Intra-heterogeneity in transcription and chemoresistant property of leukemia-initiating cells in murine Setd2−/− acute myeloid leukemia
Abstract
Background
Heterogeneity of leukemia-initiating cells (LICs) is a major obstacle in acute myeloid leukemia (AML) therapy. Accumulated evidence indicates that the coexistence of multiple types of LICs with different pathogenicity in the same individual is a common feature in AML. However, the functional heterogeneity including the drug response of coexistent LICs remains unclear. Therefore, this study aimed to clarify the intra-heterogeneity in LICs that can help predict leukemia behavior and develop more effective treatments.
Methods
Spleen cells from the primary Setd2−/−-AML mouse were transplanted into C57BL/6 recipient mice to generate a transplantable model. Flow cytometry was used to analyze the immunophenotype of the leukemic mice. Whole-genome sequencing was conducted to detect secondary hits responsible for leukemia transformation. A serial transplantation assay was used to determine the self-renewal potential of Setd2−/−-AML cells. A limiting-dilution assay was performed to identify the LIC frequency in different subsets of leukemia cells. Bulk and single-cell RNA sequencing were performed to analyze the transcriptional heterogeneity of LICs. Small molecular inhibitor screening and in vivo drug treatment were employed to clarify the difference in drug response between the different subsets of LICs.
Results
In this study, we observed an aged Setd2−/− mouse developing AML with co-mutation of NrasG12S and BrafK520E. Further investigation identified two types of LICs residing in the c-Kit+B220+Mac-1− and c-Kit+B220+Mac-1+ subsets, respectively. In vivo transplantation assay disclosed the heterogeneity in differentiation between the coexistent LICs. Besides, an intrinsic doxorubicin-resistant transcriptional signature was uncovered in c-Kit+B220+Mac-1+ cells. Indeed, doxorubicin plus cytarabine (DA), the standard chemotherapeutic regimen used in AML treatment, could specifically kill c-Kit+B220+Mac-1− cells, but it hardly affected c-Kit+B220+Mac-1+ cells. Transcriptome analysis unveiled a higher activation of RAS downstream signaling pathways in c-Kit+B220+Mac-1+ cells than in c-Kit+B220+Mac-1− cells. Combined treatment with DA and RAS pathway inhibitors killed both c-Kit+B220+Mac-1− and c-Kit+B220+Mac-1+ cells and attenuated disease progression.
Conclusions
This study identified two cell subsets enriched for LICs in murine Setd2−/−-AML and disclosed the transcriptional and functional heterogeneity of LICs, revealing that the coexistence of different types of LICs in this model brings about diverse drug response.
Abbreviations
-
- 7-AAD
-
- 7-amino-actinomycin D
-
- AKT
-
- thymoma viral proto-oncogene 1
-
- AML
-
- acute myeloid leukemia
-
- ANOVA
-
- analysis of variance
-
- Ara-C
-
- Cytarabine
-
- AT
-
- AZ628&Torin2
-
- Aurka
-
- aurora kinase A
-
- Birc5
-
- baculoviral IAP repeat-containing 5
-
- Blvrb
-
- biliverdin reductase B
-
- BM
-
- bone marrow
-
- Braf
-
- braf transforming gene
-
- C1qa
-
- complement component 1, q subcomponent, alpha polypeptide
-
- C1qb
-
- complement component 1, q subcomponent, beta polypeptide
-
- Car1
-
- carbonic anhydrase 1
-
- Car2
-
- carbonic anhydrase 2
-
- Cd55
-
- decay accelerating factor for complement
-
- Cd74
-
- cluster of differentiation 74
-
- Cdc
-
- cell division cycle 20
-
- Cdk
-
- cyclin dependent kinase
-
- Cepba
-
- CCAAT/enhancer binding protein (C/EBP), alpha
-
- CFU
-
- colony-forming unit
-
- CNVs
-
- copy number variations
-
- CRAF
-
- raf-1 proto-oncogene
-
- DA
-
- doxorubicin and cytarabine
-
- DMSO
-
- Dimethyl sulfoxide
-
- EFS
-
- event-free survival
-
- EGFP
-
- enhanced green fluorescent protein
-
- Elane
-
- elastase, neutrophil expressed
-
- ELDA
-
- Extreme Limiting Dilution Analysis
-
- FBS
-
- fatal bovine serum
-
- Fcnb
-
- ficolin B
-
- FDR
-
- false discovery rate
-
- Flt-3L
-
- fms-related tyrosine kinase 3 ligand
-
- FPKM
-
- fragments per kilobase of exon model per million mapped reads
-
- Gata1
-
- GATA binding protein 1
-
- Gata2
-
- GATA binding protein 2
-
- Gfi1
-
- growth factor independent 1 transcription repressor
-
- GO
-
- gene ontology
-
- GSEA
-
- gene set enrichment analysis
-
- H2-Aa
-
- histocompatibility 2, class II antigen A, alpha
-
- H3K36
-
- histone H3 lysine 36
-
- Hba-a1
-
- hemoglobin alpha, adult chain 1
-
- Hba-a2
-
- hemoglobin alpha, adult chain 2
-
- HGB
-
- hemoglobin
-
- HPCs
-
- hematopoietic progenitor cells
-
- HSC
-
- hematopoietic stem cell
-
- HSCs
-
- hematopoietic stem cells
-
- Id2
-
- inhibitor of DNA binding 2
-
- Ifit1
-
- interferon-induced protein with tetratricopeptide repeats 1
-
- Ikzf2
-
- IKAROS family zinc finger 2
-
- Isg15
-
- interferon-stimulated gene 15
-
- KEGG
-
- Kyoto Encyclopedia of Genes and Genomes
-
- Klf1
-
- Kruppel-like factor 1
-
- KO
-
- knockout
-
- LDA
-
- limiting dilution assay
-
- LICs
-
- leukemia-initiating cells
-
- Lmo4
-
- Lin-1, Isl1, Mec3 (LIM) domain only 4
-
- Lpl
-
- lipoprotein lipase
-
- LSCs
-
- leukemia stem cells
-
- MACS
-
- magnetic-activated cell sorting
-
- Mcm
-
- minichromosome maintenance complex component
-
- MDS
-
- myelodysplastic syndrome
-
- Mecom
-
- MDS1 and EVI1 complex locus
-
- Meis1
-
- meis homeobox 1
-
- min.pct
-
- minimum percentage
-
- mTOR
-
- mechanistic target of rapamycin kinase
-
- Nras
-
- neuroblastoma ras oncogene
-
- NS
-
- normal saline
-
- OS
-
- overall survival
-
- PAGE
-
- polyacrylamide gel electrophoresis
-
- PB
-
- peripheral blood
-
- PBS
-
- phosphate buffer saline
-
- PCA
-
- principal component analysis
-
- Pf4
-
- platelet factor 4
-
- PI3K
-
- phosphatidylinositol 3-kinase
-
- pI-pC
-
- polyinosinic–polycytidylic acid
-
- PLT
-
- platelet
-
- preGM
-
- pre-granulocyte-macrophage progenitor
-
- Pu.1
-
- spleen focus forming virus proviral integration oncogene
-
- PVDF
-
- Polyvinylidene Difluoride
-
- qRT-PCR
-
- quantitative real-time polymerase chain reaction
-
- RAS
-
- resistance to audiogenic seizures
-
- RBC
-
- red blood cell
-
- RNA-seq
-
- RNA-sequencing
-
- Rrm
-
- ribonucleotide reductase catalytic subunit M
-
- rRNA
-
- Ribosomal RNA
-
- Runx1
-
- runt related transcription factor 1
-
- S100a9
-
- S100 calcium binding protein A9
-
- SCF
-
- stem cell factor
-
- scRNA-seq
-
- single-cell RNA sequencing
-
- SDS
-
- Sodium dodecyl sulfate
-
- SETD2
-
- SET domain containing 2
-
- SNVs
-
- single nucleotide variants
-
- Tk1
-
- thymidine kinase 1
-
- tSNE
-
- t-distributed stochastic neighbor embedding
-
- UMI
-
- Unique molecular identifiers
-
- WB
-
- Western blotting
-
- WBC
-
- white blood cell
-
- WGS
-
- whole-genome sequencing
-
- WT
-
- wild type
1 BACKGROUND
Acute myeloid leukemia (AML) is a class of hematologic malignancies characterized by a distinct differentiation block and an accumulation of abnormal blasts in the bone marrow (BM) [1]. Based on cell surface markers, a study in 1994 showed that only the CD34+CD38− fraction from patients could initiate AML in immune-deficient xenograft mice [2], indicating that the differentiation path in AML, similar to normal hematopoiesis, also follows a hierarchical model [3]. Nowadays, the hierarchical leukemia stem cell/leukemia-initiating cell (LSC/LIC) model has been widely accepted in malignant hematopoiesis, in which LICs reside at the apex and give rise to their progenies with no oncogenicity [4, 5]. However, follow-up studies have shown that LICs exist in not only CD34+CD38− but also CD34− or CD38+ subsets [6, 7]. So far, there are no uniform cell surface antigens that can be used to define LICs across different patients. Nevertheless, several reports revealed that phenotypic LICs with inter-subject heterogeneity shared a common transcription signature related to hematopoietic stem cell (HSC) self-renewal and had a similar cytokine signaling pathway at the protein level [8-10]. In fact, heterogeneity is a pervasive characteristic of tumors [11, 12], which has been studied since 1978 [13]. As whole-genome sequencing (WGS) flourished in the recent two decades, tumor cells could be detected at the genomic level, leading to the disclosure of phenotypic and genetic heterogeneity within the tumor [2, 14]. Unlike normal stem cells that often reside in one population with defined cell surface markers, tumor stem cells exhibit significant diversity of surface antigens among individuals [15]. Moreover, multitype tumor stem cells were found to coexist in plenty of patients [16, 17]. For instance, LICs with different phenotypes and transcription features could coexist in the same patient [16, 18], and co-residence of multi-LIC clones also occurs in the murine AML models, though these clones differ markedly in their ability to initiate AML [19-21]. Undoubtedly, the inter-patient and intra-patient heterogeneities of tumor stem cells further complicate our understanding of cancer biology. Given that the core unit of cancer is genetically and epigenetically different, single-cell sequencing provides an extremely high resolution to unveil all biological aspects of cancers, including heterogeneity, clone evolution, tumor microenvironment, metastasis, and therapeutic resistance [22, 23]. Indeed, abundant data of single-cell RNA sequencing (scRNA-seq) have revealed tumor intra-heterogeneity and reinterpreted the composition of the tumor microenvironment [18, 24-26]. However, the pathophysiological value of the coexistent LICs in the same patient has not been completely clarified.
As is widely acknowledged, the treatment of acute leukemia mainly relies on traditional chemotherapy. Although most AML patients initially respond to standard doxorubicin and cytarabine (DA) chemotherapy, the 5-year overall survival (OS) and event-free survival (EFS) are still extremely poor [27, 28]. Given that LICs prefer to keep themselves at a quiescence state [29], and high LIC frequency at diagnosis is negatively correlated with AML prognosis [30, 31], LICs have been regarded as the inherent chemoresistant cell population [5, 32]. However, two recent studies had cast doubt on LIC-resistant theory and showed that chemotherapy displayed similar killing effects on LICs and non-LICs [33, 34]. These studies unmasked the inter-patient variability of LICs in chemotherapeutic response, which may also be true for LICs coexisting in the same individual.
The SET domain containing 2 (SETD2), a histone H3 lysine 36 (H3K36)-specific trimethyltransferase, has been shown to play critical roles in many aspects of genome regulation such as DNA repair [35], DNA methylation [36], m6A RNA modification [37], gene transcription [38], maternal epigenome [39], and genome stability [40]. SETD2 mutations are frequently detected in solid tumors [14, 41] and leukemias [42-45], and contribute to chemotherapeutic resistance [46, 47]. In our previous work, we discovered that Setd2 deficiency gave rise to a characteristic of myelodysplastic syndrome (MDS) in mice [48]. Here, by using genomic and bulk/single-cell transcriptome analysis, we investigated the secondary hits and the intra-heterogeneity of LICs in a spontaneous murine Setd2−/−-AML model to verify whether transcriptional heterogeneity could further translate into functional heterogeneity.
2 METHODS
2.1 Mice
Mice experiments were performed according to the animal care standards (DLA-MP-IACUC.06) from the Institutional Animal Care and Use Committee (IACUC) of Shanghai Jiao-Tong University School of Medicine (Shanghai, China). The Setd2 conditional knockout (KO) model was established as described previously [48]. Briefly, we inserted a LoxP-Lox2272 flanked, reversed IRES-enhanced green fluorescent protein (EGFP)-polyA cassette into the first intron of Setd2. When expressing a Cre enzyme, this cassette can be inverted, leading to the termination of Setd2 transcription and the initiation of EGFP expression driven by Setd2 promoter. Four weeks after birth, both wild-type (WT) and KO mice were intraperitoneally injected with polyinosinic-polycytidylic acid (pI-pC, tlrl-pic-5, 7 mg/kg, InvivoGen, San Diego, CA, USA) every other day for 3 times to induce the KO of Setd2. C57BL/6 mice were purchased from Charles River Laboratories China (Beijing, China). All mice used in in vivo experiments were 6 to 8 weeks old, and age- and sex-matched. Mice were bred and housed in specific pathogen-free (SPF) animal facilities.
2.2 Histological analysis
Mice were euthanized using CO2 inhalation and dissected to obtain the BM, spleen, and liver. Spleen and liver tissues were fixed with 4% paraformaldehyde, cut into 3 mm portions, then subjected to paraffin embedding. Serial sections (4 μm) were cut, deparaffinized, and stained with hematoxylin and eosin. Cell suspensions from the BM, spleen, and peripheral blood (PB) were made into cell smears, followed by Wright-Giemsa staining. The Olympus DP71 Observer microscope (Tokyo, Japan) was used to examine the histological specimens.
2.3 Flow cytometry analysis and cell sorting
Cells harvested from the PB, BM, and spleen were washed in phosphate buffer saline (PBS) with 1% fetal bovine serum (FBS) (Gibco, Waltham, MA, USA), then stained with antibodies against surface markers in 1% FBS-PBS for 30 min at 4°C, followed by flow cytometry analysis on BD LSRFortessa™ X-20 (BD Biosciences, Franklin Lakes, NJ, USA). For apoptosis analysis, each tube of cells was stained with Annexin V (556421, BD Biosciences) and 7-amino-actinomycin D (7-AAD) (559925, BD Biosciences) after surface marker labeling. Cells were then vortexed gently and incubated at 4°C for 20 min, followed by binding buffer adding and flow cytometry analysis. For cell cycle analysis, cells were fixed and permeabilized by BD Cytofix/Cytoperm buffer (554722, BD Biosciences) for at least 1 h at 4°C in dark after labeling with surface markers. Cells were then washed with Perm/Wash Buffer (554723, BD Biosciences) twice and stained with anti-Ki67 (652404, BioLegend, San Diego, CA, USA) in dark at 4°C. After 30 min, Hoechst 33342 (10 mg/mL, 62249, Life Technologies, Waltham, MA, USA) was added to each tube, followed by flow cytometry analysis. FlowJo software (version: 10.4, Tree Star, Ashland, OR, USA) was used for subsequent data analysis. As for cell sorting, spleen cells were collected under sterile conditions. The magnetic-activated cell sorting (MACS) assay was performed according to the manufacturer's instructions (76447, BioLegend) to enrich c-Kit+ cells. Next, cells were stained with antibodies against c-Kit, Mac-1, and B220 (BioLegend). c-Kit+, c-Kit+B220+Mac-1−, c-Kit+B220+Mac-1+, c-Kit+B220+Mac-1++, and c-Kit+B220−Mac-1++ cells were sorted on BD FACSAria™ III (BD Biosciences), respectively. To obtain high-purity cells, we collected sorted cells in tubes and gave a second round of sorting using the same protocol. A comprehensive list of antibodies for flow cytometry analysis and cell sorting is included in Supplementary Table S1.
2.4 Transplantation experiments
Transplantation assays were performed to determine the self-renewal potential of the leukemia cells in different subsets. About 5000 cells of c-Kit+B220+Mac-1−, c-Kit+B220+Mac-1+, c-Kit+B220+Mac-1++, and c-Kit+B220−Mac-1++ subsets of spleen cells, 2 × 104 c-Kit+ and c-Kit− spleen cells, as well as 2 × 105 total spleen cells were transplanted into sub-lethally (400 rad) irradiated C57BL/6 recipient mice through tail vein injection. Blood samples from C57BL/6 recipient mice were evaluated every two weeks to monitor the white blood cell (WBC) count and the percentage of Setd2−/− donor-derived leukemia (EGFP+) cells.
To determine the frequency of LICs, highly purified c-Kit+B220+Mac-1−, c-Kit+B220+Mac-1+, c-Kit+B220+Mac-1++, and c-Kit+B220−Mac-1++ leukemic spleen cells were injected intravenously to sub-lethally irradiated recipient mice. Cell dosages were 5, 50, 500, 5000, and 25,000 per mouse for the c-Kit+B220+Mac-1− group, 50, 500, 5000, and 25,000 per mouse for the c-Kit+B220+Mac-1+ group, and 5000 and 20,000 per mouse for the c-Kit+B220+Mac-1++ and c-Kit+B220−Mac-1++ groups. The survival rates of each group were recorded for at least 5 months. The frequency of LICs was calculated through the open-access program Extreme Limiting Dilution Analysis (ELDA) (http://bioinf.wehi.edu.au/software/elda/) [49].
2.5 Colony-forming unit (CFU) assay
c-Kit+B220+Mac-1−, c-Kit+B220+Mac-1+, c-Kit+B220+Mac-1++, and c-Kit+B220−Mac-1++ subsets were sorted from spleen cells of transplanted recipient mice using MACS and BD FACSAria™ III. About 1 mL of colony assay medium (MethoCult GF M3434, 03434, Stem Cell Technologies, Vancouver, British Columbia, Canada) containing 1000 sorted cells was planted in a 35-mm tissue culture dish. Each sample was plated in triplicate. The number of clones was counted (more than 30 cells were recognized to be a clone) with an inverted microscope after 7 days of incubation at 37°C in 5% CO2.
2.6 Quantitative real-time polymerase chain reaction (qRT-PCR)
Total BM cells were harvested from primary leukemia and WT mice and were subjected to RNA extraction with Trizol (Invitrogen, Waltham, MA, USA). c-Kit+B220+Mac-1−, c-Kit+B220+Mac-1+, c-Kit+B220+Mac-1++, and c-Kit+B220−Mac-1++ cells were sorted from the spleen of transplanted recipient mice and subjected to RNA extraction with RNeasy Plus Micro Kit (74034, Qiagen, Hilden, Germany).
For qRT-PCR, total RNA was isolated from sorted subsets and used for reverse transcription with PrimeScript™ RT reagent Kit with gDNA Eraser (RR047, Takara, Tokyo, Japan) according to the manufacturer's instructions. qRT-PCR was performed by using the TB Green® Premix Ex Taq™ (RR420, Takara), gene expression was calculated using the 2ΔΔCT method relative to housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (Gapdh). The expression variability of each gene between subsets was calculated relative to their expression level in c-Kit+B220+Mac-1− subset. The following primers were used in the qRT-PCR analyses: ribonucleotide reductase M2 (Rrm2): forward, 5’-TGGCTGACAAGGAGAACACG-3’, reverse, 5’-AGGCGCTTTACTTTCCAGCTC-3’; aurora kinase A (Aurka): forward, 5’-CTGGATGCTGCAAACGGATAG-3’, reverse, 5’-CGAAGGGAACAGTGGTCTTAACA-3’; baculoviral IAP repeat-containing 5 (Birc5): forward, 5’-GAGGCTGGCTTCATCCACTG-3’, reverse, 5’-CTTTTTGCTTGTTGTTGGTCTCC-3’; thymidine kinase 1 (Tk1): forward, 5’-AAGTGCCTGGTCATCAAGTATG-3’, reverse, 5’-GCTGCCACAATTACTGTCTTGC-3’; GATA-binding protein 1 (Gata1): forward, 5’-TGGGGACCTCAGAACCCTTG-3’, reverse, 5’-GGCTGCATTTGGGGAAGTG-3’; carbonic anhydrase 2 (Car2): forward, 5’-TGCGGCCTTTGCTAACTTC-3’, reverse, 5’-GCTGACAGTAATGGGCTCCC-3’; Kruppel-like factor 1 (Klf1): forward, 5’-AGACTGTCTTACCCTCCATCAG-3’, reverse, 5’-GGTCCTCCGATTTCAGACTCAC-3’; biliverdin reductase B (Blvrb): forward, 5’-GGCGGTGCAAGCAGGTTAT-3’, reverse, 5’-GTGCCCAGTAGCACGATGAC-3’, carbonic anhydrase 1 (Car1): forward, 5’-GACTGGGGATATGGAAGCGAA-3’, reverse, 5’-TGCAGGATTATAGGAGATGCTGA-3’; hemoglobin alpha, adult chain 1 (Hba-a1): forward, 5’-CCTCACTTTGATGTAAGCCACG-3’, reverse, 5’-GTGCTCACAGAGGCAAGGAAT-3’; elastase, neutrophil expressed (Elane): forward, 5’-AGCAGTCCATTGTGTGAACGG-3’, reverse, 5’-CACAGCCTCCTCGGATGAAG-3’; growth factor independent 1 transcription repressor (Gfi1): forward, 5’-CCTGGTCAAGAGCAAGAAGG-3’, reverse, 5’-CCTCGGTAAGCTGAGAGTCG-3’.
2.7 Bulk RNA-seq analysis
For bulk RNA-seq, ribosomal RNA (rRNA) was removed using the Ribo-Zero rRNA Removal Kit (MRZMB126, Illumina, San Diego, CA, USA). Subsequently, rRNA-depleted samples were fragmented, then subjected to first and second strand cDNA synthetization. cDNAs were ligated with sequencing adapters according to standard Novaseq 6000 platform (Illumina) protocols. For the analysis of RNA-seq data, initial quality control of raw sequencing reads was done with FastQC (version: 0.11.7, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) followed by pre-processing with Trimmomatic (version: 0.38, https://github.com/timflutre/trimmomatic). The high-quality sequencing reads were then mapped to the mouse reference genome (GRCm38) using Hisat2 (version: 2.1.0, http://daehwankimlab.github.io/hisat2/). After the procession of the alignment results by SAMtools (version: 1.7, https://github.com/samtools/), counts per gene were obtained by featureCounts (version: 1.6.3, http://subread.sourceforge.net/). The fragments per kilobase of exon model per million mapped reads (FPKM) values were calculated according to the counts and lengths of genes. Differential gene expression was calculated using the DEseq2 package (version: 3.4.3, https://bioconductor.org/packages/release/bioc/html/DESeq2.html) in R. The differentially expressed genes with the fold change (FC) ≤ 0.5 or FC ≥ 2 and P < 0.05 were selected. For gene set enrichment analysis (GSEA), normalized values of RNA-seq data (FPKM) were rank-ordered by fold change as input. The analysis was performed using GSEA (version: 3.0, https://www.gsea-msigdb.org/gsea/index.jsp) software and visualized with Cytoscape (version: 3.6.1, https://cytoscape.org/).
2.8 Gene ontology (GO) analysis
GO analysis was performed to elucidate the biological implications of unique genes in the significant or representative profiles of the differentially expressed genes in the experiment [50]. GO annotations were downloaded from NCBI (http://www.ncbi.nlm.nih.gov/), UniProt (http://www.uniprot.org/), and the Gene Ontology (http://www.geneontology.org/). Fisher's exact test was applied to identify the significant GO categories, and the false discovery rate (FDR) was used to correct the P value. The cases were selected when the calculated P value was less than 0.05. Pathway analysis was used to find out the significant pathway of the differentially expressed genes according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Fisher's exact test was used to select the significant pathway. The threshold of significance was defined by P value and FDR.
2.9 scRNA-seq
Spleen cells were harvested from the transplanted Setd2−/−-AML mice, then subjected to biotin-c-Kit antibody staining for 15 min on ice and washed with MojoSort™ Buffer (480017, BioLegend) separately. Cells were then labeled with Streptavidin Nanobeads (480016, BioLegend) on ice and washed again 15 min later. Cell pellets were resuspended in MojoSort™ Buffer, the tube was then placed in a magnet for 5 min, followed by collecting the magnetically labeled fraction twice. Subsequently, MACS isolated cells were stained with an antibody against c-Kit again and sorted by BD FACSAria™ III. After purification, the BD Rhapsody system (BD Biosciences) was used to capture the transcriptomic information of single cells. The single cells were randomly sedimented in microwells (>200,000) and captured by a limiting dilution approach. Beads with oligonucleotide barcodes were then added to reach a saturated state, ensuring that each cell can be paired with one bead. All beads were collected into a microcentrifuge tube. Cells were captured and used for scRNA-seq. Whole transcriptome libraries were constructed following the BD Rhapsody single-cell whole-transcriptome amplification workflow. Single-cell libraries were quantified using a High Sensitivity DNA chip (Agilent, Santa Clara, CA, USA) on a Bioanalyzer 2200 and the Qubit High Sensitivity DNA Assay Kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA). The library was sequenced by HiSeq Xten (Illumina) on a 150 bp paired-end run.
2.10 scRNA-seq statistical analysis
We applied fastp (version: 0.20.1, https://github.com/OpenGene/fastp) with the default parameter, filtering the adaptor sequence and removing the low-quality reads [51] to acquire the clean data. Unique molecular identifiers (UMI) were analyzed to identify the cell barcode whitelist [52]. The UMI-based clean data were mapped to the mouse genome (Ensemble, version: 92) utilizing STAR [53] with customized parameters from the UMI-tools standard pipeline to obtain the UMI count of the sample. Cells that expressed over 200 genes and had mitochondria UMI rate below 20% passed the cell quality filtering. Mitochondrial genes were removed from the expression table but used for cell expression regression to avoid the effect of the cell status for clustering analysis and marker analysis of each cluster. Seurat package (version: 2.3.4, https://satijalab.org/seurat/) was used for cell normalization and regression based on the expression table according to the UMI count of the sample and the percent of mitochondria rate to obtain the scaled data. Principal component analysis (PCA) was constructed based on the scaled data with all high variable genes, and the top 8 principles were used for t-distributed stochastic neighbor embedding (tSNE) construction. The unsupervised cell cluster result was obtained from the PCA top 8 principles by using the graph-based cluster method (resolution = 0.8). Marker genes were calculated by the function of FindAllMarkers with Wilcox rank-sum test algorithm under the following criteria: 1) logFC > 0.25; 2) P < 0.05; and 3) minimum percentage (min.pct) > 0.1.
2.11 Whole-genome sequencing (WGS) analysis
Total BM cells were harvested from the primary Setd2−/−-AML mouse. Cells were then subjected to genomic DNA extraction with Wizard® Genomic DNA Purification Kit (A1120, Promega, Madison, WI, USA), and the paired tail DNA of the primary Setd2−/−-AML mouse was used as the germline reference for WGS. DNAs were sheared by sonication to ∼300 bp, and adaptors were ligated to the fragments. Fragments were then amplified by ligation-mediated PCR, purified, and subjected to DNA sequencing on the Novaseq 6000 platform (Illumina). Sequence data were mapped to mm10 by BWA software (version 0.7.12, http://bio-bwa.sourceforge.net/bwa.shtml). Small variations, including single nucleotide variants (SNVs), were analyzed with the Mutect2 and filtered with a recommended threshold (quality ≥30 and at least 4 reads covered) for cases. Copy number variations (CNVs) were called with CNVnator_v0.3 and calculated with Fermikit (version 0.13, https://github.com/lh3/fermikit).
2.12 Western blotting (WB) analysis
Erythrocyte-depleted BM cells were lysed with 1 × sodium dodecyl sulfate (SDS) buffer (P0015, Beyotime, Shanghai, China) and boiled at 100°C for 5 min. The cell extractions were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and electrotransferred to polyvinylidene difluoride (PVDF) membranes (ISEQ00010, Sigma, Darmstadt, Germany), followed by overnight incubation with the primary antibodies at 4°C. Horseradish peroxidase-conjugated anti-rabbit antibody and anti-mouse antibody (401353, 401253, Millipore, Burlington, Massachusetts, USA) were used as the secondary antibodies, and Immobilon Western Chemiluminescent HRP Substrate (WBKLS, Sigma) was used for the detection. A comprehensive list of antibodies for WB analysis is included in Supplementary Table S1.
2.13 Cell culture and compound screening assay
A small-molecule inhibitor screening was performed on c-Kit+ spleen cells from transplanted Setd2−/−-AML mice. Cells were cultured in RPMI-1640 medium (Gibco) supplemented with 15% FBS (Gibco), 50 ng/mL stem cell factor (SCF, C775, Novoprotein, Beijing, China), 50 ng/mL Fms-related tyrosine kinase 3 ligand (Flt-3L, CC19, Novoprotein), and Plasmocin (InvivoGen, ant-mpp) at 37°C in 5% CO2 for 24 h. Cells were seeded at a density of 4000 cells per well in 384-well plates (6007688, PerkinElmer, Waltham, MA, USA) and transferred with 0.1 μL compounds (1 mmol/L) using the pin tool. Each experiment was performed in duplicate. The Bioactive Compound Library (L-2000) was purchased from Selleck (Shanghai, China) containing 2304 approved drugs. After 48 h, cell viability was determined by measuring relative ATP levels in EnVision 2105 Multimode Plate Reader (PerkinElmer) using the CellTiter-Glo® Luminescent Cell viability assay (G7570, Promega) according to the manufacture's protocol. A secondary experiment was performed for the verification of the compounds.
For doxorubicin (S1208, Selleck) treatment, sorted c-Kit+B220+Mac-1− and c-Kit+B220+Mac-1+ BM subsets were cultured in a 12-well plate containing 6 × 105 cells per well for 24 h, then planted into a 96-well plate with 4 × 104 cells per well and added with doxorubicin. The final concentrations of doxorubicin were 10, 30, 100, and 300 nmol/L, and each sample was performed in triplicate. Samples with no doxorubicin were used as the control. After 24 h treatment, the cell viability was measured with CellTiter–Glo® Luminescent Cell viability assay (G7570, Promega).
For the treatment of AZ628 [a raf transforming gene (Raf) inhibitor, S2746, Selleck] and Torin2 [a mechanistic target of rapamycin kinase (mTOR) inhibitor, S2817, Selleck], c-Kit+ spleen cells were cultured in 12-well plates containing 5 × 105 cells per well for 24 h, then added with AZ628 and Torin2, respectively. Each of the drugs was diluted to final concentrations of 100, 200, and 400 nmol/L, and each experiment was performed in triplicate. Dimethyl sulfoxide (DMSO) was used as a control. After 48 h treatment, the apoptosis was measured using 7-AAD and Annexin V as described above in the “Flow cytometry analysis and cell sorting” subsection.
2.14 Drug treatment in vivo
The spleen cells were harvested from transplanted Setd2−/−-AML mice. Total spleen cells (2 × 105) were transplanted into each sub-lethally (400 rad) irradiated C57BL/6 recipient mouse through tail vein injection. The total WBC counts and the percentage of donor-derived (EGFP+) cells in PB were measured 3 weeks after transplantation. Cytarabine (Ara-C, S1648, Selleck) and doxorubicin were dissolved in normal saline (NS) to a stock concentration of 50 mg/mL and 10 mg/mL, respectively. AZ628 and Torin2 were dissolved in DMSO to a stock concentration of 40 mg/mL. AZ628 and Torin2 in 40% polyethylene glycol 400 (PEG400, 91893, Sigma), 2% Tween80 (P8074, Sigma), and H2O were used for mouse treatment. For the Ara-C and doxorubicin (DA) combined group, the mixture of doxorubicin (3 mg/kg) and Ara-C (100 mg/kg) was injected intravenously for 3 consecutive days followed by Ara-C (100 mg/kg) alone for 2 days [46]. NS was used as a negative control. For the AZ628 and Torin2 (AT) combined group, AZ628 (10 mg/kg) and Torin2 (5 mg/kg) were injected intraperitoneally every other day for 5 days. For the DA and AT combined group, DA was injected as described in the DA alone group, and AT was administered as described in the AT alone group. DMSO was used as a negative control for all groups. The immunophenotypes of PB, BM, and spleen were analyzed before and after treatment. The survival rates of all treatment groups were recorded.
2.15 Statistical analysis
The Student's unpaired two-tailed t-test was used when there were only two groups involved. For multi-group analysis, one- or two-way analysis of variance (ANOVA) was performed. The significance of difference was considered if the P value was less than 0.05. All statistical analyses were performed with GraphPad Prism (version: 9.0.0, GraphPad Software, San Diego, CA, USA).
3 RESULTS
3.1 An aged Setd2−/− mouse developed AML
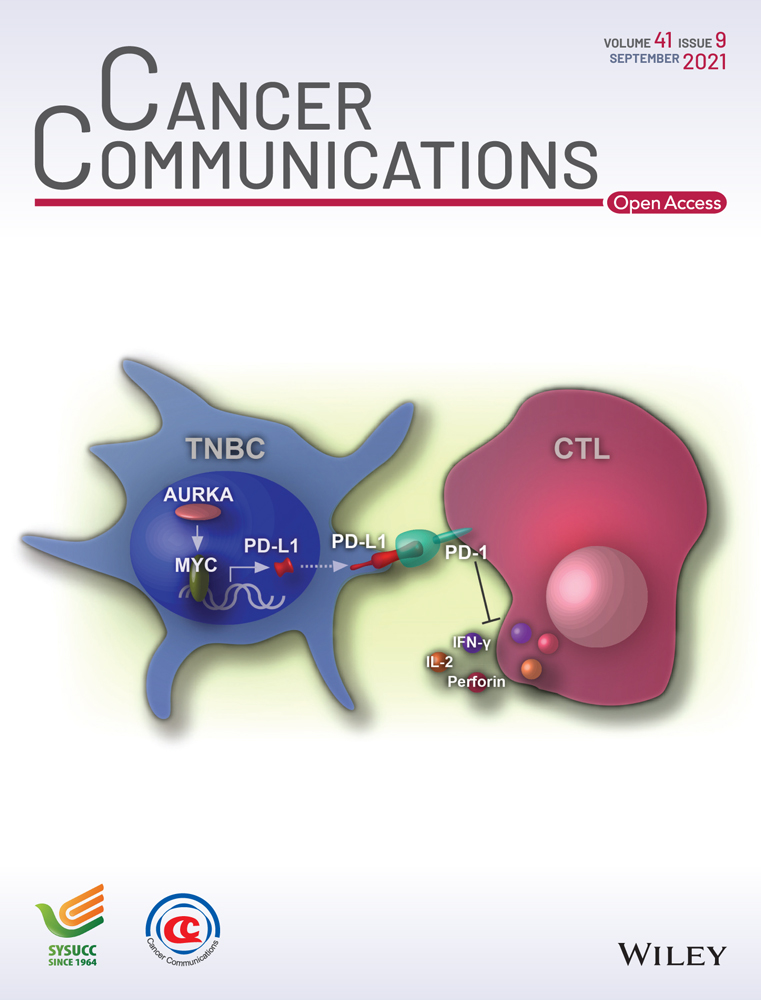
Previously, we reported that Setd2 deficiency effectively drove MDS-like transformation [48]. As of yet, it is unclear whether it also contributes to AML transformation. In this study, during a routine follow-up examination of our 122 Setd2−/− mice, except for 17 unexplained deaths, we observed a distinct phenotype with hyperleukocytosis (86.3 × 106 cells/mL) and markedly low red blood cell (RBC), hemoglobin (HGB), and platelet (PLT) counts in one mouse (Figure 1A). The BM (Figure 1B) and PB smears (Supplementary Figure S1A) showed a massive number of myeloid cells, and more than 20% of BM cells appeared to be immature. Pathological examination affirmed a destructed architecture and robust infiltration of myeloid cells in the enlarged spleen and liver from this abnormal Setd2−/− mouse (Figure 1C, Supplementary Figure S1B). Flow cytometry analysis demonstrated a dramatic increase of c-Kit+ cells in the BM (20.8%) and spleen (34.3%) as compared with the WT control (5.7% and 4.6%) (Figure 1D). All these results implied that a malignant transformation occurred in this mouse. Notably, besides B220+ and Mac-1+ cells, there were many B220+Mac-1+ cells accumulated in the BM and spleen (Figure 1D). Granulocytes (Gr-1+) and macrophages (F4/80+) were also considerably increased (Supplementary Figure S1C). However, relative to WT, HSCs and hematopoietic progenitor cells (HPCs) were remarkably decreased in leukemia BM and spleen (Supplementary Figure S1D).
Next, to gain insights into the transcriptional characteristics underlying leukemogenesis, we performed a bulk RNA-seq analysis of WT and leukemia BM cells. As a result, 1,920 upregulated and 4,078 downregulated genes were detected in the leukemia sample relative to WT control (Supplementary Table S2). MDS1 and EVI1 complex locus (Mecom), a common overexpressed or rearranged oncogene in AML [19, 54], was among the most significantly up-regulated genes (Supplementary Figure S1E), providing a reasonable explanation for our leukemia phenotype. GSEA revealed that the leukemia sample was enriched with gene sets related to HSCs and HPCs, whereas a hematopoietic mature cell-related gene set was markedly depleted, indicating that these leukemia cells might have a high self-renewal potential and were blocked in differentiation (Figure 1E). Hematopoietic fingerprint, a unique cell type-specific gene set analysis method often used to identify hematopoietic cell properties, indicated that the signature of HSC and pre-granulocyte-macrophage progenitor (preGM) was mostly enriched in the leukemia sample (Figure 1F). Based on the criteria of Bethesda proposals [55], we suggest that this mouse had developed AML.
3.2 NrasG12S and BrafK520E co-mutation occurred in the Setd2−/−-AML mouse
AML transformation knowingly follows a multi-step process. To detect the secondary hits responsible for this Setd2−/−-AML model, we performed WGS on total BM cells of the primary leukemia mouse, revealing that neuroblastoma ras oncogene (Nras) and braf transforming gene (Braf), two key cell-cycle determiners [56], displayed a high frequency of allele mutation (Figure 1G). The transcription of mutated NrasG12S and BrafK520E was confirmed by RNA-seq analysis (Supplementary Figure S1F). As reported, BrafK520E (equivalent to human BRAFK483E), a kinase-dead mutation that often co-exists with resistance to audiogenic seizures (RAS) mutations [57, 58], can form a stronger dimer with raf-1 proto-oncogene (CRAF) or mutated RAS, which can initiate tumorigenesis faster than RAS or BRAF mutation alone [58, 59] (Supplementary Figure S1G). To monitor the activity of Nras and Braf mutations, we analyzed the activation of downstream transducers in the total BM of Setd2−/−-AML transplants. Indeed, NrasG12S and BrafK520E dramatically activated the downstream mitogen-activated protein kinase (MAPK) pathway, including p-Mek/p-Erk/p-c-fos and p-Jnk/p-c-Jun (Figure 1H left and middle). Phosphatidylinositol 3-kinase (PI3K)/thymoma viral proto-oncogene 1 (AKT)/mTOR, another downstream signaling pathway of RAS that is known to be critical for ribosome/protein biogenesis and tumor proliferation [60], was also activated (Figure 1H right). Collectively, the occurrence of NrasG12S and BrafK520E mutations in this model provided a plausible explanation for malignant transformation.
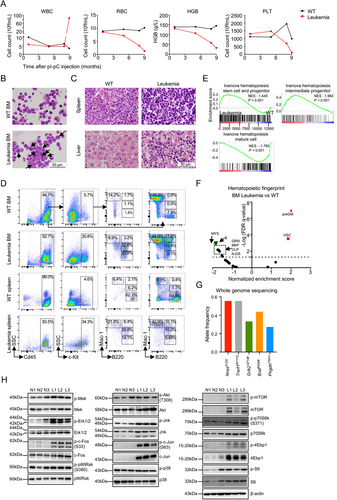
3.3 Coexistence of two types of LICs in Setd2−/−-AML
Like normal hematopoiesis, AML also displays a hierarchy with LICs residing at the apex and possessing an unlimited capacity of self-renewal [4]. To determine the self-renewal potential of leukemia cells in this Setd2−/−-AML model, we transplanted 2 × 105 total spleen cells from the primary Setd2−/−-AML mouse into sub-lethally irradiated C57BL/6 recipient mice. Immunophenotype analysis of PB, spleen, and BM cells in the transplanted mice did show a similar expression pattern of B220 and Mac-1 to the primary leukemia mouse (Figure 2A). Moreover, these properties could be kept for at least six generations (Supplementary Figure S2A-B). Also, BM cells from transplanted Setd2−/−-AML mice were injected intravenously into recipient mice, which generated the same phenotype (Supplementary Figure S2C). Previously, c-Kit+B220+Mac-1+, B220+Mac-1−, and c-Kit+B220+Gr-1− cells had been identified as LIC-enriched subsets of AML [19-21]. Thus, according to the expression levels of c-Kit, B220, and Mac-1, we sorted four subsets (c-Kit+B220+Mac-1−, c-Kit+B220+Mac-1+, c-Kit+B220+Mac-1++, and c-Kit+B220−Mac-1++) of spleen leukemia cells from the transplanted Setd2−/−-AML mice for further investigation. The morphology of c-Kit+B220+Mac-1− and c-Kit+B220+Mac-1+ subsets appeared to manifest the feature of primitive cells, whereas c-Kit+B220+Mac-1++ and c-Kit+B220−Mac-1++ subsets were rendered with a mature myeloid cell character (Figure 2B, Supplementary Figure S2D). The CFU assay exhibited a remarkable advantage of the colony-forming ability in c-Kit+B220+Mac-1− (251.3 ±7.4) and c-Kit+B220+Mac-1+ (86.7 ±3.7) subsets compared with that in the other two (Figure 2C). We then transplanted the spleen cells of these four subsets to recipient mice, respectively. The lifespan of transplanted recipient mice showed that c-Kit+B220+Mac-1− (5000 cells per mouse), c-Kit+B220+Mac-1+ (5000 cells), and c-Kit+ spleen cells (20,000 cells) were fatal to the recipient mice, which was similar to the total leukemic spleen cells (2 × 105 cells). However, c-Kit+B220+Mac-1++ (5000 cells), c-Kit+B220−Mac-1++ (5000 cells), and c-Kit− spleen cells (20,000 cells) were not lethal to the recipient mice (Figure 2D). Further, by ELDA [49], we calculated that the frequency of LICs was 1/1321 in c-Kit+B220+Mac-1− cells and 1/2109 in c-Kit+B220+Mac-1+ cells, while no LIC was detected in c-Kit+B220+Mac-1++ and c-Kit+B220−Mac-1++ cells (Figure 2E, Supplementary Figure S2E). To clarify the hierarchical relationship of these two types of LICs, we analyzed the phenotypes that were reconstituted by c-Kit+B220+Mac-1− and c-Kit+B220+Mac-1+ in transplants. The results demonstrated that both c-Kit+B220+Mac-1− and c-Kit+B220+Mac-1+ subsets could fully reconstitute leukemia cell compartments in the PB, spleen, and BM of the recipient mice, as did the total leukemic spleen cells (Supplementary Figure S2F). It is noteworthy that c-Kit+B220+Mac-1− cells effectively reestablished c-Kit+B220+Mac-1+ cells and vice versa (Supplementary Figure S2F), suggesting that these two types of LICs could be converted into each other.
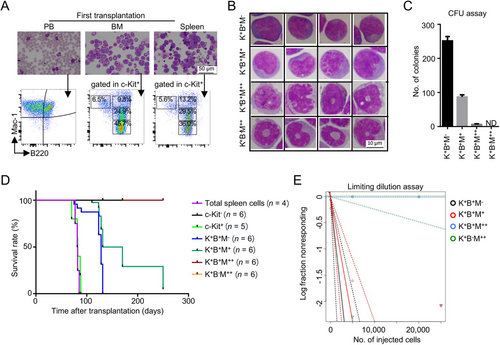
3.4 LICs of Setd2−/−-AML Exhibited Significant Intra-Heterogeneity in Transcription
Given that immunophenotypically defined subsets in leukemia were often unreliable [9], a comparative transcriptome analysis between the coexistent LICs was conducted. By the PCA and correlation analysis, we found that the transcriptomes of four c-Kit+ subsets were separated from each other, but samples within each group were converged together (Figure 3A, Supplementary Figure S3A). Transcriptome comparison showed that only 252 genes (FC ≤ 0.5 or FC ≥ 2; P < 0.05) had different expression between c-Kit+B220+Mac-1− and c-Kit+B220+Mac-1+ subsets while significant transcriptional differences were displayed between LIC-enriched subsets (c-Kit+B220+Mac-1− and c-Kit+B220+Mac-1+) and non-LIC subsets (c-Kit+B220+Mac-1++ and c-Kit+B220−Mac-1++) (Supplementary Figure S3B). GO analysis explained that c-Kit+B220+Mac-1− displayed transcriptional noise of erythroid characteristic, while transcriptional priming of the neutrophil feature was enriched in c-Kit+B220+Mac-1+ (Figure 3B). Notably, GSEA analysis exhibited that the self renewal-related gene set was highly enriched in c-Kit+B220+Mac-1− versus c-Kit+B220+Mac-1+ subset (Figure 3C), which was consistent with the fact that the former harbored more LICs than the latter.
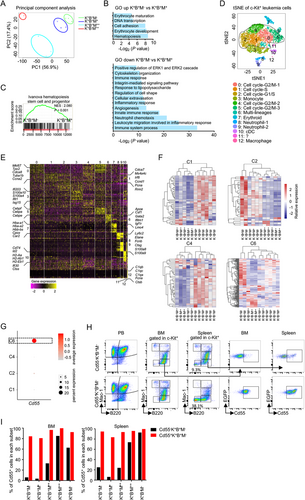
To better illuminate the transcriptional heterogeneity of LICs in this model, we performed scRNA-seq on c-Kit+ spleen cells (Supplementary Figure S3C). After initial quality control, a total of 7179 single-cell transcriptomes were acquired. Using tSNE, we identified 13 clusters (C0-C12) with distinct gene expression signatures (Figure 3D). According to the marker genes shown in Figure 3E and Supplementary Table S3, cycling cells were the most abundant components (C0, C1, C2, C4, and C5), which was in line with the highly proliferative properties of tumor cells. Non-cycling cell clusters were classified into 5 groups, including neutrophil [C8 and C9, highly expressing Elane, ficolin B (Fcnb), and S100 calcium-binding protein A9 (S100a9)], monocyte [C3, expressing lipoprotein lipase (Lpl), interferon-stimulated gene 15 (Isg15), and interferon-induced protein with tetratricopeptide repeats 1 (Ifit1)], conventional DC [C10, expressing cluster of differentiation 74 (Cd74), inhibitor of DNA binding 2 (Id2), and histocompatibility 2, class II antigen A, alpha (H2-Aa)], macrophage [C12, expressing complement component 1, q subcomponent, alpha polypeptide (C1qa) and complement component 1, q subcomponent, beta polypeptide (C1qb)], and erythroid [C7, expressing Hba-a1 and hemoglobin alpha, adult chain 2 (Hba-a2)] (Figure 3D-E). To interrogate the underlying correspondence between these single-cell clusters and four bulk subsets, we coupled the bulk RNA-seq data with representative markers within different tSNE clusters. Our results spotted that the c-Kit+B220+Mac-1+ subset was matched with C1, C2, and C4, whereas c-Kit+B220+Mac-1− was matched with C1, C2, C4, and C6 (Figure 3F), indicating these two types of LICs were transcriptionally heterogeneous at the single-cell level. Moreover, we also ascertained that the c-Kit+B220−Mac-1++ subset was matched with C8 and C9, whereas c-Kit+B220+Mac-1++ was matched with C10 and C12 (Supplementary Figure S3D). Notably, C7, an erythroid cluster, did not match any of these subsets, suggesting that we might have neglected a certain population with the erythroid transcriptional feature. Indeed, besides c-Kit+B220+Mac-1−, c-Kit+B220+Mac-1+, c-Kit+B220+Mac-1++, and c-Kit+B220−Mac-1++, we observed that c-Kit+ cells contained a minor subset of B220−Mac1− cells (Figure 1D). Subsequently, by using qRT-PCR, we clarified that c-Kit+B220−Mac1− cells highly expressed erythroid-related genes (Gata1, Klf1, Car1, Car2, Hba-a1, and Blvrb) than the other four subsets (Supplementary Figure S3E), suggesting that c-Kit+B220+Mac-1− cells, which possess erythroid transcriptional priming feature, were also capable of erythropoietic differentiation in function.
To further investigate the difference between the two types of LICs at the cellular level, we tried to distinguish them using the characteristics of C6, the differentiator between c-Kit+B220+Mac-1− and c-Kit+B220+Mac-1+. As shown in Supplementary Figure S3F, C6 presented in four subsets, displaying an apparent heterogeneity relative to other clusters. Moreover, C6 was defined as a unique population with multi-lineage differentiation potential by co-expressing markers related to erythroid (Gata1, Klf1, and Car2), megakaryocyte [platelet factor 4 (Pf4)], myelocyte [spleen focus forming virus proviral integration oncogene (Pu.1), CCAAT/enhancer-binding protein (C/EBP), alpha (Cepba), Runt-related transcription factor 1 (Runx1), and Lin-1, Isl1, Mec3 (LIM) domain only 4 (Lmo4)], and HSC [Mecom, GATA binding protein 2 (Gata2), IKAROS family zinc finger 2 (Ikzf2), and Meis homeobox 1 (Meis1)] (Supplementary Figure S3G). According to the feature of tSNE clusters, decay accelerating factor for complement (Cd55), an erythroid-associated cell surface marker [61], was partially distributed in C6 but not in C1, C2, and C4 (Figure 3G, Supplementary Figure S3H, and Supplementary Table S3). We then detected the Cd55 expression in each c-Kit+ subset by flow cytometry, showing that about 17.9% c-Kit+B220+Mac-1− cells expressed Cd55 in the leukemic spleen, whereas the proportion of Cd55+ cells was at a lower level in the c-Kit+B220+Mac-1+ subset (Supplementary Figure S3I-J). Thus, we isolated Cd55+ and Cd55− cells from the c-Kit+B220+Mac-1− subset respectively to elucidate their differentiation capacity. In vivo experiment demonstrated that Cd55−c-Kit+B220+Mac-1− subset could generate all leukemic compartments, while Cd55+c-Kit+B220+Mac-1− subset mainly differentiated to all kinds of leukemia progeny cells with the expression of Cd55 (Figure 3H-I). Moreover, we found that Cd55+c-Kit+B220+Mac-1− cells could differentiate into more c-Kit+B220−Mac1− erythroid subset cells (38.1%) relative to Cd55−c-Kit+B220+Mac-1− (9.3%) (Figure 3H). These in vivo data thus revealed a major functional difference between c-Kit+B220+Mac-1− and c-Kit+B220+Mac-1+ cells: LICs in c-Kit+B220+Mac-1− subset could differentiate into erythroid cells through C6 cluster cells under homeostasis, while c-Kit+B220+Mac-1+ could hardly do, due to the lack of C6. Taken together, these results demonstrated that the coexistent LICs in Setd2−/−-AML showed significant transcriptional and differential heterogeneity.
3.5 Different Response of the Coexistent Heterogeneous LICs to Standard Chemotherapy in Setd2−/−-AML model
Transcriptional heterogeneity often coincides with functional heterogeneity. Although GO analysis and related experiments (e.g., LDA assay) indicated that c-Kit+B220+Mac-1− cells possessed stronger self-renewal and erythroid differentiation potential than c-Kit+B220+Mac-1+ cells, the intrinsic functional difference between these two types of coexistent LICs is still unclear. We thus used GSEA to explore the probable effect, showing that a doxorubicin resistance-related gene-set [62] was enriched in the c-Kit+B220+Mac-1+ subset (Figure 4A), indicating that these two types of coexistent LICs might respond differently to anthracyclines (daunorubicin or doxorubicin) treatment. Remarkably, most of the doxorubicin resistance-related genes enriched in the c-Kit+B220+Mac-1+ subset were cell cycle-related [e.g., cell division cycle (Cdc), cyclin-dependent kinase (Cdk), ribonucleotide reductase catalytic subunit M (Rrm), and minichromosome maintenance complex component (Mcm)] (Figure 4B), suggesting that the c-Kit+B220+Mac-1+ subset might be more cycling than the c-Kit+B220+Mac-1− subset. qRT-PCR further confirmed a set of doxorubicin resistance-related genes (e.g., Aurka) highly expressed in the c-Kit+B220+Mac-1+ subset (Figure 4C). Moreover, cell cycle assay manifested that the cell cycling feature of the c-Kit+B220+Mac-1+ subset was indeed more prominent than the c-Kit+B220+Mac-1− subset (Figure 4D). The proportion of G0 phase in the BM c-Kit+B220+Mac-1− subset was statistically higher than that in the c-Kit+B220+Mac-1+ subset (49.6% ± 1.9% vs. 31.3% ± 0.9%), while the ratio of S-G2/M phase in the c-Kit+B220+Mac-1− subset were significantly lower than that in the c-Kit+B220+Mac-1+ subset (26.3% ± 3.3% vs. 44.8% ± 0.5%) (Figure 4D left). The cell cycle states of the c-Kit+B220+Mac-1− and c-Kit+B220+Mac-1+ subsets in the spleen were in line with that in BM (Figure 4D right). As the c-Kit+B220+Mac-1− and c-Kit+B220+Mac-1+ subsets were both matched with C1, C2, and C4 (Figure 3F), we split these three clusters into two subsets respectively by the expression level of Itgam (the gene encoding cell surface marker Mac-1) to address whether the c-Kit+B220+Mac-1+ cells were also more resistant to doxorubicin on the single-cell level. The Itgam+ subset represents c-Kit+B220+Mac-1+ cells and the Itgam− subset represents c-Kit+B220+Mac-1− cells. The mean expression values of the 52 doxorubicin resistance-related genes in the c-Kit+B220+Mac-1+ subset were statistically higher than those in the c-Kit+B220+Mac-1− subset in all three clusters (Figure 4E), suggesting that the two types of coexistent LICs exhibited a distinct transcriptional response to doxorubicin at both bulk and single-cell levels.
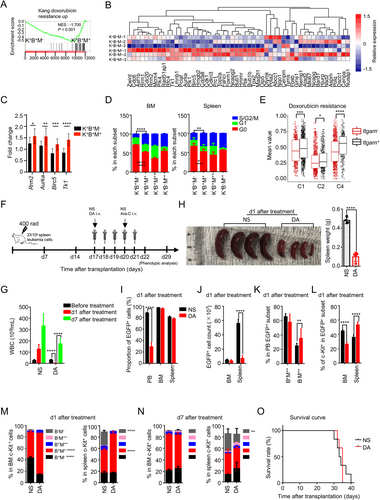
To further establish the heterogeneity of doxorubicin resistance in Setd2−/−-AML LICs at the functional level, we purified c-Kit+B220+Mac-1− and c-Kit+B220+Mac-1+ cells and treated them with different dosages of doxorubicin, followed by cell viability analysis. The results showed that the c-Kit+B220+Mac-1+ subset was indeed more resistant to doxorubicin than the c-Kit+B220+Mac-1− subset (Supplementary Figure S4A). Subsequently, DA, the standard chemotherapeutic regimen currently used in AML treatment, was applied to treat the transplanted Setd2−/−-AML mice (Figure 4F). Compared with NS, DA dramatically reduced the WBC count [(125.5 ± 11.3) × 106/mL vs. (2.4 ± 0.3) × 106/mL], as well as the proportion of donor-derived leukemia cells (EGFP+) in PB and alleviated splenomegaly on day 1 after a cycle of treatment (Figure 4G-I). Although the proportions of EGFP+ cells in the BM and spleen were not altered significantly on day 1 (Figure 4I), the absolute number of EGFP+ cells was dramatically reduced in the spleen (Figure 4J), suggesting that DA could induce partial remission of Setd2−/−-AML. Also, DA could alter leukemia cell composition in PB with an increase in the percentage of B220−Mac-1++ cells (Figure 4K). In particular, the proportions of both BM and spleen c-Kit+B220+Mac-1+ cells were increased after DA treatment, while the total c-Kit+ leukemic cells in these two tissues presented an opposite change (Figure 4L-M, Supplementary Figure S4B-D). Furthermore, in line with c-Kit+B220+Mac-1+, the percentage of c-Kit−B220+Mac-1+ cells was also fairly increased in the BM and spleen (Supplementary Figure S4E). Together these data suggest that c-Kit+B220+Mac-1+ leukemia cells are more resistant to the DA treatment than c-Kit+B220+Mac-1− cells, which corresponds with the presence of a doxorubicin-resistant transcriptomic gene signature in these cells.
Given that the c-Kit+B220+Mac-1+ subset could reconstitute Setd2−/−-AML phenotype in transplanted recipient mice, we predicted that c-Kit+B220+Mac-1+ cells that escaped from the original treatment would induce the regeneration of leukemia cells. From the result of the follow-up examination, we observed that the WBC count in the DA group was significantly increased on day 7 after treatment compared with that on day 1 (Figure 4G), and the percentage of EGFP+ cells displayed no changes between the DA and NS groups on day 7 in all three hematopoietic tissues (Supplementary Figure S4F). Immunophenotypic analysis of c-Kit+ cells disclosed that the proportion of the c-Kit+ leukemia subsets in the DA group was essentially identical to those in the NS group on day 7 (Figure 4N). In contrast, the mature cell compositions in the PB, BM, and spleen differed markedly between these two groups (Supplementary Figure S4G-J). Survival analysis indicated that DA alone could not attenuate Setd2−/−-AML progression (Figure 4O). Collectively, our results suggested that the coexistence of different types of LICs brought about diverse responses to DA and contributed to AML regeneration.
3.6 Combined DA and RAS downstream inhibitors attenuated Setd2−/−-AML progression
Next, we explored the mechanism underlying chemoresistant heterogeneity and address whether eradication of LICs in the c-Kit+B220+Mac-1+ subset can alleviate leukemia progression. Given that most of the doxorubicin resistance-related genes significantly enriched in the c-Kit+B220+Mac-1+ subset are cell cycle-related and that the RAS pathway plays a pivotal role in cell cycle regulation [63], we supposed that the chemoresistant heterogeneity of the coexistent LICs might be attributed to the abnormal activation of RAS downstream signaling pathway by NrasG12S and BrafK520E that were detected in this model. Although the allele frequencies of mutated Nras and Braf transcripts and the protein level of p-Mek showed no difference between c-Kit+B220+Mac-1− and c-Kit+B220+Mac-1+ subsets (Supplementary Figure S5), GSEA revealed that the activity of the MEK pathway was increased in c-Kit+B220+Mac-1+ cells versus c-Kit+B220+Mac-1− cells (Figure 5A). scRNA-seq also confirmed that RAS-MEK-related gene sets were enriched in Itgam+ compared with Itgam− subset in C1, C2, and C4 clusters (Figure 5B), indicating that inhibiting the RAS signaling pathway might be an efficient way to eliminate the c-Kit+B220+Mac-1+ subset. A small-molecular inhibitor screening was then performed on c-Kit+ spleen cells, in which the most robust killing occurred in cells cultured with AZ628 (Braf inhibitor) and Torin2 (mTOR inhibitor) (Supplementary Table S4). Further in vitro experiments displayed that both inhibitors dramatically augmented cell apoptosis in a dose-dependent manner (Supplementary Figure S6A).
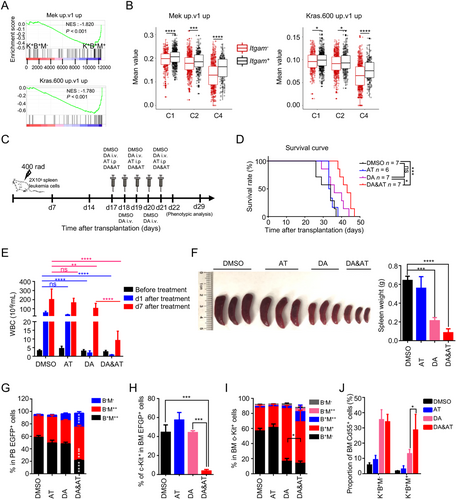
Afterward, we combined DA and AT treatment on transplanted Setd2−/−-AML mice to address the effect of LIC eradication on AML progression (Figure 5C). The combination therapeutic group (DA&AT group) not only effectively prolonged median survival (DMSO: 34 days; AT: 33.5 days; DA: 36 days; DA&AT: 43 days) (Figure 5D) but also significantly reduced the WBC count on day 1 after a cycle of treatment [DMSO: (54.8 ± 6.8) × 106/mL, AT: (31.3 ± 2.4) × 106/mL, DA: (1.7 ± 0.5) × 106/mL, DA&AT: (0.8 ± 0.1) × 106/mL] (Figure 5E). Besides, DA&AT effectively prevented the regeneration of leukemia cells on day 7 after treatment [DMSO: (199.8 ± 43.7) × 106/mL, AT: (164.3 ± 20.5) × 106/mL, DA: (102.2 ± 21.5) × 106/mL, DA&AT: (9.06 ± 2.0) × 106/mL]. Also, DA&AT reduced the proportion of donor-derived leukemia cells (EGFP+) in PB and relieved splenomegaly further than DA alone (Figure 5F, Supplementary Figure S6B). In particular, DA&AT significantly remodeled Setd2−/−-AML immunophenotype compared with DA and the other two groups (Supplementary Figure S6C-D). The proportion of B220+Mac-1++ cells was decreased in PB, accompanied by an apparent increase of B220−Mac-1++ and B220−Mac-1− subsets (Figure 5G, Supplementary Figure S6C). c-Kit+ cells were eliminated almost completely in the BM by DA&AT treatment (Figure 5H). Additionally, relative to DA, DA&AT further reduced the proportion of B220+Mac-1+ subset in BM c-Kit+ cells (Figure 5I). DA and DA&AT prominently augmented the percentage of Cd55+ cells in both c-Kit+B220+Mac-1− and c-Kit+B220+Mac-1+ subsets of the BM and spleen, indicating the induction of cell differentiation (Figure 5J, Supplementary Figure S6E). Notably, DA&AT harbored a stronger capability to induce c-Kit+B220+Mac-1+ cell differentiation relative to DA in the BM (Figure 5I, Supplementary Figure S6E). These results indicated that combination therapy of DA&AT could not only eliminate LICs in the c-Kit+B220+Mac-1+ subset of BM but also enhance their differentiation, and thus attenuating the progression of this Setd2−/−-AML model.
4 DISCUSSION
HSCs were once thought to be in a blank state without any transcription of lineage-specific genes [64]. With the development of single-cell PCR, scRNA-seq, single-cell transplantation, and in vivo single-cell tracing, the phenotypic homogeneous HSCs were identified to consist of lineage-biased HSCs. It was also revealed that lineage segregation adopted a continuous manner at HSC stage [65-69], indicating that transcriptional heterogeneity of HSCs may contribute to downstream lineage output. Notably, HSCs not only have lineage priming but also exist cell cycle-related transcriptional oscillations [70]. Therefore, transcriptional heterogeneity in proliferation and differentiation is a hallmark of HSCs [71, 72]. Actually, LICs inherit many of the characteristics of normal HSCs, such as self-renewal and hierarchical differentiation [5]. Herein, our study added further information in support of this point. By integrating traditional cell sorting, bulk RNA-seq, and scRNA-seq, we identified two types of LICs coexisting in a murine Setd2−/−-AML model, which was similar to several leukemia models with multi-type LICs [19-21]. These two types of LICs displayed conspicuously transcriptional heterogeneity, in which c-Kit+B220+Mac-1− possessed stronger self-renewal and erythroid differentiation potential, while c-Kit+B220+Mac-1+ was more prominent in transcriptional priming of neutrophil-related genes and cell cycling. Further experiments confirmed that the two types of LICs differed in erythroid differentiation potential under homeostasis. Besides differentiation heterogeneity, our results suggested that the coexistent LICs exhibited different responses to conventional chemotherapy, which may be partially ascribed to diverse cell cycle states. A series of evidence indicated that the c-Kit+B220+Mac-1+ subset was more prominent in the S/G2/M phase than the c-Kit+B220+Mac-1− subset. It is well known that DNA damage caused by chemotherapeutic drugs (e.g., doxorubicin) can trigger DNA damage response to increase tumor cell apoptosis [73]. However, overexpression of mitotic cell cycle-associated genes, such as AURKA, can impair G2-related DNA damage response [74]. Moreover, amplification of AURKA could induce Taxol chemoresistance through overriding mitotic spindle assembly checkpoint [75, 76]. Besides, it has been shown that DNA damage induced by DNA-damaging agents could not be adequately repaired during the S phase, it can be further repaired during the G2/M phase, thus the cycling leukemia cells acquiring the chemoresistant ability [46]. Given that c-Kit+B220+Mac-1+ cells expressed mitotic cell cycle-related genes and were mainly stationed in the G2/M phase, it meant that relative to c-Kit+B220+Mac-1−, c-Kit+B220+Mac-1+ had more opportunities to conquer DNA damage, thus possessing an intrinsic chemoresistant ability to doxorubicin. All along, LICs are considered as a critical factor for chemoresistance [5, 77]. However, two recent studies showed that chemotherapy displayed no killing selectivity on LSCs and non-LSCs [33, 34]. Thus, the chemotherapeutic heterogeneity of LICs in our murine Setd2−/− AML model may provide a reasonable explanation for this contradiction.
Although our results clearly showed that transcriptional intra-heterogeneity of the coexistent LICs can be effectively translated into functional heterogeneity (e.g., cell differentiation, cell cycle state, and chemotherapeutic response), the underlying mechanisms that cause transcriptional intra-heterogeneity of the coexistent LICs remain obscure. Genetic variation has been regarded as a major cause of transcriptional heterogeneity [78]. Herein, WGS on total BM cells of the primary Setd2−/- leukemia mouse detected Nras and Braf mutations, and based on the bulk RNA-seq analysis, both c-Kit+B220+Mac-1− and c-Kit+B220+Mac-1+ subsets appeared to harbor Nras and Braf mutations (Supplementary Figure S5A-B). It is necessary to carefully analyze the genetic variation between c-Kit+B220+Mac-1− and c-Kit+B220+Mac-1+ by WGS in the future. Also, a recent study discussed that KRAS mutant cancers frequently exhibited allelic imbalance of WT and mutant loci, which can promote AML clonal outgrowth and modulate drug sensitivity [79]. We thus surmised that allelic imbalance of mutant Nras and Braf might also be responsible for the intra-heterogeneity of the coexistent LICs in this Setd2−/−-AML model. However, c-Kit+B220+Mac-1− and c-Kit+B220+Mac-1+ subsets displayed similar allele frequencies of mutated Nras and Braf transcripts (Supplementary Figure S5A-B). Notably, most of doxorubicin resistance-related genes that were significantly enriched in the c-Kit+B220+Mac-1+ subset are cell cycle-related, and cell cycle analysis identified the c-Kit+B220+Mac-1+ subset with more cycling feature compared with the c-Kit+B220+Mac-1− subset. This is reminiscent of the bimodal effects of oncogenic NRASG12D, in which NRASG12D can activate different transcription pathways in distinct HSC subsets, thus assigning HSCs in different cell cycle states (quiescence or cycling) and endowing them with self-renewal or proliferation ability [80]. Hence, the bimodal effects of RAS mutation on cell cycle states might be a common feature of RAS-mutant leukemias and contribute to the drug resistance for this model. Besides, both bulk and scRNA-seq analysis indicated that the c-Kit+B220+Mac-1+ subset displayed a stronger transcriptional activation feature of RAS-MEK downstream signaling (Figure 5A-B). However, p-Mek at protein level did not show any change between c-Kit+B220+Mac-1− and c-Kit+B220+Mac-1+ (Supplementary Figure S5C). We speculate that the inconsistency in transcription and protein levels could be partly due to the sensitivity of different detection methods. As described in the Results section, the frequencies of LICs in c-Kit+B220+Mac-1− and c-Kit+B220+Mac-1+ subsets were 1:1321 and 1:2109, respectively, and the detection sensitivity of WB analysis may not be high enough. It is worth solving by a more sensitive method such as single-cell protein analysis in the future. In addition, our results demonstrated that LICs in the spleen were more resistant to chemotherapy than those in the BM, consistently, chronic myeloid leukemia stem cells in the spleen were less sensitive to imatinib than those in the BM [81], indicating that signal from niche microenvironment might also be involved in the drug response.
Mutations in epigenetic modifiers and signaling factors often co-occur in myeloid malignancies. The landscape of genetic variations in human AML was described by Alexander et al., showing that 7% (8/115) and 42% (49/115) patients present SETD2 and RAS signaling mutations (e.g., NRASG12S, 3/115), respectively. Two patients appeared to have co-mutations of SETD2 and RAS-related genes, one of them had SETD2T1761del, NRASG13R, and KRAST58I mutations simultaneously [43]. Moreover, mutant data from cBioportal revealed a co-occurrence of SETD2 mutation with NRASG13D/Q61H or BRAFV600E/V601E scattering in acute leukemia. In fact, previous researches by our group and others have indicated that SETD2-deficient cells would occur DNA replication stress [46, 48], which can alter cell cycle checkpoint, partly contributing to the chemoresistance. Therefore, we conjecture that the co-mutations of SETD2 and NRAS might cooperatively generate a disturbed cell cycle state that promotes HSC transformation and chemoresistance. However, in this study, only one Setd2−/− mouse had Nras/Braf mutations and developed AML, lacking control with Setd2+/+ and Nras/Braf mutations. Therefore, next step, we will introduce Nras and/or Braf mutations into Setd2+/+ and Setd2−/− HSCs respectively to better understand the mechanism of malignant transformation and drug response of this type of leukemia.
Finally, although all data were generated from one mouse, we think that there is still vital value to pursue. Setd2-deficient mutation has been broadly observed in hematopoietic malignancies, including AML, but how Setd2-deficient cells progress into LICs remains unclear. Herein, we found a natural secondary hit event on Setd2-mutated cells. Besides, the secondary mutations for this AML model are meaningful. As mentioned above, mutant data from cBioportal database demonstrates that co-mutation of SETD2 and NRAS is prominent (P = 0.042) in acute leukemia. Thus, this mouse may represent a type of acute leukemia with SETD2 and NRAS co-mutations. Last but not least, coexisting LICs have been frequently detected in clinical AML samples. However, the pathophysiological role of coexisting LICs in AML biology remains obscure. Interestingly, there happened to be two types of LICs coexisting in this AML model. Therefore, this model will help us to better investigate the pathophysiological role of the coexistent LICs in leukemia.
5 CONCLUSIONS
In this study, we identified two cell subsets (c-Kit+B220+Mac-1− and c-Kit+B220+Mac-1+) enriched for LICs in a murine Setd2−/−-AML model, which brought about different differentiation potentials and diverse chemotherapy responses. Combined treatment with DA and RAS pathway inhibitors could kill both kinds of LICs and attenuate disease progression. These findings may advance our understanding of the intra-heterogeneity of LICs and could stimulate the development of more efficient treatments.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Mice experiments were performed according to animal care standards, and all protocols were approved by the Animal Care Committee of Shanghai Jiao-Tong University School of Medicine (China).
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
No conflicts of interest to disclose.
ACKNOWLEDGMENTS
We thank Novel Bioinformatics Co. for the support of bioinformatics analysis. We thank Prof. Chi Wai Eric So from Leukemia Biology, King's College London, UK. for his constructive comments. We thank all colleagues in Shanghai Institute of Hematology and the National Research Center for Translational Medicine for their kind help and support. This work was supported by National Natural Science Foundation of China (81670149, 81870102), Samuel Waxman Cancer Research Foundation, and the Foundation of Key Laboratory of Veterinary Biotechnology (No. shklab202008), Shanghai, P.R. China.
AUTHORS’ CONTRIBUTIONS
YLZ and QHH designed the research. JCS, LTD, PL, YLZ, FHW, YYX, JL, and YJ performed experiments. YLZ, JCS, BZ, and GL performed bioinformatics analysis. YLZ, JCS, LTD, PL, FHW, YZ, JMZ, SJC, ZC, XJS, and QHH analyzed and discussed data. YLZ, JCS, and QHH wrote the manuscript.
Open Research
DATA AVAILABILITY STATEMENT
All sequencing data included in this study are available at National Omics Data Encyclopedia (NODE) (Access number: OEP000666). All data generated or analyzed during this study are included in this published article or uploaded as supplementary files 1-5.