KDELR2 promotes breast cancer proliferation via HDAC3-mediated cell cycle progression
Abstract
Background
Histone deacetylases (HDACs) engage in the regulation of various cellular processes by controlling global gene expression. The dysregulation of HDACs leads to carcinogenesis, making HDACs ideal targets for cancer therapy. However, the use of HDAC inhibitors (HDACi) as single agents has been shown to have limited success in treating solid tumors in clinical studies. This study aimed to identify a novel downstream effector of HDACs to provide a potential target for combination therapy.
Methods
Transcriptome sequencing and bioinformatics analysis were performed to screen for genes responsive to HDACi in breast cancer cells. The effects of HDACi on cell viability were detected using the MTT assay. The mRNA and protein levels of genes were determined by quantitative reverse transcription-PCR (qRT-PCR) and Western blotting. Cell cycle distribution and apoptosis were analyzed by flow cytometry. The binding of CREB1 (cAMP-response element binding protein 1) to the promoter of the KDELR (The KDEL (Lys-Asp-Glu-Leu) receptor) gene was validated by the ChIP (chromatin immunoprecipitation assay). The association between KDELR2 and protein of centriole 5 (POC5) was detected by immunoprecipitation. A breast cancer-bearing mouse model was employed to analyze the effect of the HDAC3-KDELR2 axis on tumor growth.
Results
KDELR2 was identified as a novel target of HDAC3, and its aberrant expression indicated the poor prognosis of breast cancer patients. We found a strong correlation between the protein expression patterns of HADC3 and KDELR2 in tumor tissues from breast cancer patients. The results of the ChIP assay and qRT-PCR analysis validated that HDAC3 transactivated KDELR2 via CREB1. The HDAC3-KDELR2 axis accelerated the cell cycle progression of cancer cells by protecting the centrosomal protein POC5 from proteasomal degradation. Moreover, the HDAC3-KDELR2 axis promoted breast cancer cell proliferation and tumorigenesis in vitro and in vivo.
Conclusion
Our results uncovered a previously unappreciated function of KDELR2 in tumorigenesis, linking a critical Golgi-the endoplasmic reticulum traffic transport protein to HDAC-controlled cell cycle progression on the path of cancer development and thus revealing a potential therapeutical target for breast cancer.
Abbreviations
-
- HDAC
-
- histone deacetylase
-
- HDACi
-
- histone deacetylase inhibitor
-
- KDELR
-
- KDEL receptor
-
- ER
-
- endoplasmic reticulum
-
- ERS
-
- ER retention sequence
-
- PP1
-
- protein phosphatase 1
-
- ISR
-
- integrated stress response
-
- NSCLC
-
- non-small cell lung cancer
-
- ATCC
-
- American Type Culture Collection
-
- DMEM
-
- Dulbecco's modified Eagle's medium
-
- FBS
-
- fetal bovine serum
-
- P/S
-
- penicillin/streptomycin
-
- STR
-
- short tandem repeat
-
- qRT-PCR
-
- quantitative reverse transcription-PCR
-
- shRNA
-
- small hairpin RNA
-
- TSA
-
- trichostatin A
-
- TDPA
-
- thailandepsin A
-
- PMSF
-
- phenylmethylsulfonyl fluoride
-
- SDS-PAGE
-
- sodium dodecyl sulfate-polyacrylamide gel electrophoresis
-
- SD
-
- standard deviation
-
- DNB
-
- DNA nanoball
-
- PI
-
- propidium iodide
-
- SEM
-
- standard error of the mean
-
- NTC
-
- non-targeting control
-
- CREB1
-
- cAMP-response element binding protein 1
-
- Co-IP
-
- coimmunoprecipitation
1 BACKGROUND
Epigenetic alterations that disrupt normal patterns of gene expression contribute greatly to tumor initiation and cancer development [1, 2]. Histone deacetylases (HDACs) act as key regulators of gene expression by removing acetyl groups from histones and repressing the transcription of target genes [3]. The aberrant expression of HDACs has been found in various types of cancers, which makes HDACs potential targets for cancer therapy [4]. HDAC inhibitors (HDACi), a novel class of small-molecule therapeutics, are approved by the U.S. Food and Drug Administration as anticancer agents because of their remarkable effects on inducing cell cycle arrest, apoptosis, and differentiation in cancer cells [5, 6]. While these therapies hold great promise for the treatment of cancers, HDACi as single agents have shown limited success in treating solid tumors in clinical studies. Thus, the mechanisms by which HDACs regulate tumor progression must be further dissected, as this may aid in the development of new anticancer strategies by synergistically targeting HDACs and other key downstream effectors.
The KDEL (Lys-Asp-Glu-Leu) receptor (KDELR) family is a key protein family involved in recycling the chaperones that maintains the dynamic equilibrium of trafficking between the Golgi and the endoplasmic reticulum (ER) [7]. Recent studies suggest that KDELRs play critical roles in the Golgi transport control system. KDELR binds to and activates the heterotrimeric signaling G-protein Gα (q/11) and in turn initiates transport through the Golgi complex [8]. As shown in a recent study by Solis et al. [], the KDELR-Gαo-Rab1/3 signaling axis controls vesicular trafficking and material delivery from the Golgi to the plasma membrane, which is critical for the elongation and stability of membrane protrusions. KDELR2 and KDELR3 mediate the secretion of endogenous ER retention sequence (ERS)-containing proteins in response to ER calcium depletion, thus suggesting that KDELRs play a role as ER stress-responsive factors in maintaining ER homeostasis [1]. In addition to their function in ER quality control, KDELRs have been reported to participate in the degradation of misfolded neurodegenerative disease-related proteins by inducing autophagy [11]. Intriguingly, according to a recent study by Henderson et al. [12], KDELRs localize at the cell surface and regulate the secretion and membrane binding of MANF, which is an ER stress-responsive protein with neuroprotective effects in animal models of neurodegeneration. KDELR1 is required for naïve T-cell homeostasis by directly regulating protein phosphatase 1 (PP1), a key phosphatase for integrated stress responses (ISR) in T-cells [13]. These studies indicate the critical role of KDELRs in cellular secretory trafficking, cell growth, autophagy, and the immune response. Recently, KDELR2 has been reported to promote glioblastoma by activating the mTOR signaling pathway in glioblastoma multiforme cells (GBM) [14] and has been identified as a prognostic biomarker related to glioma survival [15]. Bajaj et al. [16] demonstrated that KDELR2 drives non-small cell lung cancer (NSCLC) invasion and metastasis by enhancing Golgi-mediated secretion of matrix metalloproteases. However, the role of KDELR2 in tumorigenesis remains largely elusive. In this study, we aimed to identify novel downstream effectors of HDACs, and our results illustrate a previously unappreciated mechanism whereby KDELR stimulates cancer progression.
2 MATERIALS AND METHODS
2.1 Cell lines and cell culture
Human breast cancer cell lines (MDA-MB-231, T47D, MCF-7, and Hs-578t) were obtained from American Type Culture Collection (ATCC; Manassas, VA, USA) and cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; BI, Kibbutz Beit-Haemek, Israel) and 1% penicillin/streptomycin (P/S; Invitrogen, Carlsbad, CA, USA). Cells were maintained at 37°C in a humidified incubator with 5% CO2 in air. All cell lines were authenticated using short tandem repeat (STR) profiling (by GENEWIZ Co. Ltd. at Suzhou, Jiangsu, China) in 2018 and were tested for mycoplasma contamination. The cells were used within 15 passages (less than 2 months) after revival from frozen stocks.
2.2 Breast cancer specimens
Snap-frozen breast cancer tissues and corresponding paracancerous tissues that were at least 2 cm away from the edge of the tumors were taken from breast cancer patients by radical breast cancer resection in the Anhui Provincial Hospital (Anhui, Hefei, China) between May 2020 and Jan 2021. Total RNA and protein were extracted from paired tissues. The transcriptional levels and protein levels of genes in 16 pairs of breast cancer tissues and paired adjacent noncancerous tissues were analyzed by quantitative reverse transcription-PCR (qRT-PCR) and Western blotting. Prior written informed consent from the patients as well as a study approval from the Institutional Research Ethics Committee of Anhui Provincial Hospital was obtained for the use of the included patient materials. The studies were conducted in accordance with ethical guidelines of the Declaration of Helsinki.
Our results uncovered an unexpected role of HDAC3-KDELR2 axis in regulating breast cancer progression, thus revealing a therapeutically plausible target for breast cancer treatment.
2.3 Plasmids and stable cell line construction
HDAC3, KDELR2, and protein of centriole 5 (POC5) were subcloned into the pSin-3×Flag vector or pSin-HA vector (Addgene, Cambridge, MA, USA). All small hairpin RNAs (shRNAs) against HDAC1, HDAC2, HDAC3, HDAC4, HDAC5, HDAC6, HDAC8, KDELR2, CREB1, SP1, and CEBP-β in the PLKO vector were commercially purchased (Sigma-Aldrich, St. Louis, MO, USA). The target sequences of all shRNAs we used are listed in Supplementary Table S1. The subcloning primers of these shRNAs are summarized in Supplementary Table S2. HEK293T cells were cotransfected with plasmids encoding group antigen, polymerase, envelope protein, and vesicular stomatitis virus G protein using Lipofectamine 2000 (Invitrogen-Life Technologies, Waltham, MA, USA). Viral supernatant was collected 48 h post-transfection and filtered (0.22-μm pore size). MDA-MB-231 cells and T47D cells were infected with lentivirus in the presence of 8 μg/mL polybrene (Sigma Aldrich, St. Louis, MO, USA). The transduced cells were selected by puromycin (BI, Kibbutz Beit-Haemek, Israel). Trichostatin A (TSA; CAS #58880-19-6) was commercially purchased (Sigma-Aldrich, St. Louis, MO, USA). Thailandepsin A (TDPA) and FK228 were prepared from bacterial fermentation by the Cheng Group as previously described [17, 18].
2.4 Western blotting analysis
Cell lysates were prepared in RIPA buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.1% SDS, 1% NP-40) supplemented with protease cocktails (Roche, #43203100, Mannheim, Germany) and 100 μM phenylmethylsulfonyl fluoride (PMSF). The supernatant was collected after centrifugation at 13,000 × g for 10 min at 4°C. The cell lysate was quantified using Bradford (Sangon Biotech, Shanghai, China) and denatured at 100°C for 5 min. Equal amounts of protein were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to NC membranes, blocked with skimmed milk and then incubated overnight at 4°C with different primary antibodies in buffer containing 5% skimmed milk. Membranes were washed with TBS containing 0.05% Tween-20 for 30 min, blotted with secondary antibody for 1-2 h at room temperature, and then washed again three times. Western ECL Substrate (Bio-Rad, Hercules, CA, USA) was added, and the blots were imaged immediately on a Tanon-5200 Chemiluminescent Imaging System (Tanon Science and Technology, Shanghai, China). Band intensities were quantified using Image J software (Image-Processing and Analysis in Java; National Institutes of Health, Bethesda, MD, USA). Primary antibodies against the following proteins were used: HDAC3 (1:1000; 10255-1-AP; Proteintech, Wuhan, Hubei, China); SP1 (1:2000; 21962-1-AP; Proteintech, Wuhan, Hubei, China); CREB1 (1:1000; 12208-1-AP; Proteintech, Wuhan, Hubei, China); POC5 (1:1000; ab188330; Abcam, Cambridge, UK); CDK4 (Cyclin Dependent Kinase 4) (1:1000; 11026-1-AP; Proteintech, Wuhan, Hubei, China); CDK6 (Cyclin Dependent Kinase 6) (1:1000; 14052-1-AP; Proteintech, Wuhan, Hubei, China); OCT4 (POU Class 5 Homeobox 1) (1:1000; 60242-1-Ig; Proteintech, Wuhan, Hubei, China); Flag-tag (1:5000; F1804; Sigma-Aldrich, St. Louis, MO, USA); and HA-tagged HRP (1:1000; 2999; CST, MA, USA). β-Actin (1:5000; 66009-1-Ig; Proteintech, Wuhan, Hubei, China) served as the loading control. HRP-conjugated anti-rabbit and anti-mouse (Bio-Rad, Hercules, CA, USA) secondary antibodies were used.
2.5 qRT-PCR
Total RNA was isolated using TRIzol (Invitrogen-Life Technologies, Waltham, MA, USA) followed by DNase (Ambion, Austin, TX, USA) treatment and reverse transcription with an iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). qRT-PCR was performed using SYBR Green master mix (Vazyme, Najing, Jiangsu, China) and the iCycler Real-time System (Bio-Rad, Hercules, CA, USA). The annealing temperature of each primer pair was optimized by temperature gradient PCR. The relative expression of individual transcripts was normalized to 18S. The fold change of target mRNA expression was calculated based on the threshold cycle (Ct), where ∆Ct = Ct target – Ct 18S and ∆(∆Ct) = ∆Ct Control – ∆Ct indicated the condition. The data are presented as the mean ± standard deviation (SD) of three biological replicates. Briefly, we established the stably overexpression of knockdown cell lines, and collected samples and analyzed the gene expression from the cells of different passages. The primers used for analysis are shown in Supplementary Table S3.
2.6 ChIP-qPCR assay
The predicted transcriptional factors of KDELR2 were searched by the Promo (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3) and Genecards (http://www.genecards.org/). The predicted binding sites of CREB1 in the promoter region of KDELR2 gene were analyzed by JASPAR database (http://jaspar.genereg.net/).
The ChIP assay was performed using an EZ-ChIP kit (Millipore, CA, USA) following the manufacturer's instructions. Briefly, cells were fixed with 1% formaldehyde and quenched with 0.125 M glycine. Cells were sonicated using an ultrasonic cell disruptor (Scientz, Ningbo, Zhejiang, China). DNA was immunoprecipitated with either control IgG or CREB1 primary antibody (12208-1-AP, Proteintech, Wuhan, Hubei, China). RNA and protein were digested using RNase A (Fermentas, Shanghai, China) and Proteinase K (Thermo Scientific), respectively, followed by qPCR analysis. The qPCR primers are listed in Supplementary Table S4.
2.7 Immunoprecipitation/Co-Immunoprecipitation assay
The proteins that may interact with KDELR2 were predicted by the BioGRID database (https://thebiogrid.org/). For Co-Immunoprecipitation assay, cells were cotransfected with Flag-KDELR2 and HA-POC5. Cells were lysed in buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl, 2 mM EDTA, 1.5 mM MgCl2, 1% NP40, and protease inhibitors for 1 h at 4°C followed by centrifugation. The supernatants were then diluted in a buffer without NP40 and precleared with protein A/G Sepharose beads (Thermo Fisher, Rockford, IL, USA) for 30 min. The supernatants were immunoprecipitated with the indicated antibody for 12 h at 4°C, followed by incubation with protein A/G-Sepharose beads for 1 h at 4°C. After incubation, beads were then washed with IP buffer (25 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 0.5% NP40, 5% glycerol) and boiled in 2 × SDS-loading buffer (1 M Tris-HCl, 10% SDS, 0.5% bromophenol blue, 50% glycerol). Protein samples were analyzed by Western blotting.
2.8 Ubiquitination assay
HEK293T cells were cotransfected with pSIN-HA-ubiquitin and pSIN-Flag-POC5 (Addgene, Cambridge, MA, USA) in the presence of shHDAC3s or shKDELR2s as indicated. After incubation for 24 h, the 20S proteasome inhibitor lactacystin (Shanghai yuanye Bio-Technology, Shanghai, China) was added to the culture medium for an additional 24 h, followed by collection of the cells and protein lysis with SDS buffer. Equal amounts of protein lysates were immunoprecipitated with anti-Flag-M2 antibody (Sigma-Aldrich, St. Louis, MO, USA) and subjected to SDS-PAGE, followed by blotting with anti-HA-HRP antibody (Cell Signaling Technology, Danvers, MA, USA).
2.9 RNA-seq bioinformatics analysis pipelines
RNA was extracted using TRIzol Reagent (nvitrogen-Life Technologies, Waltham, MA, USA) following the manufacturer's instructions. The following sequencing and data analysis work using these RNA samples was performed by BGI-Shenzhen (Shenzhen, Guangdong, China). Briefly, total RNA was quantified using a NanoDrop and Agilent 2100 bioanalyzer (Thermo Fisher Scientific, Waltham, MA, USA). Oligo(dT)-attached magnetic beads were used to purify mRNA. Purified mRNA was fragmented into small pieces with fragment buffer at the appropriate temperature. Then, first-strand cDNA was generated using random hexamer-primed reverse transcription, followed by second-strand cDNA synthesis. Afterwards, A-Tailing Mix and RNA Index Adapters were added via incubation for end repair. The cDNA fragments obtained from the previous step were amplified by PCR, and the products were purified by Ampure XP Beads (BECKMAN, Brea, CA, USA) and then dissolved in EB solution. The product was validated on an Agilent Technologies 2100 bioanalyzer (Agilent, Santa Clara, CA, USA) for quality control. The double-stranded PCR products from the previous step were heated, denatured, and circularized by the splint oligo sequence to obtain the final library. Single-stranded circular DNA (ssCir DNA) was formatted as the final library. The final library was amplified with phi29 to make DNA nanoballs (DNBs) that had more than 300 copies of one molecule. DNBs were loaded into the patterned nanoarray, and single-end 50-base reads were generated on the BGISeq500 platform (BGI-Shenzhen, Shenzhen, Guangdong, China). The sequencing data were filtered with SOAPNUKE (V1.5.2) as follows: removing reads containing sequencing adapter; removing reads whose low-quality base ratio (base quality less than or equal to 5) was more than 20%; and removing reads whose unknown base (‘N’ base) ratio was more than 5%. Afterwards, clean reads were obtained and stored in FASTQ format. Bowtie2 (v2.2.5; Kim Lab, Bioinformatics Department on UT Southwestern's South Campus, Dallas, Texas, USA) was applied to map the clean reads to the reference coding gene set, and the expression level of the gene was calculated by the transcripts per kilobase of exon model per million mapped reads (TPM) value. Gene expression analysis was performed using Cuffdiff (v2.2.1; Trapnell Lab, University of Washington, Seattle, USA). Unsupervised clustering was performed using the cluster and tree view and visualized using heat maps.
2.10 Cell cycle analysis
Cells were harvested by trypsinization, washed twice with PBS containing 5% FBS, fixed in 70% ethanol followed by staining with 20 μg/mL propidium iodide (PI; Sigma-Aldrich, St. Louis, MO, USA) containing 20 μg/mL RNase (Fermentas, Shanghai, China). Stained cells were analyzed by flow cytometry (BD Biosciences, San Diego, CA, USA). The graph was plotted using FlowJo 7.6.5 software (FLOWJO LLC, Ashland, KY, USA).
2.11 Apoptosis assay
Cell apoptosis was measured by flow cytometry (BD LSRFortessa, USA) using the Annexin V-PI Apoptosis Detection Kit (Bestbio, Shanghai, China) according to the manufacturer's instructions. Briefly, floating and adhesive cells were collected, washed twice with PBS containing 5% FBS, suspended in binding buffer and stained with Annexin V for 15 min. After that, the cells were stained with PI for 4 min. Stained cells were analyzed by flow cytometry (BD Biosciences, San Diego, CA, USA). The graph was plotted using FlowJo 7.6.5 software (FLOWJO LLC, Ashland, KY, USA).
2.12 MTT assay
A total of 5 × 103 cells per well were incubated in 96-well plates and DMEM containing 10% FBS. Twenty-four hours later, the cells were treated with the HDACi TSA, TDPA, or FK228 at different concentrations for 72 h. Thereafter MTT (5 mg/mL; Sigma Aldrich) was added to the medium for another 4 h. Dissolving buffer (10% w/v SDS, 1% v/v isobutyl alcohol, 0.1% v/v 10M HCl) was added to the medium overnight. The OD570 was further determined using Biotek Cytation5 Microplate Reader (Biotek, SYNERGY HTX, VT, USA) and the IC50 (the half maximal inhibitory concentration) was calculated by Graphpad Prism 7.0 (GraphPad Software, San Diego, CA, USA).
2.13 Analysis of differential gene expression from the TCGA (The Cancer Genome Atlas) dataset
Gene expression data of the breast cancer were downloaded from TCGA (http://firebrowse.org). Differential gene expression between normal and cancer samples were evaluated by t test.
2.14 Kaplan-Meier survival analysis
To investigate the association between HDAC3 or KDELR2 and patient survival, we downloaded information related to the survival time of breast cancer patients on the website http://firebrowse.org, and evaluated the overall survival in all patients available by R packages (survival_3.2_10). Patients were grouped according to the optimized cutoff.
2.15 Animal studies
All animal studies were conducted with approval from the Animal Research Ethics Committee of the University of Science and Technology of China. For xenograft experiments, 5× 106 (HDACi retarding tumor growth) or 2 × 106 (KDELR2 is critical for HDAC3-regulated breast cancer progression) MDA-MB-231 cells were injected subcutaneously into 5-week-old female nude mice (SJA Laboratory Animal Company, Changsha, Hunan, China). The tumor volumes were measured using digital calipers every 3 days and calculated using the following equation: length (mm) × width (mm) × depth (mm) × 0.52.
2.16 Statistical analysis
All experimental data are presented as the mean ± SD or mean ± standard error of the mean (SEM) from at least three independent experiments. Statistical analysis was performed using Student's t-test (unpaired, two-tailed) to compare two groups of independent samples by GraphPad Prism 7 (GraphPad Software, San Diego, CA, USA). Statistical significance is displayed as P <0.05.
3 RESULTS
3.1 The expression of KDELR2 was repressed by HDAC inhibitor in breast cancer
We set out to search for potential therapeutic targets for HDACs by HDACi screening. By treating MDA-MB-231 breast cancer cells with the pan-class I/II HDACi TSA, and the class I HDACi TDPA and FK228, we found that TSA, TDPA and FK228 showed concentration-dependent cytotoxicity against MDA-MB-231 cells with IC50 values of 1.368 ± 0.324 μM, 2.763 ± 0.356 nM and 8.363 ± 0.811 nM, respectively (Figure 1A and Supplementary Figure S1A). The results of Ki67 staining confirmed that TDPA, FK228 or TSA treatment dramatically suppressed the proliferation of breast cancer cells (Figure 1B). To assess the effect of these HDACi on tumor growth in vivo, we treated mice bearing xenografts of MDA-MB-231 breast cancer cells with TDPA and FK228. The results indicated that TDPA and FK228 significantly retarded tumor growth at doses of 3 mg/kg and 0.75 mg/kg, respectively (Figure 1C and Supplementary Figure S1B). Importantly, TDPA and FK228 did not induce body weight loss (Supplementary Figure S1C) or any obvious side effects in the mice. Then, we compared the gene expression profiles of MDA-MB-231 cells in the presence or absence of HDACi. The expression levels of 949 genes were consistently reduced, and 5856 genes were upregulated in HDACi-treated MDA-MB-231 cells (Figure 1D). HDACs deacetylate nucleosomal histones, resulting in the transcriptional repression of genes [19]. Considering that the upregulated genes in HDACi-treated cells might be transactivated via histone acetylation, we focused on the genes that were transcriptionally repressed by HDACi treatment to identify the novel targets of HDACs. To narrow down the potential targets of HDACs, we then determined the clinical relevance in breast cancer patients by comparing the expression levels of those genes suppressed by HDACi in tumor tissues to normal tissues and analyzing the association of their expression levels and survival time of breast cancer patients in the TCGA program. Among the genes that were repressed by HDACi at the transcriptional level, 17 genes were significantly upregulated in breast cancer tissues compared to normal breast tissues (Log2Fc [Tumor/Normal] ≥ 0.6) and predicted a poor prognosis of breast cancer patients (P < 0.05; Figure 1E). We selected these 17 genes for further analysis.
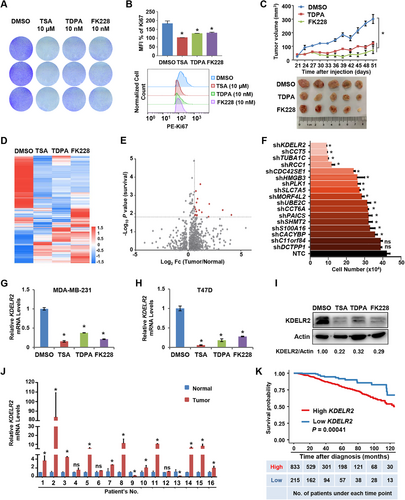
Aberrant expression of KDELR2, a novel target of HDACi, correlated with the breast cancer process. A. Crystal violet staining was used to analyze the cell proliferation ability of MDA-MB-231 cells after treatment with TSA (10 μM), TDPA (10 nM) and FK228 (10 nM) for 48 h. B. MDA-MB-231 cells were treatment with TSA (10 μM), TDPA (10 nM) and FK228 (10 nM) for 48 h. Statistical analysis of relative mean fluorescence intensities (top) and the histrogram graph (bottom) of Ki67 expression were analyzed by flow cytometry. The data are presented as the mean ± SD of three independent experiments. *, P < 0.05. C. MDA-MB-231 cells (5× 106 cells per mouse) were subcutaneously injected into female nude mice (n = 5 for each group). Mice were intraperitoneally injected with 3 mg/kg TDPA or 0.75 mg/kg FK228 every three days after inoculation. Tumor sizes were measured starting from 21 days after inoculation (top). At the end of the experiment, the tumors were extracted and compared (bottom). The data are presented as the mean ± SEM. *, P < 0.05 compared between the indicated groups. *, P < 0.05. D. The heat map from RNA-Seq analysis showed alterations in the expression of genes in breast cancer cells treated with HDAC inhibitors relative to the expression of these genes in the control group. The colors indicate the ln-transformed transcripts per kilobase of exon model per million mapped reads (TPM) values. E. Volcano plot showing the gene expression differences between breast tumor tissues and normal tissues and the statistical significance of genes related to patient survival in the TCGA database. Each point represents the gene that was consistently downregulated by 3 HDAC inhibitors from Figure 1D. The X axis represents the fold change in gene expression between breast tumor tissues and normal tissues. The Y axis represents the statistical significance of genes related to patient survival in TCGA. F. The growth of MDA-MB-231 cells with knockdown of the indicated genes identified by RNA-Seq was calculated. The data are presented as the mean ± SEM of three independent experiments. *, P < 0.05; ns, not significant. G-H. qRT-PCR analysis of the mRNA levels of KDELR2 in MDA-MB-231 (G) and T47D (H) breast cancer cells treated with HDAC inhibitors TSA (10 μM), TDPA (10 nM) and FK228 (10 nM) for 48 h. The data are presented as the mean ± SD of three independent experiments. *, P < 0.05. I. Western blotting analysis of the protein levels of KDELR2 in MDA-MB-231 cells treated with HDAC inhibitors TSA (10 μM), TDPA (10 nM) and FK228 (10 nM) for 48 h. J. qRT-PCR analysis of the mRNA levels of KDELR2 in 16 pairs of clinically matched tumor-adjacent noncancerous breast tissues (Normal) and human breast cancer tissues (Tumor). The data are presented as the mean ± SD. Group differences were analyzed by the two-tailed Student's t-test. *, P < 0.05; ns, not significant.
K. Kaplan-Meier curves from univariate analysis for patients with low versus high KDELR2 expression. The data were obtained from the website (http://firebrowse.org). Patients were grouped according to the optimized cutoff
To assess the effects of these 17 genes on cancer cell growth, we knocked down individual genes with shRNAs in MDA-MB-231 cells and found that the knockdown of KDELR2 significantly retarded the growth of breast cancer cells (Figure 1F). qRT-PCR confirmed the repressed expression of KDELR2 in breast cancer cells treated with HDACi (Figure 1G-H and Supplementary Figure S1D-E). Furthermore, Western blotting analysis revealed that the protein levels of KDELR2 were significantly repressed in MDA-MB-231 cells treated with HDACi (Figure 1I). We further analyzed the transcriptional levels of KDELR2 in 16 pairs of breast cancer lesions and paired adjacent noncancerous tissues. Consistent with the data from the TCGA database, increased mRNA levels of KDELR2 were observed in tumor tissues compared to adjacent noncancerous tissues (Figure 1J and Supplementary Figure S1F). The Kaplan-Meier test from the TCGA database indicated that KDELR2 expression in breast cancer patients was significantly associated with survival time: patients expressing high levels of KDELR2 showed worse overall survival than those with low KDELR2 expression levels (patients were grouped according to the optimized cutoff; P < 0.001; Figure 1K). Taken together, we found that KDELR2 is a potential novel regulator of breast cancer and is one of the downstream targets of HDACs.
3.2 HDAC3 was responsible for HDAC-mediated KDELR2 expression
HDACs are classified into 4 groups: class I HDACs (HDAC1, 2, 3, and 8), class II HDACs (class IIa: HDAC4, 5, 7, and 9; class IIb: HDAC6); nicotinamide adenine dinucleotide (NAD)-dependent class III HDACs (also called sirtuins); and class IV HDACs (HDAC10, 11) [20, 21]. To identify which class I and II HDACs regulate KDELR2 expression, we first infected MDA-MB-231 cells with pooled shRNAs targeting class I and class II HDACs and found that the mRNA level of KDELR2 was significantly reduced in cells with both HDAC3 and HDAC8 knockdown compared to non-targeting control (NTC) cells (Figure 2A). By further suppressing the expression levels of HDAC3 and HDAC8 by shRNAs, it was found that knockdown of HDAC3, not HDAC8, markedly reduced the mRNA level of KDELR2 in MDA-MB-231 cells (Figure 2B-C). Western blotting analysis revealed that the protein levels of KDELR2 were significantly repressed in MDA-MB-231 cells with HDAC3 knockdown (Figure 2D). Consistently, the forced expression of HDAC3 significantly increased KDELR2 mRNA expression (Figure 2E). Similar results were obtained from T47D cells in which HDAC3 expression was silenced (Supplementary Figure S2A-B). Based on these data, among class I and II HDACs, HDAC3 was identified as responsible for HDAC-mediated KDELR2 expression in breast cancer cells.
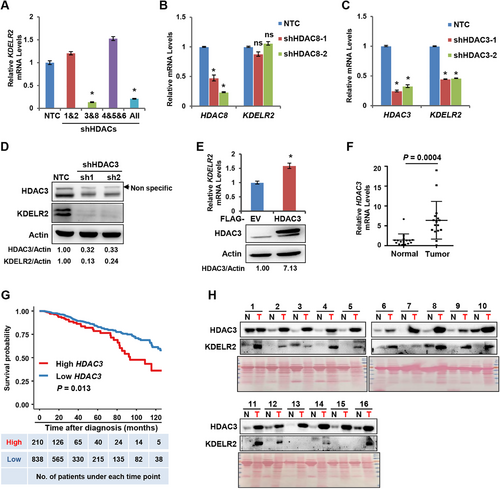
HDAC3 was responsible for HDACi-mediated KDELR2 expression. A. qRT-PCR analysis of the mRNA levels of KDELR2 in MDA-MB-231 cells transfected with pooled shHDACs and NTC cells. The data are presented as the mean ± SD of three independent experiments. *, P < 0.05. B. qRT-PCR analysis of the mRNA levels of HDAC8 and KDELR2 in HDAC8 knockdown MDA-MB-231 cells and NTC cells. The data are presented as the mean ± SD of three independent experiments. *, P < 0.05; ns, not significant. C. qRT-PCR analysis of the mRNA levels of HDAC3 and KDELR2 in HDAC3 knockdown MDA-MB-231 cells and NTC cells. The data are presented as the mean ± SD of three independent experiments. *, P < 0.05. D. Western blotting analysis of the protein levels of HDAC3 and KDELR2 in MDA-MB-231 cells stably expressing shHDAC3 and NTC cells. β-Actin served as the loading control. E. qRT-PCR analysis of the mRNA levels of HDAC3 and KDELR2 in MDA-MB-231 cells stably overexpressing 3×Flag-HDAC3 and empty vector cells. The overexpression efficiency was analyzed by Western blotting. The data are presented as the mean ± SD of three independent experiments. *, P < 0.05. F. qRT-PCR analysis of the mRNA levels of HDAC3 in 16 pairs of tumor-adjacent noncancerous breast tissues (Normal) and breast cancer tissues (Tumor). The data are presented as the mean ±SD. Group differences were analyzed by the two-tailed Student's t-test. G. Kaplan-Meier curves from univariate analysis for patients with low versus high HDAC3 expression. The data were obtained from the website (http://firebrowse.org). Patients were grouped according to the optimized cutoff. H. Western blotting analysis of HDAC3 and KDELR2 protein levels in 16 pairs of clinically matched adjacent noncancerous breast tissues (normal) and human breast cancer tissues (tumor). Ponceau staining is shown at the bottom as the loading control.
To further assess the physiological correlation between HDAC3 expression and tumorigenesis in human malignancies, we analyzed the mRNA levels of HDAC3 in 16 pairs of breast cancer lesions and adjacent noncancerous tissues. qRT-PCR analysis confirmed the increased transcriptional levels of HDAC3 in tumor tissues from breast cancer patients compared to adjacent noncancerous tissues (P < 0.001; Figure 2F), consistent with the data from the TCGA database (P = 0.001; Supplementary Figure S2C). In addition, high HDAC3 expression in tumor tissues predicted a poor prognosis of breast cancer patients according to the Kaplan-Meier test from the TCGA database (patients were grouped according to the optimized cutoff; P = 0.013; Figure 2G). Most importantly, the protein expression patterns of HDAC3 and KDELR2 exhibited a strong correlation in breast cancer lesions (Figure 2H). These data indicated a potential role for HDAC3 in regulating the expression of KDELR2 in breast cancer lesions.
3.3 HDAC3 promoted KDELR2 expression via CREB1
To obtain further insights into the mechanism by which HDAC3 enhances the transcriptional level of KDELR2, we first performed bioinformatic analysis for the potential transcription factors of KDELR2 using GeneCards and Promo and found CCAAT Enhancer Binding Protein Beta (CEBP-β), Transcription Factor Sp1 (SP1) and cAMP-response element binding protein 1 (CREB1) (Figure 3A). Then, we suppressed the expression of CEBP-β, SP1 and CREB1 in MDA-MB-231 cells using shRNAs (Supplementary Figure S3A) and found that the mRNA level of KDELR2 was decreased only when CREB1 was silenced (Figure 3B), suggesting that CREB1 is a potential transcription factor of KDELR2. Similar results were observed in T47D cells (Supplementary Figure S3B). CREB1 binds to CRE sequences in the promoter regions of downstream genes and activates their transcription [22]. We performed bioinformatic analysis of CRE sequences in the promoter regions of the KDELR2 gene using the JASPAR database to investigate whether KDELR2 is a direct target of CREB1. The results showed that KDELR2 possessed several potential CRE sequences (Figure 3C). ChIP assays validated that CREB1 directly bound to the promoter of KDELR2 in MBA-MD-231 cells (Figure 3D). Recently, CREB1 was reported to have elevated expression in breast cancer cells and promoted cell proliferation, migration, and tamoxifen resistance [23]. Here, we found that knockdown of HDAC3 markedly decreased the mRNA and protein levels of CREB1 in MBA-MD-231 cells (Figure 3E-F). Similar results were observed in T47D cells (Supplementary Figure S3C). Moreover, HDACi exerted similar effects on CREB1 expression in breast cancer cells (Figure 3G-H). To determine whether HDAC3 increases the mRNA level of KDELR2 via CREB1, we introduced lentivirus expressing shCREB1 into MBA-MD-231 cells overexpressing HDAC3. As expected, CREB1 silencing abolished the increased mRNA and protein levels of KDELR2 induced by HDAC3 overexpression (Figure 3I-J). These data indicated that HDAC3 promotes KDELR2 expression by upregulating the transcription factor CREB1.
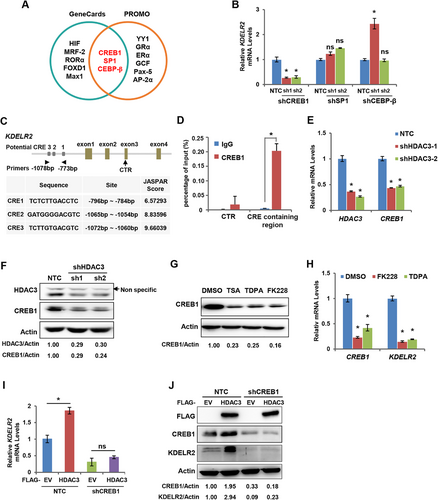
HDAC3 promoted KDELR2 expression via CREB1. A. Pie graph showing the predicted transcription factors of KDELR2 analyzed by the Genecards and Promo websites. B. qRT-PCR analysis of the mRNA levels of KDELR2 in MDA-MB-231 cells with CREB1, SP1 or CEBP-β knockdown. The data are presented as the mean ± SD of three independent experiments. *, P < 0.05; ns, not significant. C. A diagram shows the potential binding sites and sequences of CREB1 at the KDELR2 gene promoter. D. Endogenous ChIP was performed to identify the binding sites of CREB1 in the KDELR2 gene. The data are presented as the mean ± SD of three independent experiments. *, P < 0.05. E-F. qRT-PCR analysis and Western blotting analysis of the mRNA and protein levels of CREB1 in MDA-MB-231 cells with stable HDAC3 knockdown. The data are presented as the mean ± SD of three independent experiments. *, P < 0.05; β-Actin served as the loading control. G. Western blotting analysis of the expression of CREB1 in MDA-MB-231 cells treated with HDAC inhibitors TSA (10 μM), TDPA (10 nM) and FK228 (10 nM) for 48 h. β-Actin served as the loading control. H. qRT-PCR analysis of KDELR2 and CREB1 mRNA in MDA-MB-231 cells treated with HDAC inhibitors (TDPA or FK228) for 48 h. The data are presented as the mean ± SD of three independent experiments. *, P < 0.05. I. qRT-PCR analysis of the mRNA levels of KDELR2 in MDA-MB-231 cells with stable HDAC overexpression and CREB1 knockdown. The data are presented as the mean ± SD of three independent experiments. *, P < 0.05; ns, not significant. J. Western blotting analysis of the protein levels of KDELR2, CREB1 and HDAC3 in MDA-MB-231 cells with stable HDAC3 overexpression and CREB1 knockdown. β-Actin served as the loading control.
3.4 HDAC3/KDELR2 axis promoted cell cycle progression by enhancing POC5 expression
To determine the effects of the HDAC3/KDELR2 axis on the survival and proliferation of breast cancer cells, we knocked down HDAC3 or KDELR2 to observe cell growth. As expected, HDAC3 or KDELR2 knockdown markedly impaired the growth of breast cancer cells (Figure 4A-B and Supplementary Figure S4A-B). Importantly, KDELR2 silencing attenuated the HDAC3 overexpression-induced cell growth advantage (Figure 4C). KDELR2 has been reported to be responsible for ER resident proteins transport from the Golgi complex to the ER [24]. However, the mechanism by which KDELR2 promotes breast cancer progression remains unclear. We first investigated cell apoptosis to obtain insights into the mechanism by which the HDAC3/KDELR2 axis regulates cell growth. However, no significant differences in the percentages of apoptotic cells were observed either in MDA-MB-231 cells with HDAC3 overexpression and KDELR2 knockdown or in control cells (Supplementary Figure S4C). Similar results were obtained from T47D cells in which KDELR2 expression was suppressed (Supplementary Figure S4D). Next, by analyzing the cell cycle, we found that KDELR2 knockdown led to cell cycle arrest at G1 phase (Figure 4D and Supplementary Figure S4E).
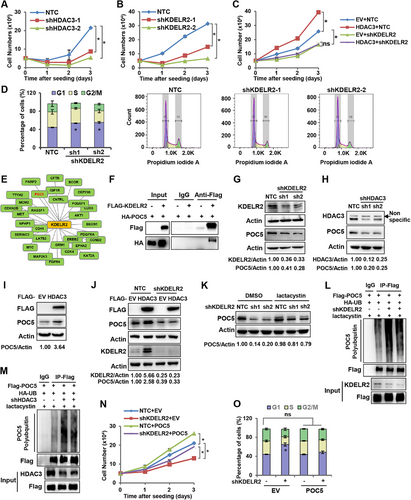
HDAC3/KDELR2 axis promoted cell cycle progression by enhancing POC5 expression. A. Cell growth analysis of HDAC3-knockdown MDA-MB-231 cells. The data are presented as the mean ± SEM of three independent experiments. *, P < 0.05. B. Cell growth analysis of KDELR2-knockdown MDA-MB-231 cells. The data are presented as the mean ± SEM of three independent experiments. *, P < 0.05. C. Cell growth analysis of MDA-MB-231 cells with stable HDAC3 overexpression and KDELR2 knockdown. The data are presented as the mean ± SEM of three independent experiments. *, P < 0.05; ns, not significant. D. Analysis of the cell cycle distribution in MDA-MB-231 cells with KDELR2 knockdown by flow cytometry. Representative histogram data and statistical results are shown. The data are presented as the mean ± SEM of three independent experiments. *, P < 0.05. E. Thirty proteins were predicted to interact with KDELR2 in the the BioGRID database. F. Coimmunoprecipitation assay of the protein interaction between KDELR2 and POC5. MDA-MB-231 cells were cotransfected with Flag-EV or Flag-KDELR2 and HA-POC5 plasmids. Cell lysates were immunoprecipitated with an anti-Flag antibody, followed by Western blotting analysis with antibodies against Flag and HA tags. G. Western blotting analysis of the expression of POC5 and KDELR2 in MDA-MB-231 cells with stable KDELR2 knockdown. β-Actin served as the loading control. H. Western blotting analysis of the expression of POC5 in MDA-MB-231 cells with HDAC3 knockdown. β-Actin served as the loading control. I. Western blotting analysis of the expression of POC5 in MDA-MB-231 cells with HDAC3 overexpression. β-Actin served as the loading control. J. Western blotting analysis of the expression of POC5 in MDA-MB-231 cells with stable HDAC3 overexpression and KDELR2 knockdown. β-Actin served as the loading control. K. Western blotting analysis of the expression of POC5 in MDA-MB-231 cells with KDELR2 knockdown treated with lactacystin (5 μM) for 24 h. β-Actin served as the loading control. L-M. Ubiquitination analysis of POC5 protein in HEK293T cells with KDELR2 (L) or HDAC3 (M) knockdown. HEK293T cells were cotransfected with HA-Ub, FLAG-POC5, and shKDELR2 and treated with lactacystin (5 μM) for 24 h before lysis. Equal amounts of proteins were used for immunoprecipitation with an anti-Flag antibody, followed by blotting with anti-HA. N. Cell growth analysis of MDA-MB-231 cells with KDELR2 knockdown and stable POC5 overexpression. The data are presented as the mean ± SEM of three independent experiments. *, P < 0.05. O. Analysis of the cell cycle distribution of MDA-MB-231 cells with KDELR2 knockdown and stable POC5 overexpression by flow cytometry. The data are presented as the mean ± SEM of three independent experiments. *, P < 0.05; ns, not significant.
To determine how KDELR2 regulates the cell cycle, we searched for proteins that might interact with KDELR2 by bioinformatics screening using the BioGRID database (Figure 4E). Thus, we focused on the proteins that are related to cell cycle regulation, including CDK4, CDK6, OCT4 and POC5, and performed an immunoprecipitation assay using HEK293T cells overexpressing Flag-KDELR2 to confirm the protein interactions. As a result, POC5 was the only protein found to have an association with KDELR2 (Supplementary Figure S4F). Furthermore, Co-IP experiments in MDA-MB-231 and HEK293T cells overexpressing Flag-KDELR2 and HA-POC5 were performed using a Flag-M2 antibody. The results of Co-IP experiments confirmed that KDELR2 interacted with POC5 (Figure 4F and Supplementary Figure S4G). POC5, which localizes on the centriole, has been reported to be associated with primary scoliosis in adolescents, and its absence can lead to cell cycle arrest [25-27]. The knockdown of either KDELR2 or HDAC3 suppressed the protein levels of POC5 (Figure 4G-H and Supplementary Figure S4H-I). Overexpression of HDAC3 enhanced the protein level of POC5 (Figure 4I). Importantly, KDELR2 knockdown abolished the increased expression level of POC5 mediated by HDAC3 overexpression (Figure 4J). Then, we found that KDELR2 upregulated POC5 protein levels without affecting its mRNA level (Supplementary Figure S4J). Intriguingly, we observed that lactacystin, a 20S proteasome inhibitor, recovered the suppressed POC5 protein levels induced by KDELR2 knockdown in breast cancer cells (Figure 4K and Supplementary Figure S4K), suggesting that KDELR2 regulated POC5 protein stability via the proteasome pathway. Moreover, ubiquitination analysis revealed that KDELR2 or HDAC3 knockdown increased the polyubiquitination of POC5 protein in HEK293T cells in the presence of lactacystin (Figure 4L-M). To determine whether KDELR2 promotes breast cancer cell growth by regulating POC5, we introduced lentivirus overexpressing POC5 into MDA-MB-231 cells with KDELR2 knockdown (Supplementary Figure S4L). As expected, forced expression of POC5 recovered the retarded cell growth induced by shKDELR2 (Figure 4N). Moreover, forced expression of POC5 released the cells arrested at G1 phase by shKDELR2, leading to a similar cell cycle distribution as the control cells (Figure 4O and Supplementary Figure S4M). Collectively, these findings indicated that POC5 was involved in KDELR2-regulated cell cycle and cell growth in cancer cells. Thus, we uncovered a novel pathway by which the HDAC3/KDELR2 axis accelerates the cell cycle of human breast cancer cells by protecting the centrosomal protein POC5 from proteasomal degradation.
3.5 KDELR2 was critical for HDAC3-regulated breast cancer progression in vivo
To address whether KDELR2 is important for HDAC3-enhanced tumor growth in vivo, we knocked down KDELR2 in MDA-MB-231 cells stably overexpressing HDAC3 for xenograft experiments (Supplementary Figure S5A). As a result, HDAC3 overexpression dramatically enhanced tumor growth and tumor mass compared to the empty vector group, and KDELR2 silencing abolished the increased tumor growth induced by HDAC3 overexpression without any influence on the body weight of mice, indicating that KDELR2 is involved in HDAC3-mediated tumor growth in vivo (Figure 5A-C and Supplementary Figure S5B). qRT-PCR analysis also confirmed that the transcriptional level of KDELR2 was increased in HDAC3-overexpressing xenograft tumor tissues (Figure 5D). Western blotting analysis using tumor tissue lysates revealed that, consistent with our in vitro results, overexpression of HDAC3 increased the protein levels of KDELR2 and POC5, and KDELR2 silencing attenuated HDAC3 overexpression-enhanced POC5 upregulation in xenograft tumors (Figure 5E). Taken together, these data demonstrate that KDELR2 is critical for HDAC3-regulated tumor growth in vivo.
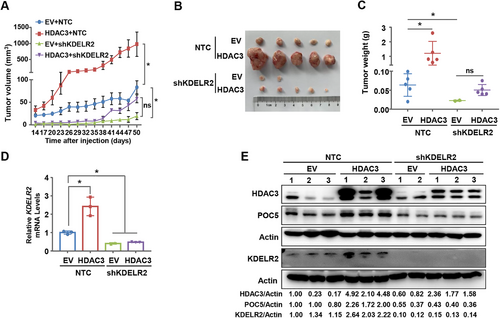
KDELR2 was critical for HDAC3-regulated breast cancer progression in vivo. A-C. MDA-MB-231 cells overexpressing empty vector or 3×Flag-HDAC3 were further infected with lentivirus expressing NTC or shKDELR2. The cell lines above were subcutaneously injected into female nude mice (n = 5 for each group). Tumor sizes were measured starting from 14 days after inoculation (A). At the end of the experiment, the tumors were extracted and compared (B-C). The data are presented as the mean ±SEM. *, P < 0.05; ns, not significant. D-E. qRT-PCR analysis of the mRNA levels of KDELR2 (D) and Western blotting analysis of the expression of HDAC3, KDELR2 and POC5 (E) in xenograft tumor tissues. The data are presented as the mean ±SEM. *, P < 0.05. β-Actin served as the loading control.
4 DISCUSSION
HDACs play pivotal roles in cell survival, homeostasis, and proliferation by regulating the deacetylation of histone proteins [19, 28]. Aberrant expression of HDACs has been observed in various types of cancers, such as hepatocellular carcinoma, colon cancer, breast cancer and prostate cancer and correlates with tumorigenesis and patient survival [29, 30]. Jiang et al. [31] assessed the efficacy and safety of the combination of the oral HDACi tucidinostat with exemestane in a large population of postmenopausal patients with advanced hormone receptor-positive breast cancer. They found that this combination improved the progression-free survival of patients with advanced hormone receptor-positive HER2-negative breast cancer compared to the placebo plus exemestane group, indicating that tucidinostat plus exemestane could represent a new clinical therapeutic option for these patients. While the inhibition of HDACs is currently marked as a feasible cancer therapeutic strategy, the underlying mechanisms remain largely unclear. Here, taking advantage of class I HDACi, which are effective against breast cancer cells at nanomolar doses in vivo, we revealed a novel mechanism by which HDAC3 drives breast cancer progression. By comparing gene expression profiles of breast cancer cells in the presence or absence of the pan-class I/II HDACI TSA, class I HDACi TDPA and FK228, and the control group, we observed dramatically decreased expression of KDELR2 in HDACI-treated breast cancer cells in vivo and in vitro. Then, we identified that among class I/II HDACs, only HDAC3 could increase KDELR2 expression by upregulating its transcription factor CREB1. Importantly, restricting KDELR2 expression in breast cancer cells abolished the cell growth advantage and tumorigenesis mediated by HDAC3 overexpression in vivo and in vitro. We further demonstrated that the mechanism driving HDAC3-KDELR2 axis-linked tumorigenesis was related to the enhanced expression of the centrosomal protein POC5 and its interaction with KDELR2 (Figure 6).
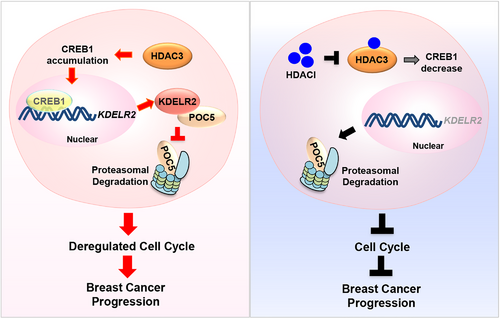
KDELR2 is required for HDAC3-regulated breast cancer progression via accelerating cell cycle.
(Left) HDAC3 transactivates KDELR2 via CREB1 and activated KDELR2 in turn protects the centrosomal protein POC5 from proteasomal degradation to accelerate cell cycle progression and breast cancer progression. (Right) HDACI treatment inactivates HDAC3, leading to the decreased expression of KDELR2 and proteasomal degradation of POC5. Consequently, blockade of HDAC3/KDELR2 axis results in cell cycle arrest and retards tumor growth in vivo.
HDAC3, a member of the Zn2+-dependent class I HDAC isoforms, has been reported recently to play crucial roles in oocyte maturation, lymphatic valve formation, chondrogenesis and diseases, such as obesity and cancer processes [29, 32]. Recent studies have revealed the correlation between the dysregulation of HDAC3 and tumorigenesis [33, 34]. However, the potential functions of HDAC3 in cancer progression and its underlying mechanisms are not well understood, given the conflicting observations in different types of tumors. Selective inhibition of HDAC3 led to growth suppression of both PTEN-deficient and SPOP-mutated prostate cancer cells through the dual inhibition of AKT-mTOR and AR signaling [35]. HDAC2 and HDAC3 induced apoptosis and retarded cell migration and invasion. Liver-specific knockout of Hdac3 led to hepatocellular carcinoma resulting from the accumulation of damaged DNA and protumorigenic transcriptional activity [36]. HDAC3 inhibition decreased PD-L1 expression in pancreatic cancer cells via its transcription factor STAT3 [37]. However, studies using B-cell lymphomas found that HDAC3 repressed the transcription of PD-L1 by directly binding to the PD-L1 promoter and increasing DNA methyltransferase 1 protein levels to indirectly suppress PD-L1 transcription [38]. In our study, HDAC3 significantly facilitated breast cancer cell growth and tumorigenesis, and more importantly, markedly increased expression levels of HDAC3 were observed in tumor tissues compared to adjacent breast tissues. These results indicate the different roles of HDAC3 in the regulation of different types of cancer. In this regard, our results might prompt more detailed studies of HDAC3 during cancer progression, which will aid in the development of new clinical therapeutic strategies targeting HDAC3.
KDELR2 is known as a regulator that mediates traffic transport from the Golgi complex to the ER. A recent study indicated that KDELR2-regulated Golgi secretion is necessary for cellular invasion and metastasis and that the inhibition of KDELR2 decreases lung cancer metastasis [16]. KDELR2 has been shown to be highly expressed in glioblastoma tissues and to regulate the phosphorylation levels of mTOR (Ser2448), which promotes glioblastoma tumorigenesis [14]. Moreover, knockdown of KDELR2 by siRNA in glioma cells reduced the protein level of CCND1 [15], but the mechanism was not determined. KDELR2 has been identified as a binding partner of FAM134B, which acts as a cancer suppressor in colon cancer, and the knockdown of FAM134B reduces the expression of KDELR2, suggesting the potential function of KDELR2 as a tumor suppressor in colon cancer [39]. This evidence points to the importance of KDELR2 in regulating the cancer progression; however, the roles of KDELR2 in different types of tumor cells and the underlying mechanism remain largely elusive. We found KDELR2 expression was significantly increased in breast cancer cells. Notably, aberrantly expressed KDELR2 in tumor cells accelerates the cell cycle to promote breast cancer progression by binding to and stabilizing the centrosomal protein POC5. POC5 has been reported to be essential for the centriole elongation and the impaired expression of POC5 led to G1 phase arrest of the cells [25], revealing its critical role for cell cycle. Therefore, we identified a novel mechanism by which KDELR2 regulates the cell cycle by binding to and increasing the protein level of POC5, linking a Golgi-ER traffic transport protein to a critical centrosomal protein during cancer progression.
Although HDACi provide new hope for cancer therapy, the limited understanding of their underlying mechanisms in different types of cancers has hampered their clinical applications. Thus, an understanding of the molecular mechanisms by which the dysregulation of HDACs influences cancer progression may hold the key to advancing applications of HDACi in a wide range of therapeutic settings.
5 CONCLUSIONS
Our study revealed a novel role for HDAC3 in regulating breast cancer progression by transactivating KDELR2, which might contribute not only to our understanding of the correlation of HDACs and tumorigenesis but also to the development of cancer therapeutic strategies targeting KDELR2.
DECLARATIONS ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study was approved by the Ethics Committee of the University of Science and Technology of China. Prior written informed consent from the patients, as well as a study approval from the Institutional Research Ethics Committee of Anhui Provincial Hospital was obtained for the use of the included patient material. The studies were conducted in accordance with ethical guidelines of the Declaration of Helsinki. All animal studies were conducted with approval from the Animal Research Ethics Committee of the University of Science and Technology of China.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Original data from RNA-Seq are available in the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/; accession numbers: GSE172182). The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.
COMPETING INTERESTS
The authors declare that they have no competing interests.
FUNDING
This work was supported in part by grants from the National Key R&D Program of China (2018YFA0800300 and 2018YFA0107103), the National Natural Science Foundation of China (82072656 and 91957203), the Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2017ZT07S054), and the Fundamental Research Funds for the Central Universities (2019MS133).
AUTHORS' CONTRIBUTIONS
P.G., X.Z., and H.Z. conceived and supervised this study. H.W., X.Z., and P.G. designed the experiments. H.W., W.M., X.L., H.L., K.L., Y.W., Z.Y., L.S., Z.H., T.P. and Z.Z. performed the experiments and analyzed the data. E.Y.C. contributed key reagents. P.G., X.Z. and H.W. wrote the manuscript. All the authors read and approved the final manuscript.
ACKNOWLEDGEMENTS
Not applicable.