Proteomic identification of new diagnostic biomarkers of early-stage cutaneous mycosis fungoides
ABBREVIATIONS
-
- BM
-
- Basement membrane
-
- BID
-
- benign inflammatory diseases
-
- DIA
-
- data-independent acquisition
-
- ECM
-
- extracellular matrix
-
- MF
-
- Mycosis fungoides
-
- TCR
-
- T cell receptor
Dear editor,
Mycosis fungoides (MF), the most common subtype of cutaneous T cell lymphoma, is a rare disease [1, 2]. Patients with early-stage MF have a 5-year overall survival between 88% and 100% [3]. In patients with advanced MF, the skin may present with tumors and erythroderma, and the median survival of patients with lymph node and visceral metastasis is 13 months [4]. Therefore, it is important to achieve an early diagnosis to improve prognosis [5].
To effectively distinguish the pathological features of early- and advanced-stage MF, as well as explore potential biomarkers for the differential diagnosis of MF and nontumor conditions (Supplementary Table S1-S3), the data-independent acquisition (DIA) proteomic technique was performed (Supplementary Figure S1), and 754 differentially expressed proteins were identified (Supplementary Table S4) in MF patients compared with patients with the benign inflammatory diseases (BID) and healthy controls (Supplementary Materials and Methods). Fourteen patients diagnosed with MF based on the World Health Organization-European Organization for Research and Treatment of Cancer (WHO-EORTC) classifications in the Department of Dermatology at Peking Union Medical Hospital from October 2018 to September 2019 were enrolled, including 4 early-stage and 10 advanced-stage MF patients. Patients who received systemic or external treatment in the previous six months were excluded. Patients included in the BID group were diagnosed with psoriasis or eczema, based on clinical manifestations and histopathological results, and those treated in the previous six months were excluded. The results of expression profile and cluster analyses revealed eight protein clusters that could characterize the distinct course of MF (Figure 1A-B). Clusters 1, 2, 3 and 4, represent proteins that were specifically upregulated in early-stage MF, gradually upregulated from early- to advanced-stage MF, both upregulated in the early- and advanced-stage MF, and specifically upregulated in advanced-stage MF, respectively, and were mostly involved in bicarbonate transport, transcription, translation, peptide and protein catabolic processes, and translation and redox. The proteins enriched in clusters 5, 6, 7, and 8, were downregulated in MF and were mostly involved in the processes of metabolism, extracellular matrix (ECM) remodeling and coagulation (including ECM organization, hemidesmosome assembly, cell-matrix adhesion, and coagulation-related processes), indicating that the wound healing ability was impaired in patients with advanced-stage MF. These findings led us to investigate the pathological features of tumor progression.
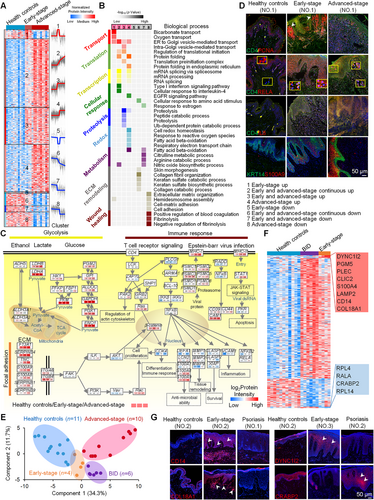
A protein-protein interaction network showed that the most substantially changed functional proteins were mainly in patients with advanced-stage MF and were related to functions such as transcription, translation, cell adhesion, innate immunity, and the T cell response (Supplementary Figure S2). In addition, abnormal angiogenesis is a common feature of tumors [6]. Our results identified expression imbalances in angiogenesis-associated proteins, indicating that they could act as therapeutic targets for abnormal angiogenesis. Interestingly, a number of virus-related proteins were upregulated in MF, suggesting that viral microbial infection was also a driving factor for tumor development. However, we found that more components of the basement membrane (BM) were downregulated in skin tissues during the development of MF (Supplementary Figure S3A), indicating that tumor cells may affect basal stem cell function by destroying the BM. We found that basal stem cell markers KRT5 (Supplementary Figure S3B, proteomics expriment), KRT14 (Supplementary Figure S3C, S4, immunofluorescence staining expriment), and associated transcription factor (P63, Supplementary Figure S3C, immunofluorescence staining expriment) were upregulated in early-stage MF. Furthermore, we found that focal adhesion signaling molecules (FAK, ILK, AKT, β-catenin, PI3K, VAV, and RAC) related to basal stem cell activity were activated during MF development, which could result in increased cell proliferation (Figure 1C). Immunofluorescence staining showed that CD4+ T cells proliferated vigorously in MF skin tissue, especially in the dermal papilla of early-stage MF (Figure 1D). These results indicated that the epidermal development was seriously altered in the skin tissues of patients with MF.
Next, the proteome results showed that proteins involved in the ethanol (ADH5), lactate (LDHB and LDHA), and glucose (HK1, PFKP, FBP1, PKM, PGM2, GPI, ALDOC, GAPDH, PGK1, and ENO1) signaling pathways were gradually upregulated from early- to advanced-stage MF (Figure 1C), suggesting possible energy metabolism remodeling to confer a selective advantage to tumor cells. In addition to metabolic reprogramming, immunosuppression is another hallmark of cancer. The proteome results also showed that T cell receptor signaling molecules (PAK2, GRB2, RAS, and PLCG1) were activated, resulting in regulation of the actin cytoskeleton, cell differentiation or immune response. Furthermore, we found that proteins involved in NF-κB signaling (IKKβ, IKKγ, NFκB1, RELA, and NFκB2), which is also downstream of T cell receptor signaling were activated during MF development, involved with several biological processes including anti-microbial ability (DEFA1, LCN2, S100A7, S100A8, and S100A9, Figure 1C, Supplementary Figure S4), tissue remodeling (MMP1, MMP3 and MMP9, Figure 1C), survival (CLASP1, CLASP2 and XIAP, Figure 1C), and inflammatory responses (VCAM1 and IL8, Figure 1C-D, Supplementary Figure S3D) in the MF tissues. It has been reported that the canonical NF-κB pathway contributes to cell death resistance in MF cells [7]. Our results showed that the transcription factor RELA of the canonical NF-κB pathway was localized to the nucleus of epidermal cells and CD4+ T cells, indicating that the canonical NF-κB pathway of both types of cells was activated in MF skin tissue of patients with MF (Figure 1D).
Furthermore, eight upregulated proteins and four downregulated proteins in the early-stage MF group were identified compared with those in the healthy controls and BID groups through proteomic analysis (Figure 1E-F). These 12 proteins could be considered potential biomarkers to distinguish early-stage MF and BID. The four proteins with the greatest differences in expression between early-stage MF and BID were selected for verification; they include three upregulated proteins (DYNC1I2, CD14, and COL18A1) and one downregulated protein (CRABP2). Immunofluorescence staining of skin tissues from 9 healthy people, 15 early-stage MF patients, and 11 BID (2 eczema and 9 psoriasis) patients, showed that these four biomarkers effectively distinguished early-stage MF and BID (Figure 1G and Supplementary Figure S5, S6), indicating that they are useful diagnostic markers of MF.
In conclusion, the pathological features of early- and advanced-stage MF were analyzed through proteomics techniques, which provided clues for the pathogenesis of MF as well as identified markers that could distinguish early-stage MF and BID. These diagnostic biomarkers of early-stage MF could help prevent delayed therapy due to misdiagnosis.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Medical Ethics Committee of Peking Union Medical College Hospital (No. JS-2547), and written informed consent was obtained from the patients.
AVAILABILITY OF DATA AND MATERIAL
All mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the iProX partner repository with the dataset identifier PXD022798. All equipment, reagents and supplies are available in Supplementary Table S5.
COMPETING INTERESTS
The authors declare that they have no competing interests.
FUNDING
This work was supported by the National Natural Science Foundation of China (No. 82173449), the Beijing Nature Science Foundation (No. 7182127), the Nonprofit Central Research Institute Fund of Chinese Academy of Medical Sciences (No. 2019XK320024), the National Key Research and Development Program of China (No. 2016YFC0901500), and the Medical and Health Science and Technology Innovation Project of Chinese Academy of Medical Sciences (2019-I2M-1-001).
AUTHORS’ CONTRIBUTIONS
JL and YHL raised clinical problems and designed a patient group experiment for proteomics research. LL conceived the overall study and designed the experiments. LL, JM, and YPZ performed the proteomics experiments and bioinformatics analyses. ZRL, SYZ, YKW, and JCW participated in tissue preparation. LL, LYL, DQG, and YJW performed most of the biological and functional experiments and analyzed the data. LL, JM, JL, and ZRL wrote and edited the manuscript. All authors made important comments regarding the manuscript. All authors and corresponding authors are fully accountable for the accuracy of all data and descriptions presented in this work
ACKNOWLEDGMENTS
We thank Dr. Mansheng Li (Beijing Institute of Lifeomics) for his useful discussion on proteomics data analysis and Bintao Qiu (Peking Union Medical College Hospital) for his help in proteomics experiments.




