated with CT26 cells stably transfected with shRNA against Bcl2 (shBcl2). After 72 h, we observed an increased number of apoptotic cells (Supplementary Figure S2D) and CD206+ macrophage accumulation (Figure 1I). One dose of intraperitoneal chemotherapy (1 mg/kg DNR) induced tumor cell apoptosis in the cavity (Supplementary Figure S2E). In the later stage of the CT26 mouse malignant ascites model (7 days after inoculation), one dose of intraperitoneal DNR treatment resulted in M2-like (CD206+) TAM accumulation in the peritoneal cavity (Figure 1J). We further explored the relationship between elevated apoptosis and M2-like TAM accumulation after chemotherapy by obtaining DNR-induced apoptotic cells (DNR-CT26) (Supplementary Figure S2F). As detected by flow cytometry (Figure 1K), inoculation of DNR-CT26 cells induced CD206+ macrophage accumulation in the peritoneal cavity, which suggests that apoptotic cells play a critical role in inducing M2-like macrophages and immunosuppression.
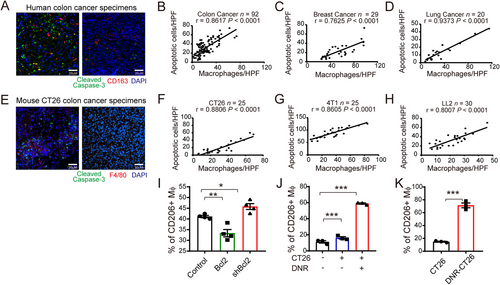
Apoptotic tumor cells correlated with M2-like macrophages in tumor. (A) Representative immunofluorescence images of apoptotic tumor cells (Cleaved Caspase-3+, green) and macrophages (CD163+, red) in human colon cancer specimens are shown (left: more apoptotic tumor cells; right: fewer apoptotic cells). (B-D) Correlation analyses for the number of apoptotic tumor cells and macrophages in human colon cancer (B), breast cancer (C), and lung cancer (D) are shown. (E) Representative immunofluorescence images of apoptotic cells (Cleaved Caspase-3+, green) and macrophages (F4/80+, red) in mouse CT26 colon cancer specimens are shown (left: more apoptotic tumor cells; right: fewer apoptotic cells). (F-H) Correlation analyses for the number of apoptotic tumor cells and macrophages in CT26 (F), 4T1 (G), or LL2 (H) subcutaneous tumor are shown the number of apoptotic tumor cells or macrophages per specimen were calculated by the mean number counted in ten HPFs. The Pearson correlation coefficieI(r) and significance levels (P) are presented. (I) 72 h after peritoneal injection of different CT26 cells (Control, Bcl2, or shBcl2), the accumulation of M2 macrophages (CD45+CD11b+ F4/80+ CD206+) in the peritoneal cavity were detected by flow cytometry. (J) Infiltration of M2 type macrophages (CD11b+ F4/80+ CD206+) in CT26 malignant ascites with or without DNR treatment (1 mg/kg, once) are shown. (K) Chemotherapy-induced apoptotic CT26 cells (DNR-CT26) elevated CD206+ macrophage accumulation in peritoneal cavity. Data are presented as the mean ± SEM. * P < 0.05, ** P < 0.01, and *** P < 0.001 calculated using a two-tailed unpaired Student's t-test
Abbreviations: HPF, high power field; DNR, daunorubicin.
3.2 Phosphatidylserine derived from apoptotic tumor cells induced macrophage polarization to M2-like phenotype in vitro
We examined the contents of apoptotic cells to determine the reason for the increase in the number of M2-like macrophages in tumor tissue. Primary tumor cell culture supernatants or tumor-derived factors, such as lactic acid, macrophage colony-stimulating factor (M-CSF), and noncoding RNA [20-22], can induce the M2 phenotype of macrophages. Thus, we hypothesized that there might be unidentified apoptosis-associated molecules that are responsible for the M2-like phenotype transition of macrophages. To exclude the effect of residual chemotherapeutics, we used heat-induced apoptotic tumor cells instead of chemotherapy-induced apoptotic tumor cells (DNR-induced apoptotic tumor cells). Our results showed that both apoptotic tumor cells (ACs) and apoptotic cell culture medium (ACM) induced arginase 1 (Arg1) transcription in macrophages (Figure 2A), which is an important marker of the M2 phenotype [21, 23]. To further characterize the molecule category, apoptotic cell culture medium was boiled at 100°C or digested with DNase I or trypsin. After boiling or digestion, apoptotic cell culture medium retained its ability to induce Arg1 expression in macrophages (Figure 2B). Thus, we ruled out the possibility that proteins and nucleic acids induced this effect, and we focused our attention on the lipid composition. We found that crude lipids extracted from both apoptotic tumor cells and apoptotic cell culture medium were sufficient to induce Arg1 expression in macrophages (Figure 2C).
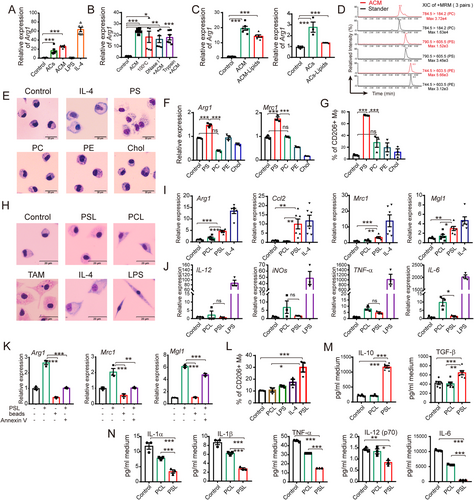
Phosphatidylserine derived from apoptotic tumor cells induced M2-like phenotype of peritoneal macrophages in vitro. (A) Relative expression of Arg1 in peritoneal macrophages was detected after incubation with apoptotic tumor cells (ACs) or apoptotic cell culture medium (ACM) for 24 h. LPS (100 ng/mL) and m-IL4 (20 ng/mL) stimulation were used as positive controls. (B) Relative expression of Arg1 in peritoneal macrophages after being treated with ACM, which was boiled at 100°C for 20 min or digested with DNAse I (100 U/mL) or trypsin (10 μg/mL) for 8 h was detected. (C) Relative expression levels of Arg1 induced by crude lipids extracted from ACs (ACs-Lipids) and ACM (ACM-Lipids) in peritoneal macrophages were detected. (D) PS, PC, and PE in ACM were quantified by LC-MS. (E) Representative morphological images of peritoneal macrophages cultured with commercial lipids (PS, PC, or PE) and Chol for 24 h are shown. The m-IL4-treated (20 ng/mL) group was used as a M2 macrophage positive control. (F) The increased expression of Arg1 and Mrc1 induced by commercial lipids in peritoneal macrophages is shown. (G) The elevated expression of CD206 induced by commercial lipids in peritoneal macrophages was detected by flow cytometry. (H) Representative morphological images of peritoneal macrophages cultured with m-IL4 (20 ng/mL), LPS (100 ng/mL), PCL (50 μg/mL), or PSL (50 μg/mL) for 24 h are shown. (I-J) The relative expression of M2 marker genes (I) and M1-related genes (J) in peritoneal macrophages co-cultured with PCL or PSL for 24 h is shown. (K) Latex beads did not induce M2-like polarization. Annexin V protein (2.5 μg/mL) abrogated PSL-induced M2 marker gene expression in peritoneal macrophages. (L) The expression level of CD206 in peritoneal macrophages after being co-cultured with LPS (100 ng/mL), m-IL4 (20 ng/mL), PCL (50 μg/mL), or PSL (50 μg/mL) for 24 h (detected by flow cytometry). (M-N) Expression of M2-related (M, by ELISA) or M1-related cytokines (N, by Luminex) in macrophage culture medium with or without PS stimulation for 48 h. All the relative gene expression data were detected by RT-PCR and normalized to GAPDH. The fold changes were relative to the control group without treatment or with DMSO (F). The data are presented as the mean ± SEM. * P < 0.05, ** P < 0.01, *** P < 0.001 calculated using a two-tailed unpaired Student's t-test. Scale bar, 2 μm
Abbreviations: AC, apoptotic tumor cell; ACM, apoptotic cell culture medium; PS, phosphatidylserine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; Chol, cholesterol; LC-MS, liquid chromatography-mass spectrometry; PCL, phosphatidylcholine containing liposome; PSL, phosphatidylserine containing liposome; ELISA, enzyme-linked immunosorbent assay; SEM, standard error of mean; Mϕ, macrophage; LPS, lipopolysaccharide; Arg1, Arginase 1; Mrc1, Mannose Receptor C Type 1; TNF-α, Tumor necrosis factor alpha; Mgl1, macrophage galactose-type C-type lectin 1; CCL2, CC motif chemokine ligand 2; IL, interleukin; iNOs, inducible nitric oxide synthase; TAM, Tumor-associated macrophages; TGF-β, Tumor growth factor-beta; XIC, Extracted ion chromatogram; MRM, multiple reaction monitoring.
The main cell constituent phospholipids in the crude lipid extracts of apoptotic cell culture medium were then characterized using LC-MS (Figure 2D). Then, mouse peritoneal macrophages were treated with commercial lipids (phosphatidylserine, PS; phosphatidylcholine, PC; and phosphatidylethanolamine, PE) or cholesterol (Chol) for 24 h. We evaluated M2-like morphological changes (Figure 2E) as well as Arg1/Mrc1 transcription and CD206 expression levels (Figure 2F-G) and found that PS played a critical role in macrophage polarization.
PS is one of the most important markers of cell apoptosis. It is normally located on the extracellular membrane surface of apoptotic cells and membrane-bound vesicles [24], and is involved in the clearance of apoptotic cells [25, 26]. To mimic the apoptotic cells with PS on the surface, we prepared PSLs and used PCLs as controls. PSLs triggered vacuolated and foamy cytoplasm morphological changes in the macrophages, which was similar to those of TAMs isolated from mouse ovarian cancer ascites (ID8) (Figure 2H). The transcription of M2 marker genes (Arg1, Mgl1, Mrc1, and Ccl2), but not the M1 marker genes (IL-6, IL-12, tumor necrosis factor alpha [TNF-<], and iNOs) in peritoneal macrophages was remarkably elevated by PSLs (Figures 2I and 2J). Furthermore, M2-like polarization of macrophages could not be induced by nonspecific phagocytosis of other particles, such as PCLs (Figure 2I) or latex beads (Figure 2K). Annexin V, a PS mask protein, blocked the PSL-induced M2-like polarization (Figure 2K), suggesting that PS binding is critical. In agreement with the above results, PSLs enhanced the expression of the M2 biomarker CD206 (Figure 2L), as well as the secretion of immunosuppressive cytokines, such as IL-10 and TGF-β (Figure 2M). The expression of pro-inflammatory cytokines IL-1α, IL-1β, IL-12(p70), IL-6, and TNF-α was decreased by PSL stimulation (Figure 2N). These results were confirmed in the macrophages isolated from BALB/c mice (Supplementary Figure S3A and B) and human peripheral blood-derived macrophages (Supplementary Figure S3C and D).
3.3 PS induced accumulation of M2-like macrophages in vivo and promoted tumor growth
We further investigated whether PS could induce M2-like macrophage accumulation and promote tumor growth in animal models. After intraperitoneal injection of PSLs (10 mg/kg/day) into C57 mice continuously for 3 days, significant accumulation of CD206+ M2-like (Figures 3A and 3B) and IL-10+/TGF-β+ immunosuppressive macrophages (Supplementary Figure S3E and F) in the peritoneal cavity was detected by flow cytometry. Consistent with the accumulation of IL-10+/TGF-β+ immunosuppressive macrophages, high levels of IL-10 and TGF-β in the peritoneal lavage fluids of the PSL-stimulated group were detected by ELISA (Figures 3C and 3D), which may contribute to the M2-like macrophage-induced immunosuppressive microenvironment of the peritoneal cavity. The foamy peritoneal macrophages isolated from PSL-treated mice showed similar morphology as that of the TAMs (Figure 3E). Additionally, the expression of M2 phenotype-associated genes (Arg1, Mgl1, Mrc1, and Ccl2) in the PSL-treated group was higher than that in the controls (both NS and PCL, Figure 3F). Similar results were observed in BALB/c mice (Supplementary Figure S3G and H).
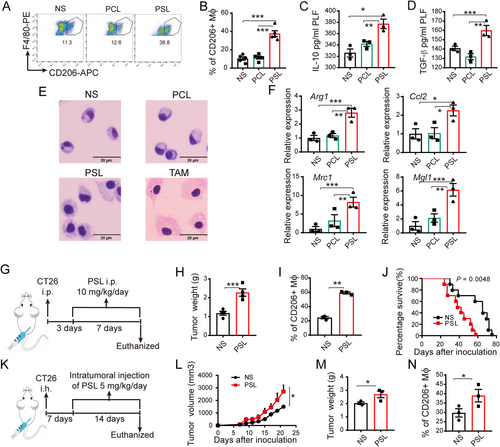
Phosphatidylserine induced M2-like macrophages in vivo and promoted tumor growth. Peritoneal macrophages were isolated and characterized 72 h after intraperitoneal injection of PSL (10 mg/kg) or PCL (10 mg/kg). (A) Representative images of the gating strategy used to define CD45+ CD11b+ F4/80+CD206+ M2-type macrophages in NS, PCL and PSL intraperitoneal injected mice. (B) Accumulation of CD206+ M2-like macrophages in the cavity of NS, PCL and PSL intraperitoneal injected mice. (C-D) I10 (C) and TGF-β (D) expressions in mouse peritoneal lavage fluids were measured byIISA. (E) Representative morphological images of peritoneal macrophages are shown. TAMs isolated from the ascites of ID8 tumor-bearing mice were used as a positive control. Scale bar, 20 μm. (F) Relative expressions of M2- related genes were detected by RT-PCR. (G) Schematic diagram of PSL treatment in a mouse CT26 malignant ascites model. (H-I) After sustained PSL treatment for seven days, tumor weight and CD206+ macrophage infiltration (gating from CD45+CD11b+ F4/80+) were measured. (J) An 80-day survival study was performed (n = 10), P = 0.0048 by the log-rank (Mantel-Cox) test. (K) Schematic diagram of PSL treatment in a mouse CT26 subcutaneous tumor model. (L-N) Tumor volume, tumor weight, and CD206+ macrophage infiltration (gating from CD45+CD11b+ F4/80+) in the CT26 subcutaneous tumor model with sustained PSL treatment were assessed. All of the relative gene expression data were analyzed by RT-PCR and normalized to GAPDH. The fold changes were relative to those in the control group treated with NS. Bars indicate mean ± SEM. * P < 0.05, ** P < 0.01, *** P < 0.001 calculated using a two-tailed Student's t-test
Abbreviations: NS, normal saline; PCL, phosphatidylcholine containing liposome; PSL, phosphatidylserine containing liposome; Mϕ, macrophage; LPS, lipopolysaccharide; Arg1, Arginase 1; Mrc1, Mannose Receptor C Type 1; Mgl1, macrophage galactose-type C-type lectin 1; IL, interleukin; i.p., intraperitoneal injection; i.h., hypodermic injection; TAM, Tumor-associated macrophages; RT-PCR, Real time-polymerase chain reaction; ELISA, enzyme-linked immunosorbent assay; SEM, standard error of mean.
We next determined how PS affects tumor growth. PSLs were administered to mouse CT26 peritoneal metastasis or subcutaneous tumor models. The experimental design is illustrated in Figure 3G-K. Notably, we discovered that sustained PSL treatment remarkably promoted abdominal tumor growth (Figure 3H), with an elevated accumulation of CD206+ (Figure 3I) and IL-10+ macrophages (Supplementary Figure S4A) in malignant ascites fluid. Moreover, the results of the survival study suggested that intraperitoneal administration of PSL significantly shortened the survival of mice in the peritoneal metastasis model (Figure 3J). Furthermore, in the subcutaneous tumor model, intratumoral injection of PSL also led to a significant increase in tumor volume and weight (Figures 3L and 3M), accompanied by increased infiltration of CD206+ M2-like macrophages (Figure 3N) and IL-10+ macrophages (Supplementary Figure S4B) in tumor tissue.
To further examine the significance of these results and confirm the specificity of PSL to promote tumor growth, the tumor volumes of CT26, MC38, and 4T1 subcutaneous tumors with either PSL and PCL treatment were recorded. We found that PSL, but not PCL, induced tumor growth (Supplementary Figure S4C-E). These results demonstrated that PS could induce macrophage polarization towards the M2-like phenotype in tumor tissues in vivo, which may contribute to the local immunosuppressive microenvironment and result in tumor progression.
3.4 Phosphatidylserine receptors were essential for induction of M2-like macrophage by PS
PS externalized on apoptotic cell membranes was reported to bind to PS receptors (PSRs), such as MERTK, TIM-4, RAGE, CD36, BAI1 and Stabilin-2, to induce phagocytosis [27-30]. Our results showed that PS binding was critical for PSL-induced M2-like macrophage polarization (Figures 2I and 2K). Thus, we examined the involvement of PSRs in the induction of M2-like macrophages by PS. The expression level of PSRs on the surface of peritoneal macrophages was determined, suggesting a relatively high expression of MERTK (90%-98% of cells) and TIM4 (80%-90% of cells; Figure 4A). Interestingly, in Mertk-deficient mice (Mertk−/−), the polarization of M2-like macrophages induced by PSL was markedly impaired (Figure 4B). Similar results were found when macrophages were co-cultured with an anti-TIM4 blocking antibody (100 μg/mL) or the MERTK inhibitor UNC569 (3 μmol/L; Figure 4C). In addition, blocking MERTK and TIM4 abrogated the transcription of M2 marker genes (Arg1 and Mgl1) (Figure 4D-E).
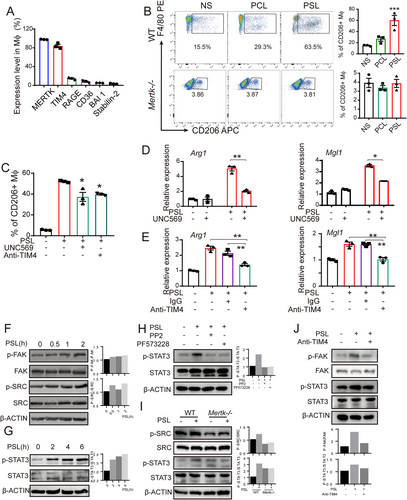
PS receptors and the STAT3 axis were essential in the PSL-induced M2-like macrophage polarization. (A) The expression of the main PSRs on mice peritoneal macrophages were detected by flow cytometry. (B) CD206+ macrophages in Mertk−/- and wild-type mice after peritoneal injection of PSL were detected by flow cytometry. Representative images and a histogram of the data are shown. (C-E) The abrogation of CD206+ macrophage accumulation I and expression of Arg1 and Mgl1 (D-E) in macrophages was observed by pre-treatment of macrophages with a specific inhibitor (MERTK inhibitor, UNC569, 3 μmol/L) or antibody (anti-TIM4, 100 μg/mL) for 30 min. (F) The activations of FAK and SRC in macrophages were observed at 120 min after PSL stimulation. (G) The phosphorylation of STAT3 was observed at 4-6 h after PSL stimulation. Western blotting analysis of FAK/SRC-STAT3 activation was performed 4 h after PSL stimulation. (H) The activation of STAT3 was abrogated by pre-treating macrophages with the SRC inhibitor PP2 (10 μmol/L) or the FAK inhibitor PF573228 (30 μmol/L). (I) SRC and STAT3 activation in wild-type or Mertk−/− macrophages after being incubated with PSL for 4 h were detected. (J) Treatment with an anti-TIM4 antibody (100 μg/mL) abrogated the activation of FAK and STAT3 induced by PSL in peritoneal macrophages. All of the relative gene expression data were detected by RT-PCR and normalized to GAPDH. The fold changes were relative to those of the control Group with Ns. mean ± SEM. *P < 0.05, ** P < 0.01, *** P < 0.001 calculated using a two-tailed Student's t-test
Abbreviations: NS, normal saline; PCL, phosphatidylcholine containing liposome; PSL, phosphatidylserine containing liposome; Mϕ, macrophage; Arg1, Arginase 1; Mgl1, macrophage galactose-type C-type lectin 1; RAGE, receptor for advanced glycation end products; MERTK, C-MER proto-oncogene tyrosine kinase; BAI 1, Brain-specific angiogenesis inhibitor 1; TIM, T-cell immunoglobin mucin; RT-PCR, Real time-polymerase chain reaction; SEM, standard error of mean.
Several studies have reported that FAK and SRC played important roles in the anti-inflammatory phenotype [31, 32] and migration in macrophages [33, 34]. We investigated whether FAK and SRC are involved in PS-induced M2-like polarization of macrophages. As detected by Western blotting, FAK and SRC were activated after 2 h of PSL treatment (Figure 4F). FAK and SRC activation was reported to be involved in the subsequent activation of STAT3 in many immune cells [32, 35]. Likewise, we observed STAT3 activation in the PSL-treated macrophages (Figure 4G). Similar results were observed in human peripheral blood-derived macrophages (Supplementary Figure S4F). Pre-treatment of macrophages with an SRC inhibitor (PP2, 10 μmol/L) or FAK inhibitor (PF573228, 30 μmol/L) attenuated the PSL-induced activation of STAT3 (Figure 4H). Moreover, in Mertk-deficient peritoneal macrophages (isolated from Mertk−/− mice), the activation of SRC and STAT3 was partly impaired (Figure 4I). Pre-treating peritoneal macrophages with an anti-TIM4 antibody (100 μg/mL) attenuated the activation of both FAK and STAT3 (Figure 4J). Thus, we suggest that PSRs, namely MERTK and TIM4, are involved in PS-induced macrophage polarization, which leads to the activation of the FAK/SRC-STAT3 signaling pathway.
3.5 Phosphatidylserine induced M2-like macrophage polarization via the STAT3-JMJD3-IRF4 signaling axis
After the identification of related phosphatidylserine receptors, we aimed to determine the transcriptional events that occurred after the injection of PS in macrophages. Stimulation with PSL resulted in an immediate increase in the levels of Jmjd3 and Irf4 in macrophages (Figures 5A and 5B and Supplementary Figure S4G). JMJD3, a histone 3 Lys27 (H3K27) demethylase, is known to be crucial for parasite-induced M2-like macrophage polarization and Th17 cell differentiation in arthritis [36, 37]. IRF4 was identified as a JMJD3 target gene [36]. We found that the transcription levels of Jmjd3 and Irf4 peaked 1 h after PSL treatment (Figures 5A and 5B). Consistent with the change in mRNA expression, the level of JMJD3 protein was increased 4-6 h after PSL treatment (Figure 5C). JMJD3 is known to promote the conversion of H3K27me3 (trimethylated) to H3K27me1 (monomethylated) [38]. We observed demethylation of H3K27me3 in macrophages at approximately 6 h after PSL treatment (Figure 5C). Importantly, knockout of Jmjd3 in macrophages could effectively abrogate PS-induced M2 gene transcription (Arg1 and Mgl1). The knockout of Stat3 showed similar results (Figure 5D). Such impairment of M2-like macrophage gene transcription may be owing to the decrease in Jmjd3 and Irf4 expression caused by Jmjd3 and Stat3 deficiency (Figure 5E). In addition, pre-treatment of macrophages with the JMJD3 inhibitor GSK-J1 (30 μmol/L) or S3I-201 (DNA-binding inhibitor of STAT3, 200 μmol/L) also notably impaired M2-related gene transcription induced by PSL (Figure 5F-G). The elevated demethylation of H3K27me3 and the expression of Jmjd3 and Irf4 induced by PSL were abrogated by treatment with GSK-J1 (Figure 5H) and S3I-201 (Figure 5I), respectively, which also supports the above observation. These results demonstrated that PS derived from apoptotic cells might be a key factor for the induction of M2-like phenotype macrophages in the tumor microenvironment through the PSRs-STAT3-JMJD3 signaling axis. It is critical to know whether targeting this axis would inhibit tumor growth by reducing the accumulation of M2-like macrophages and reshaping the microenvironment. We administered small-molecule inhibitors (MERTK inhibitor, UNC569; STAT3 inhibitor, S3I201; JMJD3 inhibitor, GSK-J4) or an anti-TIM4 blocking antibody in mice bearing CT26 peritoneal tumors. The results showed that inhibiting the function of PSRs (MERTK, TIM4), STAT3, and JMJD3 remarkably inhibited CT26 tumor progression, which was quantified by tumor weight and ascites volume, which may be caused by the impaired accumulation of CD206+ macrophages (Figure 6).
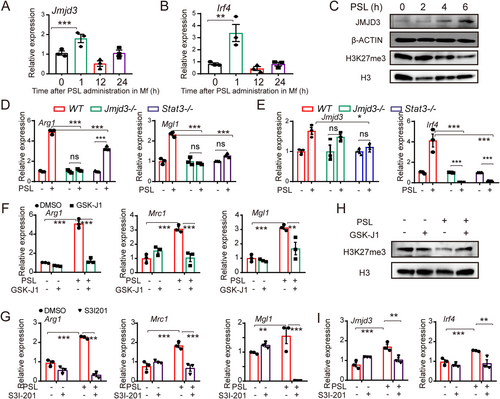
PSL induced M2-like macrophage polarization via STAT3-JMJD3-IRF4 axis. (A-B) Relative expressions of Jmjd3 (A) and Irf4 (B) in peritoneal macrophage after PSL stimulation. (C) JMJD3 protein expression and H3K27 demethylation were elevated after PSL treatment, as detected by western blotting. H3 was the loading control. (D) PSL-induced expression of Arg1 and Mgl1 was abrogated by Jmjd3 or Stat3 deficiency. (E) Jmjd3 and Irf4 expressions induced by PSL stimulation was likewise abrogated by Jmjd3/Stat3 deficiency. (F) Pre-treating macrophages with GSK-J1 (30 μmol/L) successfully impaired the PSL-induced transcription of M2 marker genes (Arg1, Mrc1, and Mgl1). (G) Inhibiting the function of STAT3 with S3I-201 (200 μmol/L) abrogated the PSL-induced M2-related gene transcription (Arg1, Mrc1, and Mgl1) in macrophages. (H) The elevated demethylation of H3K27me3 induced by PSL in macrophages was abrogated by treating them with GSK-J1. (I) The PSL-induced expression of Jmjd3 and Irf4 in macrophages was abrogated by treating them with S3I-201. All of the relative gene expression data were detected by RT-PCR and normalized to GAPDH. The fold changes were relative to those of the macrophages treated with NS or with DMSO (F, G). The data are presented as the mean ± SEM, * P < 0.05, ** P < 0.01, *** P < 0.001 calculated using a two-tailed Student's t-test
Abbreviations: Jmjd3, Jumonji domain-containing protein 3; Irf4, interferon-regulatory factors 4; WT, wild type; H3, histone 3; STAT3, Signal Transducer and Activator of Transcription 3; Arg1, Arginase 1; Mrc1, Mannose Receptor C Type 1; Mgl1, macrophage galactose-type C-type lectin 1; DMSO, dimethylsulfoxide.
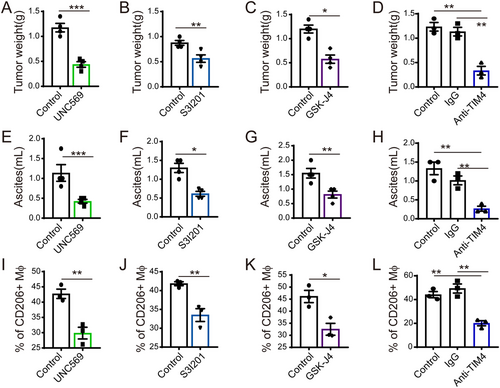
Potential anti-tumor treatment by targeting the PSR-STAT3-JMJD3 axis. After treated with MERTK inhibitor (UNC569, 20 mg/kg/day), a STAT3 inhibitor (S3I-201, 5 mg/kg/day), a JMJD3 inhibitor (GSK-J4, 0.5 mg/kg/day), or an anti-TIM4 antibody (5 mg/kg/3 days). Mice bearing CT26 mouse peritoneal tumor were sacrificed. Tumor weight (A-D) and ascites volume (E-H) was recorded. Percentages of CD206+ macrophages in ascites (gating from CD45+CD11b+ F4/80+ cells) are shown by flow cytometry (I-L). The data are presented as the mean ± SEM. * P < 0.05, ** P < 0.01, *** P < 0.001, calculated using a two-tailed Student's t-test
Abbreviations: Mϕ, macrophage; MERTK, C-MER proto-oncogene tyrosine kinase; JMJD3, Jumonji domain-containing protein 3; STAT3, Signal Transducer and Activator of Transcription 3; TIM 4, T-cell immunoglobin mucin 4; SEM, standard error of mean.
4 DISCUSSION
In this study, we revealed that phosphatidylserine released by apoptotic tumor cells was responsible for the immunosuppressive microenvironment in tumor tissues and resulted in promoting tumor progression. Several observations have been made and revealed that PS significantly enhanced M2-like polarization of macrophages via the PSRs-STAT3-JMJD3 pathway.
Macrophages, as the most abundant immune cells in the tumor microenvironment, adopt an M2-like phenotype, and have been reported to be associated with tumor progression and contribute to immune escape through the production of vascular endothelial growth factor (VEGF), IL-10, and TGF-β [39]. Multiple signals are involved in M2-like polarization of macrophages [40]. In proliferating malignant tumors, apoptotic tumor cells might be beneficial for tumor growth [9, 10, 41]. Efferocytosis of apoptotic tumor cells may trigger wound healing cytokines in the tumor microenvironment to promote metastasis of breast cancer [42], and cause deleterious consequences in residual tumors [43]. In the present study, we found that apoptotic tumor cells were co-localized with macrophages in tumor tissues. In addition, the numbers of apoptotic tumor cells and macrophages were correlated in many types of tumors. Elevated M2-like macrophage (CD45+CD11b+F4/80+CD206+) infiltration in ascites was observed in mice inoculated with apoptotic tumor cells induced by disruption of Bcl2 expression or chemotherapeutic drugs. Given that macrophages play major roles in apoptotic tumor cell efferocytosis, we speculate that M2-like macrophages, which exhibit elevated Arg1 transcription, may be associated with apoptotic tumor cell clearance. However, the mechanism of apoptotic tumor cell-mediated M2-like macrophage accumulation in the tumor microenvironment remains unclear.
In recent years, many apoptosis-mediated factors have been shown to be associated with M2-like macrophages. Sphingosine-1-phosphate derived from apoptotic tumor cells can increase ARG1 activity via activation of the S1P1 receptor on macrophages [44]. Moreover, apoptotic tumor cell debris induced by chemotherapy is considered to contribute to tumor promotion via activation of M2-like macrophages [41]. However, the apoptotic tumor cell-derived factors associated with M2 phenotype remain unidentified.
In the present study, lipids extracted from apoptotic tumor cells and apoptotic tumor cell-CM were found to be sufficient to increase Arg1 expression in macrophages. Among the main lipids detected in apoptotic tumor cell-CM (PE, PC, and PS), only PS caused macrophages to exhibit a typical M2-like phenotype at both the RNA (Arg1 and Mrc1) and protein (CD206) levels. Administration of PSL in vivo and in vitro effectively switched macrophages to a M2-like phenotype, with increased expression of CD206 and M2-related genes. Administration of PSL elevated the production of immunosuppressive cytokines (TGF-β and IL-10) in macrophages, which were detected in both macrophage culture medium and mouse peritoneal lavage fluid. Given that apoptotic tumor cells induced by anti-tumor treatment have been reported to promote tumor progression [10, 45, 46], we speculate that the promotion of tumor progression may be due to the release of PS by apoptotic tumor cells in the tumor microenvironment. As expected, the pro-tumor growth effect of PS was observed in both the CT26 malignant ascites models and the subcutaneous tumor models.
The redistribution of PS on the extracellular side of the plasma membrane, which aids in the clearance of apoptotic cells by macrophages, is a biomarker of cell apoptosis [47]. However, the effect of PS on macrophage recruitment and polarization is unclear, and a better understanding of the role of PS in M2-like macrophages is needed. As a classical “eat me” signal, PS can mediate efficient phagocytosis of apoptotic cells by binding with PSRs [48, 49]. In this study, both the accumulation of M2-type macrophages in the peritoneum and PSL-induced M2-related gene and protein expression was attenuated by the inhibition of PSRs. FAK and SRC have been reported to contribute to phagocytosis downstream of PSRs. In this study, we focused on the rapid activation of FAK and SRC in PSL-stimulated macrophages. The inhibition of these kinases affected the subsequent activation of STAT3, which has been shown to contribute to M2-like macrophage polarization in different physiological processes [50, 51].
JMJD3, an H3K27 demethylase, has been reported to regulate M2-like macrophage polarization by controlling the H3K27me3 levels of M2 marker genes, such as Arg1, in macrophages [36, 52]. The transcription factor Irf4 is a target gene of JMJD3 and contributes to the expression of M2 marker genes in chitin-induced M2-like macrophages. Enhanced transcription of Jmjd3 and Irf4 was observed 1 h after the administration of PSL, and a consequent decrease in H3K27me3 was detected by Western blotting. Therefore, we speculate that a change in epigenetic status might play a key role in PS-associated macrophage polarization. We then found that administration of a JMJD3 inhibitor successfully blocked PS-induced M2 marker gene transcription and H3K27me3 demethylation in macrophages. Pre-treatment of macrophages with the STAT3 inhibitor S3I-201 blocked the increase in the transcription of Jmjd3 and Irf4 induced by PSL stimulation. These results suggest that STAT3 may play an important role in PS-mediated M2-like polarization of macrophages by affecting the transcription of Jmjd3 and Irf4, which was supported by subsequent experiments in Jmjd3−/− and Stat3−/− mice. These findings suggest that PS derived from apoptotic tumor cells can induce M2-like polarization in macrophages via the PSR-STAT3-JMJD3 axis. Thus, PSRs, STAT3, and JMJD3 were evaluated as potential targets, and inhibiting the expression of these factors successfully attenuated tumor growth in the CT26 malignant ascites model.
5 CONCLUSIONS
Taken together, our results suggest PS activated PSR-STAT3-JMJD3 axis is a novel mechanism for the M2-like polarization of macrophages, which may contribute to the immunosuppressive tumor microenvironment. These findings extend our understanding of the pro-tumor effects of the interactions between macrophages and apoptotic tumor cells, providing a potential rationale for tumor cell apoptosis-mediated promotion of tumor progression or relapse. Although the molecular mechanisms critical for the PS-induced M2-like polarization of macrophages require further evaluation, the present work identified several targets, such as PSRs (including MERTK and TIM4) and the STAT3-JMJD3 axis, as candidates for antitumor treatment.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
All animal studies involved were approved by the Animal Care and Use Committee of Sichuan University. The tumor tissue microarrays were purchased from Shanghai Outdo Biotech Company and the use of patient samples in this study was approved by the Ethics Committee of Shanghai Outdo Biotech Company.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data that support the findings of this study are available from the corresponding author upon reasonable request.
COMPETING INTERESTS
The authors declare that they have no competing interests.
ACKNOWLEDGEMENTS
We thank Prof. Charlie Degui Chen (Chinese Academy of Sciences) for sharing Jmjd3-flag knock-in mice and Jmjd3flox/flox mice with C57BL/6 background. This work was supported by the Project of the National Science Foundation for Excellent Young Scholars (32122052), National Natural Science Foundation Regional Innovation and Development (U19A2003), and National Natural Science Foundation of China (81902662).
AUTHORS' CONTRIBUTIONS
Xia-Wei Wei and Yu-Quan Wei designed the experimental approach and supervised the study. Xiao Liang, Min Luo, Bin Shao and Ren Jun performed the animal experiments. Jing-Yun Yang, Xiao Liang, An Tong, and Yan-Tong Liu performed flow cytometry analysis. Xiao Liang, Rui-Bo Wang, Tao Yi, and Ting Liu performed knockout mice identification and LC-MS/MS sample preparation and analysis. Xiao Liang, Xia-Wei Wei, and Xia Zhao analyzed data and wrote the paper. Authors declare no competing interests. All authors read and approved the final manuscript.




