DNA crosslinking and recombination-activating genes 1/2 (RAG1/2) are required for oncogenic splicing in acute lymphoblastic leukemia
Abstract
Background
Abnormal alternative splicing is frequently associated with carcinogenesis. In B-cell acute lymphoblastic leukemia (B-ALL), double homeobox 4 fused with immunoglobulin heavy chain (DUX4/IGH) can lead to the aberrant production of E-26 transformation-specific family related gene abnormal transcript (ERGalt) and other splicing variants. However, the molecular mechanism underpinning this process remains elusive. Here, we aimed to know how DUX4/IGH triggers abnormal splicing in leukemia.
Methods
The differential intron retention analysis was conducted to identify novel DUX4/IGH-driven splicing in B-ALL patients. X-ray crystallography, small angle X-ray scattering (SAXS), and analytical ultracentrifugation were used to investigate how DUX4/IGH recognize double DUX4 responsive element (DRE)-DRE sites. The ERGalt biogenesis and B-cell differentiation assays were performed to characterize the DUX4/IGH crosslinking activity. To check whether recombination-activating gene 1/2 (RAG1/2) was required for DUX4/IGH-driven splicing, the proximity ligation assay, co-immunoprecipitation, mammalian two hybrid characterizations, in vitro RAG1/2 cleavage, and shRNA knock-down assays were performed.
Results
We reported previously unrecognized intron retention events in C-type lectin domain family 12, member A abnormal transcript (CLEC12Aalt) and chromosome 6 open reading frame 89 abnormal transcript (C6orf89alt), where also harbored repetitive DRE-DRE sites. Supportively, X-ray crystallography and SAXS characterization revealed that DUX4 homeobox domain (HD)1-HD2 might dimerize into a dumbbell-shape trans configuration to crosslink two adjacent DRE sites. Impaired DUX4/IGH-mediated crosslinking abolishes ERGalt, CLEC12Aalt, and C6orf89alt biogenesis, resulting in marked alleviation of its inhibitory effect on B-cell differentiation. Furthermore, we also observed a rare RAG1/2-mediated recombination signal sequence-like DNA edition in DUX4/IGH target genes. Supportively, shRNA knock-down of RAG1/2 in leukemic Reh cells consistently impaired the biogenesis of ERGalt, CLEC12Aalt, and C6orf89alt.
Conclusions
All these results suggest that DUX4/IGH-driven DNA crosslinking is required for RAG1/2 recruitment onto the double tandem DRE-DRE sites, catalyzing V(D)J-like recombination and oncogenic splicing in acute lymphoblastic leukemia.
Abbreviations
-
- AGAP1
-
- ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
-
- ALL
-
- Acute lymphoblastic leukemia
-
- BCR-ABL
-
- Breakpoint duster region fused with Abelson protooncogene
-
- BLI
-
- Biolayer interferometry
-
- C6orf89
-
- Chromosome 6 open reading frame 89
-
- C6orf89alt
-
- Chromosome 6 open reading frame 89 abnormal transcript
-
- CD19
-
- Cluster of differentiation 19
-
- ChIP-seq
-
- Chromatin immunoprecipitation high-throughput sequencing
-
- CLEC12A
-
- C-type lectin domain family 12, member A
-
- CLEC12Aalt
-
- C-type lectin domain family 12, member A abnormal transcript
-
- Co-IP
-
- Co-immunoprecipitation
-
- COL9A1
-
- Collagen type IX alpha 1 chain
-
- CY5
-
- Cyanine dye 5
-
- DEGs
-
- Differentially expressed genes
-
- DRE
-
- DUX4-resposive-element
-
- DUX4
-
- Double homeobox 4
-
- ERG
-
- E-26 transformation-specific (ETS) family related gene
-
- ERGalt
-
- E-26 transformation-specific family related gene abnormal transcript
-
- ETV6-RUNX1
-
- E-26 transformation-specific (ETS) variant transcription factor 6 fused with RUNX family transcription factor 1
-
- FLT3L
-
- FMS-like tyrosine kinase 3 ligand
-
- FSHD
-
- Facioscapulohumeral muscular dystrophy
-
- GEO
-
- Gene expression omnibus database
-
- GSEA
-
- Gene set enrichment analysis
-
- HD
-
- Homeobox domain
-
- IgG
-
- Immunoglobulin G
-
- IL-3/6/7
-
- Interleukin 3/6/7
-
- IPTG
-
- Isopropyl β-d-1-thiogalactopyranoside
-
- PAX5
-
- Paired box 5
-
- PDGFRA
-
- Platelet derived growth factor receptor alpha
-
- PLA
-
- Proximity-ligation-assay
-
- PTPRM
-
- Protein tyrosine phosphatase receptor type M
-
- RAG1/2
-
- Recombination-activating genes 1/2
-
- RNA-seq
-
- RNA sequencing
-
- RSS
-
- Recombination signal sequences
-
- RT-PCR
-
- Quantitative real-time polymerase chain reaction
-
- SAXS
-
- Small angle X-ray scattering
-
- SDS-PAGE
-
- Sodium dodecyl sulfate polyacrylamide gel electrophoresis
-
- TCF3-PBX1
-
- Transcription factor 3 fused with PBX homeobox 1
-
- WT
-
- Wild type
1 BACKGROUND
Acute lymphoblastic leukemia (ALL) is the most common pediatric cancer associated with the estimated cumulative risk of ∼1 in 2,000 among children [1]. Recently, using the second-generation sequencing and RNA-sequencing (RNA-seq) profiling technologies, our group [2-4] and other research groups [5, 6] have identified an axis-of-leukemogenesis, double homeobox 4 fused with immunoglobulin heavy chain (DUX4/IGH)-E-26 transformation-specific family-related gene abnormal transcript (ERGalt) deregulation, in B-cell ALL. In our previous studies, ∼7% Chinese B-ALL patients displayed DUX4/IGH deregulation and often accompanied with the biogenesis of ERGalt, an E-26 transformation-specific family-related gene (ERG) alternative splicing isoform [2-4]. Supportively, two independent investigations in Japan [5] and US reported the similar observations [6]. This reiterates that the abnormal expression of DUX4/IGH and the subsequent ERGalt biogenesis controlled by the DUX4/IGH-driven alternative splicing are the major drivers that lead to a full-fledged leukemogenesis [5, 6].
Using structural and cellular approaches, we had shown that the DNA-binding activity mediated by the homeobox domain 1 and 2 (HD1-HD2) double homeobox was essential to the DUX4/IGH-driven transactivation [2, 4]. Inhibition of the recognition between DUX4/IGH and DUX4-resposive-element (DRE) not only prevented the oncogenic transactivation but also significantly impaired the inhibitory effects of DUX4/IGH on B-cell differentiation in mouse progenitor cells [2]. In addition, the aberrant expression of wild-type (WT) DUX4 protein was considered as the leading cause of another human disease, facioscapulohumeral muscular dystrophy (FSHD) [7]. This, together with the observations of the zygotic genome activation in placental mammals [8-10], echoes the importance of DUX4-DRE recognition. However, in spite of recent breakthroughs [2, 4, 11], it remains unclear how DUX4/IGH triggers ERG alternative splicing.
In this study, we aimed to understand DUX4/IGH-driven splicing: 1) whether there are more aberrant splicings in DUX4/IGH subtype; 2) how the repetitive DRE-DRE sites might contribute to oncogenic splicing; 3) whether DUX4/IGH requires a helper protein.
2 MATERIALS AND METHODS
2.1 Intron retention in DUX4/IGH target genes
The RNA-seq data of 135 B-cell acute lymphoblastic leukemia (B-ALL) samples from a published work [12] were used to screen for the intron retention events in the DUX4/IGH subtype. The data contained 47 DUX4-fusion samples and 88 other oncogenic fusions. The other subtypes of B-ALL samples [i.e., E-26 transformation-specific variant transcription factor 6 fused with RUNX family transcription factor 1 (ETV6-RUNX1), transcription factor 3 fused with PBX homeobox 1 (TCF3-PBX1), breakpoint duster region fused with Abelson protooncogene (BCR-ABL)] were used as controls. The differential intron retention analysis was conducted by using IRFinder software (version 1.2.5) [13] and R package IntEREst (version 1.8.0) [14]. Events with adjusted P value below 0.05 supported by both IRFinder and IntExRet were chosen for further validation in Integrative Genomics Viewer software (version 2.4.10) [15].
2.2 DRE repeats in DUX4/IGH target genes
Three chromatin immunoprecipitation high-throughput sequencing (ChIP-seq) datasets were used to analyze the revised DRE 5′-TAGT/TTA-3′: the human WT DUX4 [Gene Expression Omnibus database (GEO) number: GSE75791] [16], the mouse WT DUX (a human DUX4 homolog, GEO number: GSE87279) from myoblasts [17], and the human DUX4/IGH (European Genome-phenome Archive accession: EGAS00001001923) from leukemia cell lines NALM-6 and Reh [6]. The program HOMER[18] (v4.10, 04-01-2019, University of California, San Diego, CA, USA) was used for De novo motif-discovery analysis. The revised DRE cross-validated by structural and ChIP-seq investigation was used as a template to search for DUX4/IGH target genes in the genomic region (hg19/GRCh37): exons and introns [2, 19, 20].
2.3 Protein expression, purification, and DNAERG preparation
The DNA fragment encoding the HD1-HD2 domain (i.e., residues 1-150) of human DUX4 protein, termed DUX41-150, was cloned into a modified pET15b (Youbio, Changsha, Hunan, China) using Nde I and Xho I restriction sites. An N-terminal SUMO tag was engineered to enhance the solubility of DUX41-150. Then the constructs were transformed into Escherichia coli BL21 (DE3) cells (Sangon, Shanghai, China) for DUX41-150 production. In brief, the cells were grown in LB Borth (Sangon) at 37°C for 6 h and induced with 500 µmol/L IPTG (Sangon) for 14 h at 16°C when OD600 reached 0.8-1.0. Cells were harvested by centrifugation (4,000 rpm, 20 min) and resuspended in buffer containing 20 mmol/L 4-hydroxyethyl piperazine ethyl sulfonic acid (HEPES), 100 mmol/L NaCl, pH = 7.4, prior to a French press treatment (JNBIO, Guangzhou, Guangdong, China). The clear lysate was separated from cell debris by centrifugation (22,000 rpm, 90 min) at 4°C before it was applied to a pre-equilibrated nickel column (His-Trap HP, GE Healthcare, Chicago, IL, USA). The non-specific bindings were washed off with buffer containing 20 mmol/L HEPES, 500 mmol/L NaCl, 40 mmol/L imidazole, pH = 7.4. The DUX41-150 fused with 6 × histidine and SUMO tag, termed HIS-SUMO-DUX41-150, was eluted from the column with buffer containing 20 mmol/L HEPES, 100 mmol/L NaCl, 1M imidazole, pH = 7.4. Then the His-SUMO tag was removed by digestion with thrombin enzyme (Sigma, Milwaukee, WI, USA) at 4°C for 12 h accompanied with a dialysis treatment with buffer containing 20 mmol/L HEPES, 20 mmol/L NaCl, 1mmol/L dithiothreitol, 10% glycerol, pH = 7.4. The protein samples were further purified by a cation exchange SP column (GE HealthCare) in which the buffer A was 20 mmol/L HEPES, 20 mmol/L NaCl, pH = 7.4 and the buffer B was 20 mmol/L HEPES, 1 mol/L NaCl, pH = 7.4. The eluents were concentrated and loaded onto a gel filtration S100 column (GE Healthcare) pre-equilibrated with the buffer containing 20 mmol/L Tris, 100 mmol/L NaCl, pH = 8.0. The final purified DUX41-150 was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and mass spectrum analysis.
Concerning DNAERG, two synthetic oligonucleotides (5′- CAGTCTAATCTCATCAAGTCG-3′, 5′-CGACTTGATGAGATTAGACTG-3′, DRE site in bold) were resuspended in sterile water, respectively. To obtain double-stranded DNA, the oligonucleotides were mixed at a 1:1 molar ratio and annealed under 95°C for 10 min. The mixture was then cooled to 4°C. The annealed DNAERG was concentrated to a final concentration of 141 mg/mL as monitored by the absorbance at 260 nm.
2.4 Crystallization, data collection, and structural determination
Before the co-crystallization of DUX41-150-DNAERG (DUX4/IGH binding sequence/site-derived from ERG gene), the DUX41-150 (9 mg/mL) and the double-stranded DNAERG were incubated at a 1:1 molar ratio at 4°C for 30 min in buffer of 20 mmol/L HEPES, pH = 7.4, 100 mmol/L NaCl. Then the DUX41-150-DNAERG was mixed at 1:1 (v/v) ratio with the buffer containing 0.1 mol/L Bis-Tris propane, 20% polyethylene glycol 3350, and 0.2 mol/L sodium bromide, pH = 7.5. The crystals were flash-cooled in liquid nitrogen under a cryo-protection by Paratone-N oil (Hampton Research, Aliso Viejo, CA, USA). The diffraction data were recorded in BL19U1 at Shanghai Synchrotron Radiation Facility (Shanghai, China). Following diffraction, the data were processed, integrated and scaled using MOSFLM/SCALA [21]. The statistics of the data collection are shown in Supplementary Table S1.
The DUX41-150-DNAERG was initially phased by molecular replacement using HD2-ERG structure (Protein Data Bank ID: 6A8R) as a search template. The programs REFMAC5 [22] and PHENIX.REFINE [23, 24], together with manual building implemented in COOT [21], were used to improve the phases, in particular in the regions of HD1, HD1-HD2 linker, and ERG duplex. The final model contained 3915 atoms from residues/nucleotides and 8 water molecules. Ramachandran statistics estimated by PROCHECK [25] showed that 95.3% and 4.7% of the atoms were in the most favored and allowed regions, respectively. The detailed structure refinement statistics are reported in Supplementary Table S1. The DUX41-150-DNAERG coordinates had been deposited into the Protein Database Bank (https://www.rcsb.org/) with the entry code of 7DW5.
2.5 Small angle X-ray scattering (SAXS)
The purified WT/mutant DUX41-150 complexed with/without DNAERG were concentrated to 2 mg/mL in the buffer of 20 mmol/L Tris, 100 mmol/L NaCl, pH = 8.0. The SAXS experiment was carried out at Beamline station BL19U2 (Shanghai Synchrotron Radiation Facility). The measurements were implemented with 1 s exposure time and repeated for 20 times to avoid radiation damage. Data subtraction and analysis were performed with PRIMUS [26]. Crystal data fitting was carried out using the MIXTURE algorism implemented in CRYSOL [27]. The atomic models presented here and published elsewhere [28] were used in this characterization.
2.6 Analytical ultracentrifugation
Sedimentation experiment was conducted using a Beckman XL-1 analytical ultracentrifuge (Beckman Coulter, Brea, CA, USA) equipped with a 6-hole rotor. Ultraviolet (UV)-vis absorbance optics was used to monitor the sedimentation and diffusion processes of recombinant DUX41-150 and the linker mutant H78A/E93R. The protein samples kept in the buffer of 20 mmol/L Tris, 100 mmol/L NaCl (pH = 8.0) were spun at the speed of 42,000 rpm at 20°C for at least 12 h. The collected data were analyzed by SEDFIT using a continuous distribution c (s) model [29]. The sedimentation coefficient (s = v/ω2r) was extrapolated to water at 20°C.
2.7 Biolayer interferometry (BLI) experiment
BLI experiment was measured by using the Octet Red 96 instrument with SA biosensor at 30°C (ForteBio, Gottingen, Niedersachsen, Germany). The whole experiment was carried out with a buffer consisting of 10 mmol/L HEPES, 150 mmol/L NaCl and 0.005% Twain-20 (v/v), pH = 7.4. Before the assay, the recombinant DUX41-150 protein or mutants were exchanged into the HBST buffer above by gel filtration. A 96-well microporous plate was used for this assay. For immobilizing, the SA biosensor was immersed in the well of DUX41-150 (or mutants) with gradient concentration from 0.5 to 16 µmol/L for 200 s. Then 0.1 mol/L NaOH was used to terminate the reaction. The ForteBio Data Analysis software (version 7.1) was used to estimate the binding constant (KD).
2.8 Patients and samples
A total of 165 Chinese B-ALL patients were enrolled under the Shanghai Institute Hematology protocol (Chinese Clinical Trial Registry, number ChiCTR-RNC-14004969) and Shanghai Children's Medical Center protocol (Chinese Clinical Trial Registry, number ChiCTR-ONC-14005003) [6, 30]. Other patients’ clinical information was obtained from TARGET/COG ALL project [31-34], Singapore Lund University Hospital cohort [35], Malaysia MaSpore cohort [35], and Japan Adult Leukemia Study Group (JALSG) cohort [5]. The data and samples from these patients were only used in the following RNA-seq and prognosis analysis.
2.9 Plasmids, viruses packaging, and cell culture
The cDNA fragment encoding full-length DUX4/IGH was amplified from B-ALL patients mentioned above and engineered into MigR1-IRES-GFP vector (Addgene, Watertown, MA, USA) or LEGO-iG2 vector (gift from Dr. Jianqing Mi, Shanghai JiaoTong University, Shanghai, China). HA tag was in frame with 5′ DUX4/IGH coding for further detection by anti-HA antibody (ab9110, Abcam, Cambridge, UK). The short hairpin RNA (shRNA) sequences for knock-down assays were designed to subclone into the PLVX-shRNA2 vector (Clontech, Mountain View, CA, USA). The site-directed mutagenesis technology (KOD-401, TOYOBO, Osaka, Japan) was used for DUX4/IGH mutants. All the primers are shown in Supplementary Table S2.
For lentivirus packaging, the plasmids WT/mutant LEGO-iG2-HA-DUX4/IGH, psPAX2, pMD2.G, and RSV were co-transfected into 293T cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). In the shRNA knock-down assays, the plasmid PLVX-shRNA2 was used. Concerning B-cell differentiation assay, the plasmid MigR1-HA-DUX4/IGH or mutants, as well as the plasmid Ecopack, were co-transfected into 293T cells for retrovirus packaging. The 293T and Reh cells used in study were derived from Shanghai Institute of Hematology (Shanghai, China) and cultured in the dulbecco's modified eagle medium (DMEM) and RPMI-1640 medium, respectively. Both mediums were supplemented with 10% fetal bovine serum (FBS), and mycoplasma contamination was detected as required.
2.10 Abnormal variant biogenesis induced by DUX4/IGH
The lentiviruses of LEGO-iG2-HA-DUX4/IGH or mutants were transduced into Reh cells. On the fourth day after transfection, the cells were harvested and lysed by sonication in the radio-immunoprecipitation assay (RIPA) buffer. For Western blotting analysis, the clear lysate was resolved in a 12% SDS-PAGE before it was transferred onto a polyvinylidene fluoride (PVDF) membrane. The ERGalt production was detected using antibody against ERG (ab92513, Abcam). For cross-validation, the ERGalt, chromosome 6 open reading frame 89 abnormal transcript (C6orf89alt), and C-type lectin domain family 12, member A abnormal transcript (CLEC12Aalt) biogenesis at mRNA level was further monitored by the real-time PCR technique using ABIPRISM 7500 (Applied Biosystems, Foster City, CA, USA). Similarly, DUX4/IGH-driven transactivations in ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 (AGAP1), platelet-derived growth factor receptor alpha (PDGFRA), collagen type IX alpha 1 chain (COL9A1), and protein tyrosine phosphatase receptor type M (PTPRM) were also monitored by quantitative real-time polymerase chain reaction (RT-PCR). The primers are shown in Supplementary Table S3.
2.11 Primary B-cell differentiation experiment
A lineage depletion kit (Miltenyi Biotec, Bergisch Gladbach, Germany) was used to isolate mouse bone marrow lineage-negative (Lin−) cells. The flow cytometry (FACS) was used to obtain Lin−/c-Kit Low cells. The bone marrow cells were then infected with the retroviruses containing MigR1-HA-DUX4/IGH or mutants. The transfected cells were co-cultured with an OP9 monolayer for 5 days in Iscove's Modified Dubecco's Medium (IMDM) containing 20% FBS, 50 ng/mL FMS-like tyrosine kinase 3 ligand (Flt3L), 10 ng/mL Interleukin 3 (IL-3), 10 ng/mL IL-6, 50 ng/mL IL-7, and 20 ng/mL recombinant human stem cell factor (SCF). The lymphoid lineage differentiations by DUX4/IGH and mutants were evaluated by FACS using antibodies against mouse CD19 (561737, BD Pharmingen, Franklin Lakes, NJ, USA) and Mac-1 (48-0112-82, eBioscience, San Diego, CA, USA).
2.12 Structure-based RNA-seq characterization
The LEGO-iG2-HA-DUX4/IGH and structure-based derivatives (i.e., N69A/144A, H78A/E93R, R76A/R79A/R80A) were used for expression in Reh cells. The total RNA was extracted using Trizol kit (Invitrogen), followed by quantification with Agilent 2100 bioanalyzer (Thermo Fisher Scientific, Waltham, MA, USA). The sequencing libraries were prepared using the Beijing Genomics Institute (BGI) protocols, and sequencing was performed on the BGIseq-500 (The Beijing Genomics Institute, Beijing, China).
The raw unfiltered RNA-seq reads were aligned to human reference genome hg19 using hierarchical indexing for spliced alignment of transcripts (Hisat2) [36] (version 2.0.5, http://www.ccb.jhu.edu/software/hisat/), with default parameters, and imported to differential expression analysis for sequence count data (DESeq2, http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html) [37] for differential gene expression analysis (|log2(fold change)| > 1, P < 0.01). Gene Ontology analysis [38] of differentially expressed genes (DEGs) were conducted using the R package, clusterProfiler [39]. Gene-set enrichment and pathway analysis were performed by gene set enrichment analysis (GSEA) [40] (version 4.0.3, https://www.gsea-msigdb.org/gsea/index.jsp). The published prognosis data derived from 421 B-ALL patients [30] were used to estimate the 5-year overall survival (OS, calculated after treatment) rates of patients expressing DUX4/IGH target genes. Survival curves were estimated with the Kaplan–Meier method, compared by two-sided log-rank test, and plotted by R packages (version 3.6.2, http://r-project.org/) survival (version 3.2-3, https://github.com/therneau/survival) and survminer (version 0.4.8, https://rpkgs.datanovia.com/survminer/index.html). The least absolute shrinkage and selection operator (LASSO) regression analysis [41] and signature gene scores were calculated with R package glmnet (version 4.0-2, https://glmnet.stanford.edu).
2.13 In situ proximity ligation assay
In situ proximity ligation assay (PLA) is suitable for quantification of protein expression and for characterization of modifications and interactions of proteins [42, 43]. PLA assay was performed using Duolink® PLA Fluorescence (Merck, Darmstadt, Hessen, Germany) according to manufacturer's instruction. In brief, the Reh cells expressing DUX4/IGH or mutants were transfected with LEGO-iG2 vector containing recombination-activating gene 1/2 (RAG1/2). The cells were harvested and attached to coverslips through centrifugation (700 rpm, 5 min). The antibodies of DUX4/IGH (bs-12369R, 1:500 dilution, Bioss, Shanghai, China; ab124699, 1:1500 dilution, Abcam) and RAG1/2 (sc377127 and sc517209, 1:500 dilution, Santa Cruz Biotechnology, Santa Cruz, CA, USA) were incubated, then staining with the PLA probes (secondary antibody, one PLUS and one MINUS). If the target proteins interacted with each other, the DNA could be amplified and visualized by fluorescently labeled complementary oligonucleotide probes. The number and intensity of the dots were detected by fluorescence microscopy.
2.14 Co-immunoprecipitation (co-IP)
The Reh cells with expression of HA-DUX4/IGH1-431, HA-DUX4/IGH1-150, or HA-DUX4/IGH151-431 were cultured for 48 h. Then cells were harvested and lysed with pre-cooled RIPA buffer. The clear lysate was then incubated with beads coated with anti-HA antibody overnight. The precipitant pulled down by HA-DUX4/IGH were further analyzed using anti-RAG1 (sc377127, 1:500 dilution, Santa Cruz Biotechnology) and anti-RAG2 antibodies (sc517209, 1:500 dilution, Santa Cruz Biotechnology). In the reverse co-IP, the beads coated with anti-RAG1 antibody were used as bait. The co-IP of HA-DUX4/IGH and mutants were detected by using anti-HA antibody (ab9110, 1:3000 dilution, Abcam). Anti-mouse IgG (sc-2025, Santa Cruz Biotechnology) and anti-rabbit IgG (Cell Signaling Technology, 2729, Boston, MA, USA) were used as negative controls.
2.15 Mammalian two-hybridization assay
This assay was performed using the CheckMateTM Mammalian Two-Hybrid system (Promega, Madison, WI, USA) in 293T cells. To detect the interaction between DUX4/IGH and RAG1/RAG2, the cDNA of WT/mutant DUX4/IGHs and paired box 5 (PAX5) were engineered into pACT vectors (Promega), in which the latter was used as a positive control. The cDNA of RAG1 and RAG2 were cloned into pBIND vectors (Promega). The 293T cells were co-transfected with pG5-luc, pBIND-RAG1/2, and WT/mutants pACT-DUX4/IGH mixtures at a molar ratio of 1:1:1 using Lipofectamine 2000 (Invitrogen). The transfected 293T cells were harvested after 48 h. The relative luciferase activities were determined by using the Dual-Luciferase Reporter Assay System (Promega). In brief, the harvested cells were lysed with the lysis buffer in the above kit for 15 min, then centrifuged for 5 min at 12,000 rpm to collect the supernatant and discarded the cell debris. The biofluorescence of different samples was measured by using a luminometer (Titertek-Berthold, Huntsville, AL, USA). The relative reaction values of all samples were normalized against the empty vector.
2.16 In vitro cleavage assays
In brief, the cleavage activity of RAG proteins was evaluated using an in vitro assay developed in the Gellert laboratory [44]. The recombinant human RAG1/2 proteins were obtained and purified from the embryonic cell line 293. The 20 µL cleavage reactions contained the newly identified recombination signal sequence (RSS)-like substrates labeled with 100 nmol/L 5′-Cy5, 100 nmol/L RAG1/2, 100 nmol/L high mobility group box 1 (HMGB11-163) and the buffer of 25 mmol/L 3-(N-Malindai) propane sulfonic acid-potassium hydroxide (MOPS-KOH), 60 mmol/L potassium glutamate, 100 µg/mL bovine serum albumin, and 1 mmol/L MgCl2, pH = 7.0. The reactions were incubated at 37°C for 1 h. Sample loading solution (8 mol/L urea, 20 mmol/L ethylene diamine tetraacetic acid (EDTA) was added with a volume of 5 µL, prior to the final termination by heating at 95°C for 10 min. Then 7 µL of the reaction products obtained from the ERG, C6orf89, and CLEC12A sequences were respectively fractionated on a 15% (19:1 acrylamide/bisacrylamide) Tris-borate-EDTA (TBE)-urea polyacrylamide gels in 90 mmol/L Tris-borate (pH = 8.0) and 0.2 mmol/L EDTA. The classical and published 12-RSS and 23-RSS substrates [44] were used as positive control. The cleavage products were detected by a fluorescence imager using a 635 nm laser and 670 ± 30 nm filter (Typhoon, GE Healthcare).
2.17 Cleavage under targets and tagmentation (CUT & Tag assays)
CUT & Tag assay was performed as described previously with modifications [45]. Briefly, 1 × 105 cells were washed twice gently with wash buffer (20 mmol/L HEPES pH = 7.5, 150 mmol/L NaCl, 1 × protease inhibitor cocktail, 0.5 mmol/L spermidine) before mixing with 10 µL concanavalin A-coated magnetic beads (Diagenode, Liège, Belgium) and then incubated at about 25°C for 10 min. The unbound supernatant was removed, and bead-bound cells were resuspended with 100 µL dig wash buffer (20 mmol/L HEPES pH = 7.5, 150 mmol/L NaCl, 0.5 mmol/L spermidine, 1 × protease inhibitor cocktail, 0.05% digitonin, 2 mmol/L EDTA). After overnight incubations with a primary antibody (DUX4/IGH antibody, ab124699, 1:50 dilution, Abcam; RAG1 antibody, ab172637, Abcam; or normal rabbit IgG antibody, 12-370, Millipore, Billerica, MA, USA) on a rotating platform and followed by a 0.5–1 h incubation with secondary antibody (anti-Rabbit IgG antibody, AP132, Millipore) in dig wash buffer, the beads were washed and resuspended in a 1:100 dilution of pA-Tn5 adapter complex in dig-med buffer (0.05% digitonin, 20 mmol/L HEPES, pH = 7.5, 300 mmol/L NaCl, 0.5 mmol/L spermidine, 1 × protease inhibitor cocktail) at about 25°C for 1 h. Cells were washed 2-3 times in 1 mL dig-med buffer to remove unbound pA-Tn5 protein, followed by resuspending in tagmentation buffer (with 10 mmol/L MgCl2 in dig-med buffer) and then incubated at 37°C for 1 h.
Next, DNA was purified using phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation. To make and amplify libraries, The 21 µL DNA was mixed with 2 µL of a universal i5 primer and a uniquely barcoded i7 primer (New England Biolabs, Ipswich, MA, USA). A volume of 25 µL NEBNext HiFi 2 × PCR Master mix (New England Biolabs) was added and mixed. The sample was placed in thermocycler with a heated lid using the following cycling conditions: 72°C for 5 min (gap filling); 98°C for 30 s; 14 cycles of 98°C for 10 s and 63°C for 30 s; final extension at 72°C for 1 min and hold at 8°C. Post-PCR clean-up was performed using XP beads (Beckman Coulter). Amplified libraries were determined and sequenced on Agilent 4200 TapeStation (Agilent, Santa Clara, CA, USA) and Illumina Novaseq 6000 (150 bp paired-end) (Illumina, San Diego, CA, USA), respectively. Paired-end reads were aligned to human genome hg19 (GRCh37) using the BWA program (Cambridge, MA, USA) [46]. For annotation, MACS2 software (Boston, MA, USA) [47] was used for peak calling with a cutoff q value < 0.05. Peaks were annotated by using Homer (v4.10, 04-01-2019, San Diego, CA, USA) [18].
2.18 Statistical analysis
All data were presented as mean ± standard deviation (SD). Statistical analyses were conducted using the GraphPad Prism 7.0 (GraphPad Software, San Diego, CA, USA), including unpaired two-tailed Students’ t-test and analysis of variance (ANOVA). ANOVA was conducted to compare between 2 or 3 means in independent experiments on repeated measures. The data were expressed as mean ± 95% confidence interval (CI). P < 0.05 was considered statistically significant.
3 RESULTS
3.1 Crystal structure of DUX41-150-DNADRE hetero-tetramer revealed an unprecedented crosslinking activity in oncogenic driver
Until this was reported, ERGalt was the only splicing ever reported in DUX4/IGH subtype leukemia [6] (Figure 1A). Here, we used a published RNA-seq dataset derived from 135 B-ALL patients [12] to screen the intron retention events exclusively associated with the DUX4/IGH subtype (identified in 47 patients). The other B-ALL subtypes (identified in 88 patients), including ETV6-RUNX1, TCF3-PBX1, and BCR-ABL, were used as control. The differential intron retention testing algorithm implemented in programs IRFinder [13] and IntEREst [14] were used in this analysis. Events with adjusted P value < 0.05 supported by both IRFinder and IntExRet were chosen for further validation in Integrative Genomics Viewer [15]. Intron retention events (i.e., alternative splicing) were clearly observed in CLEC12A, C6orf89, AGAP1, and PTPRM (Figure 1B and Supplementary Figure S1). This was further supported by RT-PCR analysis, in which the Reh cells harboring DUX4/IGH showed alternative splicing, leading to the biogenesis of CLEC12Aalt and C6orf89alt (Figure 1B). To our surprise, similar to ERGalt [6], repetitive arrangement of the DRE sites were also detected in CLEC12A and C6orf89 (Figure 1B), prompting the investigation of whether and how tandem DRE-DRE might control alternative splicing. To check whether these DRE repeats were available in the DUX4/IGH target genes, we performed ChIP-seq analysis in Reh cells with DUX4/IGH expression. The repetitive DRE sites were consistently observed within the aberrantly splicing genes (ERG, CLEC12A, and C6orf89). Similar results were obtained in the ChIP-seq analysis of Nalm6 cells with endogenous DUX4/IGH expression [6] (Supplementary Figure S1A-C). Consistent with the results of ERG, CLEC12A, and C6orf89, repetitive DREs were observed in close proximity of AGAP1alt and PTPRMalt splicing sites (Supplementary Figure S1D and S1E). This observation, together similar results in other splicing sites, supports the hypothesis that DRE-DRE arrangement and DUX4/IGH-driven crosslinking might be important factors for oncogenic splicing.
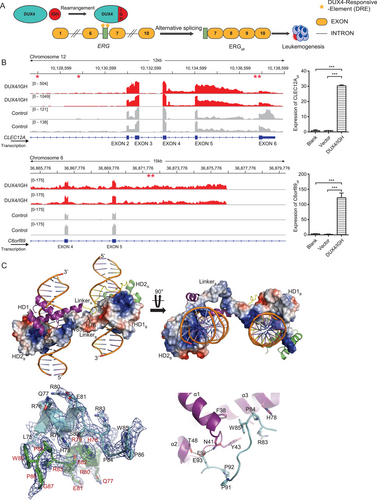
To understand how DUX4/IGH induced ERG alternative splicing, we had determined the crystal structure of DUX41-150-DNAERG at 2.8 Å resolutions (Figure 1C and Supplementary Table S1). The structural complex contained a HD1-HD2 dimer and two DNAERG duplexes, i.e., 5′-C-4GACTT1GATGAGATTA11GACTG16-3′ (forward chain) and 3′-G4′CTGAA1′CTACTCTAAT11′CTGAC16-5′ (reverse chain), in which the DRE sites were bold and underscored. The two DUX41-150 molecules packed against each other in a head-to-tail configuration. The double-kiss of two DRE sites at the sides of the DUX4 dimer gave rise to a remarkable dumbbell-shape architecture (Figure 1C). The dimerization was mainly mediated by the residues 76-98 (termed HD1-HD2 linker) and two inter-molecular hydrogen bonds, T48-E93 and N41-E93 (Figure 1C). Via domain-swap, the inter-molecular chimera HD1A-HD2B or HD1B-HD2A (A/B for different DUX4 monomers) were located on the sides of the dimer, primed for the recognition of two adjacent DRE sites.
In our previous reports, we had proposed that DUX4 double homeobox can bind to DNA sequences with TGAT- and TAAT-like repeats [2, 4]. The new DUX41-150-DNAERG structure showed that this is indeed the case (Supplementary Figure S2A-E). Different sets of charged residues were mobilized to form direct hydrogen bonds with TAAT- and TGAT-binding. In HD1, the engagement was mainly mediated between R20/R23/N69 and 5′-T9TA11-3′/3′-A9′AT11′-5′ (Supplementary Figure S2A and S2B). The invariant N69 laid in the heart of this interaction and forming two direct hydrogen bonds with nucleotide A9′ in the major groove. On the opposite side, the conserved R20 and R23 contributed another two hydrogen bonds to the nucleotide reading. As a result, the RRKR loop and the α3 helix acted as like two clamping hands ensuring the reading of 3′-end of the DRE site by HD1. In HD2-DNA binding, similar clamping mechanism could be observed (Supplementary Figure S2C and S2D). The conserved N144 and R148 in the α6 helix were utilized to form four direct hydrogen bonds with 5′-T1GAT4-3′. The interaction between HD2 and the 5′-end of DRE was further cemented by two extra direct hydrogen bonds between R95/R98 and the complementary DNA 3′-A1′CT3′-5′. Furthermore, as shown in Supplementary Figure S2E and S2F, the structural alignment between HD1 and HD2 might explain why DUX4 homeobox modules prefer TAAT and TGAT, respectively. The E70/Q74 in HD1 could pull the guanidinium head group of R73 away from the TAAT. In marked contrast, R145/H149 in HD2 stabilized R148 side chain to make double hydrogen bond with TGAT.
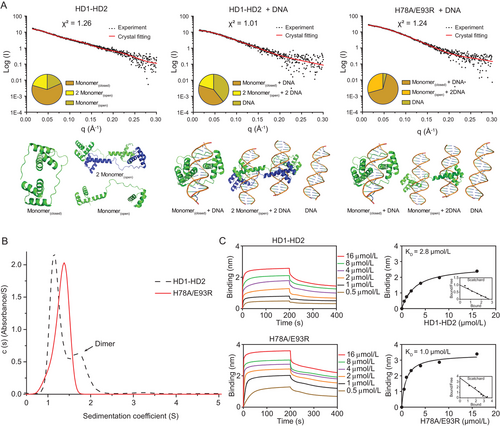
3.2 HD1-HD2 linker
In previous studies, our group [2] and other research groups [28, 48] had proposed a two-step clamp-like mechanism, in which the DUX4 HD1-HD2 folds into a circular structure upon DNA binding. However, the DUX41-150-DNAERG presented here displayed a completely different configuration, suggestive of a versatile DUX4/IGH-DRE binding/crosslinking controlled by the HD1-HD2 linker (i.e., residues 76-98). To characterize this further, the HD1-HD2 protein and DNAERG were subjected to SAXS and analytical ultracentrifugation analysis in solution (Figures 2A and 2B). For WT HD1-HD2 alone, the X-ray scattering data were fitting well with various DUX4 monomer/dimer structures (χ2 = 1.26). The good match between the experimental data and the crystal structures suggested that DUX4 HD1-HD2 could undergoes significant conformational changes and dimerization via the HD1-HD2 linker (Figure 2A). Consistently, when the WT HD1-HD2 were mixed with DNAERG, HD1-HD2-linker-driven dimerization and DRE crosslinking (20.4%, χ2 = 1.01) were observed (Figure 2A). This was further supported by H78A/E93R mutation (Figure 2A). As monitored by SAXS and analytical ultracentrifugation, the HD1-HD2 linker perturbation disrupted dimerization in solution (Figures 2B and 2C). Interestingly, these mutations did not impair its DNA binding capacity, as characterized by BLI analysis [49] (Supplementary Figure S3).
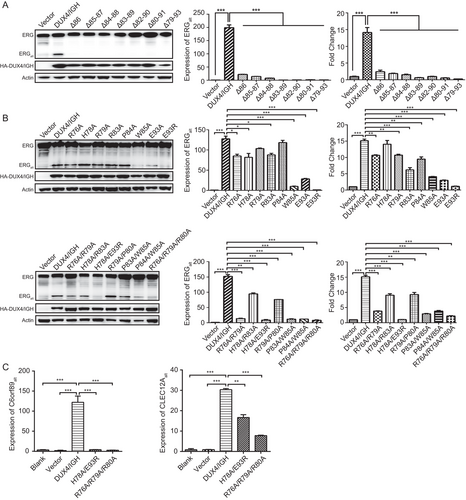
We next checked whether the HD1-HD2 linker was essential to DUX4-driven alternative splicing. Firstly, the linker loop was subjected to deletion analysis. The Western blotting and RT-PCR techniques were used to monitor the disruptive impact on ERGalt biogenesis. As shown in Figure 3A, a single amino acid deletion was sufficient to abrogate DUX4/IGH's alternative splicing activity. Similar results were observed in luciferase assay using the DRE site derived from ERG (Figure 3A). Secondly, to characterize this further, we had done a more vigorous mutation scanning over the linker region (Figure 3B). The single amino acid mutations (i.e., W85A, E93A, and E93R) appeared to be the most devastative perturbations, resulting in significant loss in ERGalt (Figure 3B). Other residues/positions in the HD1-HD2 linker, although causing minor disruption in single mutations, were also critical when tested by poly amino acid mutations (Figure 3B). This was further supported by the CLEC12Aalt and C6orf89alt biogenesis assay, in which the structure-based perturbation consistently disrupted the DUX4/IGH-driven alternative splicing (Figure 3C).
3.3 Structure-based RNA-seq analysis uncovered new prognosis marker in B-ALL
Consistently, the importance of the HD1-HD2 linker/dynamics was also highlighted by B-cell differentiation assay. H78A/E93R and R76A/R79A/R80A, which had little impact on DNAERG binding (Figure 2C and Supplementary Figure S3), significantly abrogated ERGalt production in Reh cells (Supplementary Figure S4A) and oncogenic activity of DUX4/IGH upon B cell arrest (Figure 4A). When monitored by FACS, the overexpression of WT DUX4/IGH led to B cell arrest (CD19+ cells, 8.2%). In comparison, the mouse progenitor (Lin−/c-KitLow) cells expressing H78A/E93R and R76A/R79A/R80A regained B-cell differentiation (CD19+ cells, 40.6% and 42.0%, respectively). This was further supported by RNA-seq and GSEA using the GO gene set “B_CELL_DIFFERENTIATION” (Figure 4A). Agreeable with the functional characterization above, the genes that were frequently associated with B cell development were enriched in empty vector and HD1-HD2 linker mutants, but not in WT DUX4/IGH. Altogether, these results had led to the hypothesis that the HD1-HD2 linker, although not essential for DNA binding (Figure 2C and Supplementary Figure S3), was critical for DUX4/IGH dimerization, DRE crosslinking, and subsequent alternative splicing.
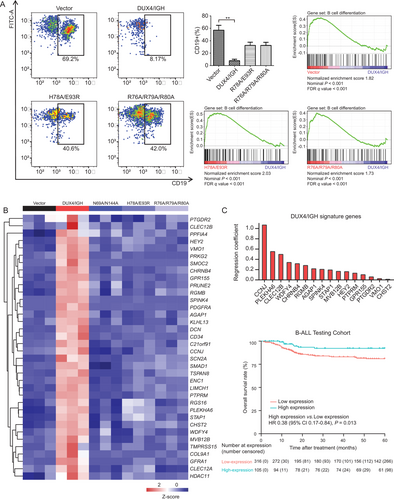
Furthermore, in light of the recent breakthrough in structural biology presented here and elsewhere [2, 4], we wanted to design a structure-based RNA-seq mining/cross-validation to pin-down the DUX4/IGH target gene with great precision. In brief, the Reh cells expressing WT DUX4/IGH or mutants (which target HD-DRE recognition and HD1-HD2-driven crosslinking, respectively) were subject to RNA-seq. In each cluster, we used the intersection/overlap between the upregulation (DUX4/IGH vs. vector) and downregulation (mutant vs. DUX4/IGH) to predict DUX4/IGH target genes that might underpin B-ALL leukemogenesis. A total of 36 DUX4/IGH target genes were identified, 26 of which were not reported before (Figure 4B).
To further verify the structure-based RNA-seq mining results, the newly observed DUX4/IGH target genes were checked against the published RNA-seq datasets derived from B-ALL patients [30] (Supplementary Figure S4B), followed by RT-PCR confirmation (Supplementary Figure S4C-F). Compared to other B-ALL subtypes, 36 DUX4/IGH target genes, which were uncovered by the RNA-seq mining using WT/mutant Reh cells, were abnormally transactivated (Supplementary Figure S5). In line with other investigations [30, 50, 51], AGAP1, cyclin J (CCNJ), PTPRM, CLEC12A, CLEC12B, prostaglandin D2 receptor 2 (PTGDR2), G protein-coupled receptor 155 (GPR155), sodium voltage-gated channel alpha subunit 2 (SCN2A), ectodermal-neural cortex 1 (ENC1), pleckstrin homology domain containing A6 (PLEKHA6), and carbohydrate sulfotransferase 2 (CHST2) were also highlighted in this report. In addition, COL9A1, signal transducing adaptor family member 1 (STAP1), PTPRF interacting protein alpha 4 (PPFIA4), hes related family bHLH transcription factor with YRPW motif 2 (HEY2), vitelline membrane outer layer 1 homolog (VMO1), protein kinase cGMP-dependent 2 (PRKG2), SPARC related modular calcium binding 2 (SMOC2), cholinergic receptor nicotinic beta 4 subunit (CHRNB4), prune homolog 2 with BCH domain (PRUNE2), repulsive guidance molecule BMP co-receptor b (RGMB), serine peptidase inhibitor Kazal type 4 (SPINK4), platelet derived growth factor receptor alpha (PDGFRA), kelch like family member 13 (KLHL13), decorin (DCN), cluster of differentiation 34 (CD34), chromosome 21 open reading frame 91 (C21orf91), SMAD family member 1 (SMAD1), tetraspanin 8 (TSPAN8), LIM and calponin homology domains 1 (LIMCH1), regulator of G protein signaling 16 (RGS16), WDFY family member 4 (WDFY4), multivesicular body subunit 12B (MVB12B), transmembrane serine protease 15 (TMPRSS15), GDNF family receptor alpha 1 (GFRA1), and histone deacetylase 11 (HDAC11) were new DUX4/IGH target genes uncovered by structure-based RNA-seq mining in this report. More importantly, this had paved a way for interesting data mining to check whether these DUX4/IGH target genes might be used as prognosis markers to predict the OS rates of B-ALL patients [30]. Most of these patients were treated with the modified VDLCP chemotherapy using vincristine, daunorubicin, PEG-asparaginase, cyclophosphamide, and prednisone [3, 12]. To extract the signature target genes of DUX4/IGH, we applied the LASSO regression analysis to a published RNA-Seq dataset of 1,223 B-cell precursor acute lymphoblastic leukemia (BCP-ALL) cases [3, 5, 12, 30] (Supplementary Figure S6A). The BCP-ALL cases were thus divided into two cohorts: 421 with survival data were used as the testing set, whilst the rest 802 as the training set. The 36 DUX4/IGH target genes were subjected to LASSO analysis, and 16 signature genes were identified, which can be calculated for each patient as the weighted sum of DUX4/IGH-driven deregulation (Figure 4C). We then evaluated the association of the signature gene score with survival data in the testing cohort and observed that high signature gene score was frequently associated with high OS rate (P = 0.013, Figure 4C). At individual gene level, we observed that the low expression levels of CCNJ and PTPRM were often associated with the relatively low 5-year OS rate in all B-ALL patients. In marked contrast, the high expression levels of these marker genes resulted in a high therapeutic response rate (>85%, Supplementary Figure S6B). CCNJ and PTPRM were shown to be important in leukemogenic development [50, 52, 53]. Moreover, the low expression of PTPRM was associated with poor prognosis in breast cancer [52, 53]. These observations were consistent with the DUX4/IGH signature genes reported here. The DUX4/IGH-driven transactivation of these signature genes might increase the sensitivity to the VDLCP chemotherapy in B-ALL patients and improve prognosis [5, 54, 55]. However, based on current data, it was not yet clear how these genes might interplay with each other, and what molecular mechanisms/networks were responsible for the leukemia treatment. This would remain an interesting direction for our future investigation.
3.4 RAG1/2 recruitment by DUX4/IGH
Endonuclease RAG1 and its co-factor RAG2 were frequently associated with genomic instability in lymphomagenesis [56]. In ALL, the RAG1/2-mediated recombination was considered as the predominant driver of oncogenic rearrangement [57-64]. In DUX4/IGH subtype leukemia, it had been speculated that RAG1/2 might play roles in ERG alternative splicing [6]. In line with this hypothesis, a DUX4-like transcription factor PAX5, which shared significant overlapping in deregulation and DNA-binding mode (Supplementary Figure S7A-C), was shown to make direct interaction with RAG1/2 and trigger V(D)J rearrangement in B lineage cells [65]. Supportively, the DUX41-150-DNA structure presented here indeed allowed the envision of RAG1/2 recruitment upon DRE-DRE crosslinking. The highly positively charged pocket derived from DUX4 dimer might serve as a putative binding site for RAG1/2 (Supplementary Figure S7D). This was further supported by the duolink PLA technology [42, 43]. In brief, Reh cells were stained with immunofluorescence-compatible primary antibodies to the target proteins (i.e., DUX4/IGH and RAG1/2, respectively). The two primary antibodies used in the present study were generated from different species (rabbit for DUX4/IGH and mouse for RAG1/2). Cells were then stained with secondary antibodies known as the PLA probes. The PLA probes that bound to the constant regions of the primary antibodies contain a unique DNA strand. If the proteins of interest interacted with each other, the DNA probes hybridized to make circular DNA, which could be amplified and visualized by fluorescently-labelled complementary oligonucleotide probes. The number and intensity of the dots, which were visualized by fluorescence microscopy, were used to monitor the direct interaction between RAG1/2 and WT/mutant DUX4/IGHs. Consistent with the structural observation, the N-terminal HD1-HD2, but not the C-terminal moiety, was responsible for the DUX4/IGH-RAGA1/2 engagement (Figure 5A).
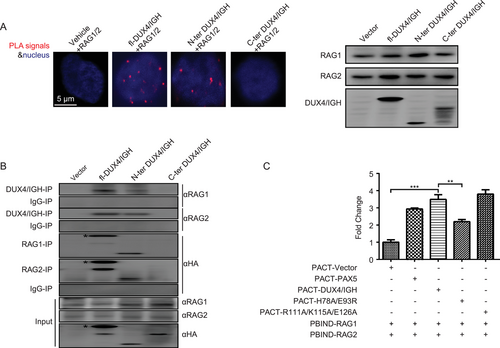
To cross-validate the direct interaction between RAG1/2 and DUX4/IGH, we had performed co-IP in Reh cells. When the WT HA-DUX4/IGH and mutants were pulled-down using antibody against HA tag, a co-precipitation of endogenous RAG1/2 with WT DUX4/IGH and DUX4/IGH1-150, but not DUX4/IGH151-431, were observed (Figure 5B). Vice versa, when RAG1/2 was used as bait, similar results were obtained (Figure 5B). To understand whether DUX4 dimerization and HD1-HD2 linker dynamics were important for RAG1/2 recruitment, a more sensitive technique, known as mammalian two hybrid assay, was used. In this experiment, the PAX5 protein, which was known to interact with RAG1/2 [65], was used as a positive control. The empty vector was used as a negative control. The R111A/K115A/E126A (i.e., the randomly chosen residues/positions that are far away from the DNA-binding site and dimeric interface) was used as an extra level of control to monitor the perturbation upon the overall fold/structure of DUX4/IGH. In good agreement with the PLA and co-IP experiments described above, WT DUX4/IGH, like PAX5, displayed interaction activity against RAG1/2. In marked contrast, H78A/E93R that targeted the DUX4/IGH dimerization impaired RAG1/2 recruitment (Figure 5C and Supplementary Figure S7E and S7F), echoing the importance of DUX4/IGH-driven crosslinking in RAG1/2 recruitment.
3.5 RAG1/2 cleavage in DUX4/IGH-mediated splicing
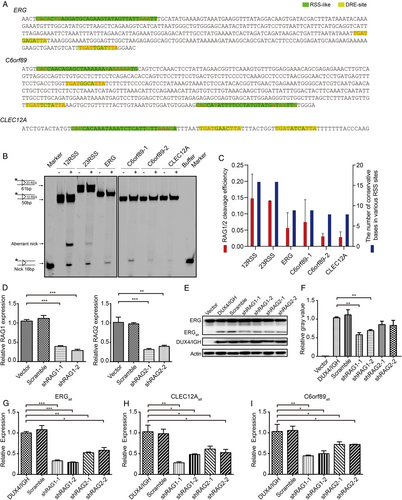
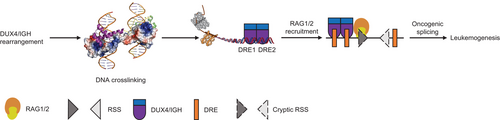
It was well established that RAG1/2 could recognize RSS sequences and trigger V(D)J recombination that led to alternative splicing variants [59, 66]. In this study, the RSS-like sequences were observed in ERG, CLEC12A, and C6orf89 (Figure 6A and Supplementary Figure S7G). To check whether RAG1/2 might recognize these putative RSSs, we had performed in vitro RAG1/2 cleavage assay. As expected, the 16-nucleotide nicked product (a signature of RAG1/2 reaction) was observed in the classical RSS substrates. Interestingly, the RSS-like sites derived from DUX4/IGH-driven splicing variants were also cleaved by RAG1/2 (Figure 6B). Consistently, the reduced cleave efficiency was in good agreement with their number of conservative bases in the RSS and RSS-like sequences (Figure 6C). Furthermore, the RAG1/2 involvement in DUX4/IGH-driven splicing was further supported by shRNA knock-down assay. The shRNA knock-down experiments were carried out with two random chosen shRNA sequences for RAG1 and RAG2, respectively (Figure 6D). As shown in Figure 6E-G, the RAG1/2 knock-down significantly disrupted ERGalt biogenesis both in mRNA and protein levels. Consistently, we obtained similar results in CLEC12Aalt and C6orf89alt. Compared with the control, the expression levels of CLEC12Aalt and C6orf89alt were significantly reduced in RAG1/2 knock-down cells (Figures 6H and 6I). To further characterize the RSS-like site, CUT & Tag analysis with RAG1/2 antibody and DUX4/IGH antibody were used. In line with the previous report [67], RAG1/2 tended to bind to the promoter region (62.7%) in the absence of DUX4/IGH. However, when DUX4/IGH was introduced, the RAG1-binding preference was markedly shifted from the promoter region to the intron region (43.0%) (Supplementary Figure S8A). More importantly, RAG1/2 started to bind to the splicing genes, including ERG, CLEC12A, and C6orf89, in the presence of DUX4/IGH. Altogether, these results reiterated the importance of RAG1/2 engagement in DUX4/IGH-mediated oncogenic splicing (Figure 7 and Supplementary Figure S8).
4 DISCUSSION
The current results highlighted a previously unrecognized crosslinking activity in DUX4/IGH double homeobox. As recently discussed and observed here [2, 5], DUX4 HDs were unique in two-fold: 1) it could bind multiple DNA signatures such as TGAT, TAA, and chimeric TGAT/TAA repeats; 2) unlike other HD family proteins, double tandem arrangement was exclusively observed in DUX4 and DUX4/IGH. The competent DNA engagement might be a critical step underpinning the DUX4-driven diseases [5, 6, 68, 69]. In leukemia, it was well known that DUX4/IGH could trigger alternative splicing, leading the abnormal expression of ERGalt that was critical for leukemogenesis [6]. Consistently, the DUX4/IGH-driven alternative splicing was highlighted in this report. In addition to ERGalt, we reported two new splicing isoforms in B-ALL patients: CLEC12Aalt and C6orf89atl. Interestingly, all these DUX4/IGH-driven variants harbored repetitive DRE arrangements closed to their splicing site. This had prompted the re-think of DUX4/IGH-driven transactivation and alternative splicing. In the published DUX4-DNA complex [28, 48], the double homeobox adopted a close configuration, clamping on a consensus DRE site. Here, the crystal structure of DUX41-150-DNAERG revealed a completely different DRE engagement. Two HD1-HD2 molecules adopted an open configuration, forming a trans dimer. The HD1A/HD2B and HD1B/HD2A (in which A and B stand for different DUX4 monomers) could bind to two sets of DREs. As characterized by using SAXS, analytical ultracentrifugation, and ERGalt biogenesis assays, the HD1-HD2 linker was critical for DUX4/IGH dynamics, dimerization, and function. Supportively, four proline residues were observed in the HD1-HD2 linker (i.e., residues 76-98). The poly-proline content might increase the intrinsic protein flexibility, enabling versatile intra- and inter-molecular dynamics (Supplementary Figure S9) that were required for DNA clamping and crosslinking for transactivation and alternative splicing, respectively.
Cancer-specific alternative splicing was widely observed and increasingly recognized as the driving factor in carcinogenesis [70-72]. Based on a pan-cancer analysis (from 8,705 patients), it was clear that tumors tend to have up to 30% more alternative splicing events than normal samples [73]. Furthermore, it had been demonstrated that the RAG1/2 complex was not only a critical factor for V(D)J recombination but also a great threat to genomic stability [67]. In several cases of ALL, RAG1/2 was thought to interplay with the major oncogenic fusion/driver, leading to full-fledged leukemogenesis [57-64]. As a homolog to DUX4 protein, it had been shown that PAX5 might recruit RAG1/2 for subsequent V(D)J recombination [65]. Here, we demonstrated a previously unrecognized DUX4/IGH-RAG1/2 axis in oncogenic splicing. As demonstrated by the duolink PLA, immunoprecipitation, and mammalian two hybrid assays, DUX4/IGH HD1-HD2, but not its C-terminal domain, was required for RAG1/2 recruitment. Furthermore, when the dimerization and crosslinking activities were perturbed, the DUX4/IGH-RAG1/2 engagement was also impaired. Consistently, when RAG1/2 was knock-down by shRNA, ERGalt, CLEC12Aalt, and C6orf89alt biogenesis was significantly disrupted. Furthermore, several RSS-like sequences could be observed near the DRE-DRE sites of ERGalt, CLEC12Aalt, and C6orf89alt. More importantly, the successful cleavage of these RSS-like sequences indeed supported the direct involvement of RAG1/2 in DUX4/IGH-mediated DNA crosslinking and subsequent alternative splicing (Figure 7). However, based on current data, it was not yet clear how RAG1/2 cleavage in the genomic sequences might trigger V(D)J-like recombination. Instead, the mis-targeting of RAG1/2 by DUX4/IGH recruitment might induce genomic instability by creating illegitimate DNA nicks. In turn, this lesion might embark the aberrant assembling of transcription initiation complex and spliceosome, which might lead to the abnormal biogenesis of ERGalt, C6orf89alt, and CLEC12Aalt (Figure 7).
Finally, the prognosis prediction results presented here not only help to cross-validate the DUX4/IGH target genes but also highlighted a novel DUX4/IGH signature gene scoring system that might be used to predict the overall B-ALL treatment outcome. While applying this score system to a cohort of 421 BCP-ALL patients, those with high gene score (n = 105) displayed a high 5-year OS rate. Of note, 48 patients who did not harbor DUX4/IGH fusion also belonged to this high score subgroup, suggestive of common disease mechanism shared among B-ALL subtypes. This finding had led to the proposal that the newly identified scoring system might be of prognosis value in patients with BCP-ALL.
5 CONCLUSION
Here, we report a previously unrecognized molecular mechanism, in which DRE-DRE crosslinking by DUX4/IGH was a critical step in DUX4/IGH-driven alternative splicing. In addition, we demonstrated that RAG1/2 recruitment was also important for the production of various transcript variants, including the secondary leukemogenic hit ERGalt.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The B-ALL patients were enrolled under the Shanghai Institute Hematology protocol (Chinese Clinical Trial Registry, number ChiCTR-RNC-14004969) and Shanghai Children's Medical Center protocol (Chinese Clinical Trial Registry, number ChiCTR-ONC-14005003). Mouse studies were conducted under a protocol approved by the Animal Care and Use Committee of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, P. R. China.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data and materials of the study are available from the corresponding author upon reasonable request.
COMPETING INTERESTS
The authors have declared that no conflict of interest exists.
ACKNOWLEDGEMENT
We thank Dr. Na Li and other personnel in beamlines BL17U/18U1/19U1 (Shanghai Synchrotron Radiation Facility/National Facility for Protein Science, Shanghai, China) for help during data collection. This work was supported by research grants 81970132, 81770142, 81800144, and 31800642 from National Natural Science Foundation of China, a research grant 20JC1410600 from Shanghai Science and Technology Committee, Shanghai Guangci Translational Medical Research Development Foundation, a research grant 20152504 from “Shanghai Municipal Education Commission—Gaofeng Clinical Medicine Grant Support”, “The Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institute of Higher Learning”, Samuel Waxman Cancer Research Foundation.
AUTHORS’ CONTRIBUTIONS
Conceived and designed the experiments: GM. Performed the experiments: HZ, NC, ZL, LB, CF, YL, WZ, XD, and MJ. Analyzed the data: HZ, NC, ZL, LB, CF, YL, WZ, XD, MJ, YL, SZ, JM, JZ, YZ, SJC, YZ, XQW, WH, ZC, JH, GM. Preparation of figures manuscripts: HZ, NC, ZL, LB, WH, ZC, JH, GM. Project supervision and wrote the paper: GM. All authors read and approved the final manuscript.




