RNF114 suppresses metastasis through regulation of PARP10 in cervical cancer cells
Abbreviations
-
- HCC
-
- hepatocellular carcinoma.
-
- PARP10
-
- poly(ADP-ribose) polymerase 10
-
- RNF114
-
- RING finger 114
Dear editor,
ADP-ribosylation is a post-translational modification catalysed by poly(ADP-ribose) polymerases (PARPs) and plays important roles in diverse biological processes, such as DNA repair, chromatin remodelling, telomere homeostasis, transcription, chromosome segregation, cell proliferation, and cell death [1, 2]. While much is known about the cellular roles of PARPs that catalyse polyADP ribosylation, the function of the PARPs that catalyse monoADP ribosylation is substantially less understood.
PARP10 is a mono-ADP-ribosyltransferase and has been found to be involved in diverse biological processes, such as nuclear factor-κB (NF-κB) signalling and tumor development [3, 4]. More recently, we identified PARP10 as a tumor metastasis suppressor [5]. We demonstrated that PARP10 interacted with and mono-ADP-ribosylates Aurora A, and then inhibited its kinase activity, thereby regulating Aurora A downstream signalling to suppress tumor cell epithelial-mesenchymal transition processes and tumor metastasis. Moreover, the expression level of PARP10 was lower in intrahepatic metastatic hepatocellular carcinoma (HCC) than in corresponding primary HCC and adjacent non-tumor tissues [5]. Although PARP10 plays important roles in physiological and pathological processes, the regulation and modification mechanisms of PARP10 remain largely unclear.
In addition to Aurora A, RING finger 114 (RNF114), a ubiquitin E3 ligase [6], was also found in the PARP10 complex [5], suggesting that RNF114 might also be a PARP10-interacting protein. Then, we performed exogenous and endogenous reciprocal immunoprecipitation assays to verify the interaction between PARP10 and RNF114. As expected, exogenously expressed HA-RNF114 interacted with S-Flag-SBP (SFB)-PARP10, and vice versa (Supplementary Figure S1). Endogenous PARP10 also interacted with RNF114, as determined by a co-immunoprecipitation assay using an anti-PARP10 antibody (Figure 1A). Interestingly, the G888W mutant of PARP10, which abolished the enzymatic activity of PARP10 [7], did not interact with RNF114 (Supplementary Figure S2A), suggesting that the enzymatic activity of PARP10 is required for its association with RNF114. Moreover, His-PARP10 purified from E. coli did not interact with SFB-RNF114 expressed in 293T cells. However, after auto-mono-ADP-ribosylation in vitro, the mono-ADP-ribosylated His-PARP10 efficiently pulled down SFB-RNF114 (Supplementary Figure S2B), suggesting that the interaction between RNF114 and PARP10 is dependent on the auto-mono-ADP-ribosylation of PARP10.
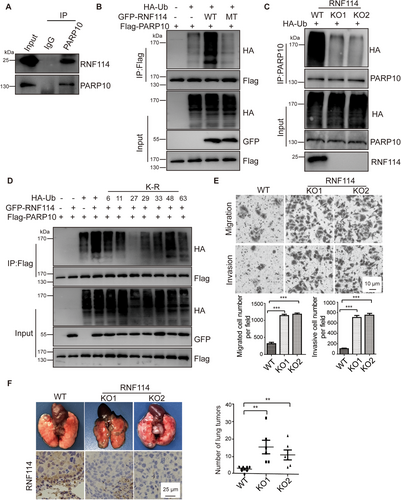
Both PARP10 and RNF114 are enzymes, raising the possibility that RNF114 is a substrate of PARP10 or PARP10 is a substrate of RNF114. To test this hypothesis, we first performed in vitro mono-ADP-ribosylation assays using biotin-nicotinamide adenine dinucleotide+ (NAD+) as a donor to examine whether PARP10 can modify RNF114 via mono-ADP-ribosylation. As shown in Supplementary Figure S3, PARP10, not RNF114, was ADP-ribosylated by PARP10, suggesting that RNF114 may not be a PARP10 substrate in vitro. Therefore, we performed ubiquitination assays to determine whether PARP10 is an RNF114 substrate. As shown in Figure 1B, co-expression of wild-type GFP-RNF114, not C29G/C32G mutant of RNF114 that abolishes the E3 ligase activity of RNF114, dramatically increased the ubiquitination level of Flag-PARP10 in 293T cells, suggesting that RNF114 may mediate the ubiquitination of PARP10 in a manner dependent on the RNF114 E3 ligase activity. Previously, we have demonstrated that PARP10 deficiency suppressed the metastasis of HeLa cells [5]. Then, we generated RNF114-knockout HeLa cells by CRISPR-Cas9 editing and used these cells to examine the effect of RNF114 deficiency on PARP10 ubiquitination. As shown in Figure 1C, the ubiquitination level of PARP10 was dramatically decreased in RNF114-deficient cells compared with the level in wild-type cells, further suggesting that RNF114 promotes the ubiquitination of PARP10. However, the total level of PARP10 was not obviously changed in RNF114-deficient cells compared with wild-type cells (Figure 1C), suggesting that the ubiquitination of PARP10 mediated by RNF114 may not regulate its stability. Next, an in vivo ubiquitination assay was performed by using a panel of ubiquitin mutants with single K/R mutations at the sites of the seven lysine residues in ubiquitin. As shown in Figure 1D, compared with wild-type and other lysine mutants, the ubiquitin K27R mutant abolished the ubiquitination of PARP10 mediated by RNF114, suggesting that RNF114 ubiquitinates PARP10 through K27-linked ubiquitination. More recently, porcine RNF114 was also found to inhibit classical swine fever virus replication via the K27-linked polyubiquitination of classical swine fever virus nonstructural 4B (CSFV NS4B) protein although RNF114 has been found to promote substrate degradation [8], indicating that RNF114-mediated polyubiquitination is complicated and RNF114 may regulate both substrate stability and activity in different biological processes.
Emerging evidence suggests that K27-linked polyubiquitination plays important roles in the regulation of substrate function instead of stability [9]. Therefore, we examined whether RNF114-mediated ubiquitination of PARP10 affected its function. First, we examined the mono-ADP-ribosylation levels of PARP10 and its substrate Aurora A in wild-type and RNF114-knockout cells by pull-down assay using the glutathione S-transferase (GST)-Macro 2 domain of PARP14, which specifically associates with mono-ADP-ribosylated proteins [10]. As shown in Supplementary Figure S4A, PARP10 and Aurora A were pulled down by the GST-Macro 2 domain from wild-type cells more efficiently than by that from the RNF114-knockout cells. We also used the anti-ADP-ribose antibody to examine the ADP-ribosylation of PARP10 and Aurora A in wild-type and RNF114-knockout cells. As shown in Supplementary Figure S4B, compared with wild-type cells, the ADP-ribosylation of PARP10 and Aurora A was decreased in RNF114-knockout cells, suggesting that RNF114 regulates the ADP-ribosylation of PARP10 and its downstream substrates. Next, we examined the phosphorylation levels of Aurora A and its substrate Akt in wild-type and RNF114-knockout cells. As shown in Supplementary Figure S5, compared with those in wild-type cells, the phosphorylation levels of Aurora A and Akt were increased in RNF114-knockout cells, suggesting that RNF114 deficiency increases Aurora A kinase activity. Taken together, these results indicated that RNF114 positively regulated PARP10 activity, which may in turn inhibit Aurora A activity and downstream signalling.
Recently, we demonstrated that PARP10 is a tumor metastasis suppressor [5]. RNF114 regulates PARP10 activity, raising the possibility that RNF114 may also play important roles in cancer cell migration and tumor metastasis. To test this hypothesis, we performed cell migration and invasion assays using wild-type and RNF114-deficient HeLa cells. As shown in Figure 1E, compared with these processes in wild-type cells, the extent of the migration and invasion of RNF114-deficient cells were dramatically increased. Re-expressed wild-type RNF114, not C29G/C32G mutant of RNF114, suppressed the migration of RNF114-deficient HeLa cells (Supplementary Figure S6), suggesting that RNF114 regulates cancer cell migration depending on its ubiquitin ligase activity. Moreover, when wild-type HeLa cells and two RNF114-deficient cell lines were injected into BALB/c nude mice, the number and size of the lung tumors derived from RNF114-deficient HeLa cells were significantly increased comparing with the tumors established by wild-type HeLa cells, suggesting that RNF114 deficiency promotes tumor metastasis in vivo (Figure 1F). In addition, PARP10 overexpression inhibited the migration and invasion of RNF114-deficient cells, indicating that RNF114-mediated cell migration and invasion might be dependent on the regulation of PARP10 (Supplementary Figure S7). We analyzed the correlation between PARP10/RNF114 expression and overall survival of cervical cancer patients in the Human Protein Atlas database (http://www.proteinatlas.org/). As shown in Supplementary Figure S8, the overall survival was longer in cervical cancer patients with high expression of PARP10 or RNF114 than in those with low expression of PARP10 or RNF114, although the difference was not significant (P = 0.059). These results further indicated that PARP10 and RNF114 might function as cervical cancer suppressors.
Our findings identify RNF114 as a novel functional regulator of PARP10 and provide evidence of crosstalk between the components of K27-linked polyubiquitination and mono-ADP ribosylation. Our observations also provide a new insight into the physiological function of RNF114 in tumor metastasis and make RNF114 as a potential tumor metastasis suppressor.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The animal work was performed after approval was granted by the Ethics Review Committee for Animal Experimentation of Fudan University. Approval number: BE1909.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
All data generated or analyzed during this study are included in this published article and its supplementary information files. The materials used in the current study are available from the corresponding author on reasonable request.
AUTHOR'S CONTRIBUTIONS
Y.Z. designed the study. Y.Z., X.L. and L.W. conducted most of the experiments, and wrote the manuscript. Y.L., J.L., H.F., F.Z. and Y.W. analyzed the data and contributed to the manuscript completion. J.M. revised the manuscript. JW conceived the study, provided overall guidance, and contributed to the manuscript completion.
FUNDING
The National Natural Science Foundation of China (81972712 to J. W. and 81502106 to H.M.) supported this work.
CONFLICT OF INTEREST
The authors declare no competing financial interests.




