Gene signatures associated with genomic aberrations predict prognosis in neuroblastoma
Abstract
Background
Neuroblastoma (NB) is a heterogeneous disease with respect to genomic abnormalities and clinical behaviors. Despite recent advances in our understanding of the association between the genetic aberrations and clinical features, it remains one of the major challenges to predict prognosis and stratify patients for determining personalized therapy in this disease. The aim of this study was to develop an effective prognosis prediction model for NB patients.
Methods
We integrated diverse computational analyses to define gene signatures that reflect MYCN activity and chromosomal aberrations including deletion of chromosome 1p (Chr1p_del) and chromosome 11q (Chr11q_del) as well as chromosome 11q whole loss (Chr11q_wls). We evaluated the prognostic and predictive values of these signatures in seven NB gene expression datasets (the number of samples ranges from 94 to 498, with a total of 2120) generated from both RNA sequencing and microarray platforms.
Results
MYCN signature was a more effective prognostic marker than MYCN amplification status and MYCN expression. Similarly, the Chr1p_del score was more prognostic than Chr1p status. The activity scores of MYCN, Chr1p_del and Chr11q_del were associated with poor prognosis, while the Chr11q_wls score was linked to good outcome. We integrated the activity scores of MYCN, Chr1p_del, Chr11q_del, and Chr11q_wls and clinical variables into an integrative prognostic model, which displayed significant performance over the clinical variables or each genomic aberration alone.
Conclusions
Our integrative gene signature model shows a significantly improved forecast performance with prognostic and predictive information, and thereby can be served as a biomarker to stratify NB patients for prognosis evaluation and surveillance programs.
Abbreviations
-
- AUC
-
- under the curve
-
- Chr11q_del
-
- chromosome 11q deletion
-
- Chr11q_wls
-
- chromosome 11q whole loss
-
- Chr17q_gain
-
- chromosome 17q gain
-
- Chr17q_wgn
-
- chromosome 17q whole gain
-
- Chr1p_del
-
- chromosome 1p deletion
-
- INRG
-
- International Neuroblastoma Risk Group
-
- INSS
-
- International Neuroblastoma Staging System
-
- LOH
-
- loss of heterozygosity
-
- NB
-
- neuroblastoma
1 BACKGROUND
Neuroblastoma (NB), the most common solid pediatric tumor, accounts for approximately 7%-10% of pediatric malignancies and 15% of all children cancer death [1-3]. NB displays extensive heterogeneity both biologically and clinically [1, 4], ranging from spontaneous regression in neonates to fatal progression in older children despite intensive multimodality treatment. According to the International Neuroblastoma Risk Group (INRG) classification system by incorporating clinical factors such as the patient age at diagnosis and tumor stage (the International Neuroblastoma Staging System, INSS), as well as tumor histopathology, DNA index, and MYCN gene status as prognostic factors, patients with NB can be classified into three risk groups (low-risk, intermediate-risk, and high-risk) and be given therapeutic stratification accordingly [5]. Generally, children with NB diagnosed before the age of 1.5 years have a better prognosis than those diagnosed at an older age, and patients at the early stage (stage 1, 2 or 4S) have a better outcome than those at the advanced stage (stage 3, 4) [1, 3, 6]. NB patients with MYCN amplification tend to have a worse prognosis than those without MYCN amplification [4, 6]. Although the prognosis of NB has been greatly improved, less than 40% of patients with high-risk NB achieved long-term survival even with multiple, aggressive therapies and the recent inclusion of immunotherapy with antibodies targeted at NB-specific antigens, such as GD2 [7-9]. Due to the broadly divergent outcomes, it remains a challenge to develop methods for prognostic prediction so that the most effective therapeutic and management strategies can be designed for diverse subsets of NB.
Accumulated evidence has indicated that many genomic abnormalities, such as loss of chromosome 1p and 11q, gain of chromosome 17q, and MYCN gene amplification, are powerful prognostic markers and are strongly associated with clinical outcome [10-12]. However, multiple controversial findings on the prognostic value of gene markers have been observed. For example, MYCN amplification does not automatically trigger the increased expression of MYCN [13, 14], while the overexpression of MYCN also occurs in the absence of MYCN amplification in NB patients [13-15], although MYCN amplification is usually considered to be linked to high expression of MYCN; segmental chromosomal aberrations, such as chromosome 11q deletion (Chr11q_del) and chromosome 17q gain (Chr17q_gain), are associated with poor prognoses, while numerical chromosomal aberrations, such as chromosome 11q whole loss (Chr11q_wls) and chromosome 17q whole gain (Chr17q_wgn), tend to be associated with good prognoses [16-18]. This highlights the importance of validating the predictive ability of these existing genetic markers and integrating newly identified prognostic markers into more sophisticated algorithms to further improve the predictive accuracy for clinical applications.
Gene expression profiling has been widely used for biomarker identification. Several well-documented studies have demonstrated that multigene signatures predict the prognosis of NB patients more effectively than traditional individual prognostic marker [19-22]. Gene signatures were usually defined as a set of selected genes identified from statistical analysis. However, many of these signatures are limited by their low reproducibility with poor predictive performance across different datasets; due to interpatient heterogeneity and complex interrelationships between genes. Notably, bioinformatics approaches have been developed to identify significant gene sets or pathway profiles involved in biological processes and diseases, including cancer, based on gene expression data, which allow for quantitatively measuring the activation of intracellular signaling pathways [23-25]. Our group has previously defined a gene signature to quantify p53 pathway activity and verified that the p53 gene signature was a better predictor of recurrence-free survival compared with p53 mutation status in patients with early-stage lung adenocarcinoma [26] based on a rank-based statistical algorithm [27]. Additionally, multiple studies have shown that chromosome aberrations can lead to dramatic expression changes of the genes within the abnormal region and can be detected by gene expression profiles [28-30]. Unfortunately, for NB only a few copy number variation datasets have been generated, which makes it difficult to investigate the prognostic association of chromosomal loss/gain events in NB. In contrast, a large number of gene expression datasets have been published with large sample size and high-quality clinical information including patient outcome information. Therefore, it is desirable to develop computational methods to infer the activity of signaling pathway and the status of chromosome abnormalities for prognosis prediction based on these gene expression data.
In this study, we integrated different computational methods to infer MYCN activity and the statuses of several critical chromosome band aberrations, including chromosome 1p deletion (Chr1p_del), Chr11q_del, and Chr11q_wls, by using gene expression data and attempted to develop a computational model for predicting prognosis of NB patients.
2 RESOURCES AND METHODS
2.1 Neuroblastoma gene expression datasets
A summary of the NB gene expression datasets used in this study is provided in Supplementary Table S1. Raw data are available from the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo; accession: GSE73517, GSE62564, GSE49710), the ArrayExpress database (www.ebi.ac.uk/arrayexpress/; accession: E-TABM-38, E-MTAB-179), the PREdiction of Clinical Outcomes from Genomic Profiles (PRECOG) portal (https://precog.stanford.edu/; accession: Berwanger_NB), and the Children's Oncology Group (COG) Therapeutically Applicable Research to Generate Effective Treatments (TARGET) data matrix portal (http://target.nci.nih.gov/dataMatrix/TARGET_DataMatrix.html; accession: TARGET_ NB).
The GSE73517 dataset was used to define gene signatures associated with MYCN amplification and five chromosome band aberrations (Chr1p_del, Chr11q_del, Chr11q_wls, Chr17q_gain, and Chr17q_wgn). The dataset contain 105 NB samples, providing gene expression profiles, patient clinical information and the statuses of MYCN amplification, Chr1p, Chr11q, and Chr17q [31].
The prognostic value of the resulting signatures was assessed in 6 NB gene expression datasets: the GSE62564, GSE49710, E-TABM-38, E-MTAB-179, Berwanger_NB, and TARGET_NB datasets. Among them, the GSE62564 and GSE49710 datasets contain gene expression profiles for the same set of 498 NB patients measured by two different methods with RNA sequencing and microarray analyses, respectively. The other datasets are independent microarray datasets, containing data from 198, 478, 94, and 249 patients, respectively. The data on MYCN amplification status are available for GSE49710, E-TABM-38, and E-MTAB-179 datasets. The data on Chr1p status are only available for the E-MTAB-179 dataset. In this article, we labelled datasets according to the first author of the published studies, as followings: Henrich for GSE73517 [31], Su for GSE62564 [32], Wang for GSE49710 [33], Westermann for E-TABM-38 [34], Oberthuer for E-MTAB-179 [35], Berwanger for Berwanger_NB [36], and Pugh for TARGET_NB [37].
2.2 Defining MYCN gene signature and chromosome aberration signatures based on the Henrich dataset
The MYCN amplification gene signature was defined based on the Henrich dataset by comparing differential expression of genes between MYCN amplification and non-amplification samples while considering confounding variables [26, 27]. For each gene, a logistic regression model was constructed using MYCN amplification as the response variable. We denote P as the probability of MYCN amplification, and the model is formulized as ∼ gene + age + gender + stage, where the predictor variables are the gene expression level, age at the time of diagnosis (age), gender, and INSS stage (stage 1, 2, 3, 4, or 4S). By applying these models to the Henrich data, we estimated the coefficients (β) and their statistical significance (P) for all genes. Given (β, P) values for all genes; we subsequently defined the MYCN signature, which was represented as a pair of weight profiles, and, each containing the weights of all genes. For each gene i, and were assigned in the following way: = − and = −. To avoid extreme values, the weights were trimmed at 10, and then transformed into a value within [0, 1], by subtracting the minimum value and then dividing by the range. If a gene i is more significantly up-regulated in MYCN amplification versus non-amplification samples, it will associate with a higher but of zero. Conversely, a more significantly down-regulated gene will associate with a higher but of zero. We selected all genes with > 0.5 and > 0.5 as up- and down-regulated genes in MYCN amplification versus non-amplification samples, respectively.
According to the statuses of Chr1p, Chr11q, and Chr17q provided by the Henrich dataset, differentially expressed chromosome subbands associated with each chromosome deletion/gain event were obtained (P < 0.05). We then merged all genes located in the chromosome subbands as gene sets that can recapitulate the genomic statuses of Chr1p_del, Chr11q_del, Chr11q_wls, Chr17q_gain, and Chr17q_wgn, respectively. After filtering out genes that do not reflect chromosome aberration event (e.g., genes not down-regulated or even up-regulated in Chr11q_del vs. Chr11q normal) and shared by two incompatible events for the chromosome band (e.g., down-regulated genes in both Chr11q_del and Chr11q_wls, and up-regulated genes in both Chr17q_gain and Chr17q_wgn), we defined five gene sets for Chr1p_del, Chr11q_del, Chr11q_wls, Chr17q_gain, and Chr17q_wgn. If the number of genes in a set was less than 10, the gene set was halted. Finally, we obtained three final band gene sets to infer the statuses of Chr1p_del, Chr11q_del, and Chr11q_wls.
2.3 Calculating patient-specific scores for MYCN activity and chromosome aberration events
Given the expression profiles for a group of NB samples, patient-specific MYCN activity scores were calculated based on the MYCN gene signature by using a previously published method called binding association with sorted expression (BASE) [27].
To calculate chromosome aberration scores, we used a method similar to the one for calculating the MYCN activity score, but with minor modification. Specifically, we replaced the weight profile pair (for MYCN) with a single profile containing all genes, with = 1 if gene i is include in the gene set associated with a specific chromosome aberration (e.g., Chr1p_del), and otherwise = 0. These chromosome band aberration-specific profiles were used as inputs to the BASE algorithm, which output patient-specific scores that indicate the likelihood of the corresponding chromosome aberration event.
2.4 Predicting patient survival using MYCN, Chr1p_del, Chr11q_del, and Chr11q_wls scores
Cox proportional hazard models were constructed to examine the effectiveness of the activity scores of MYCN, Chr1p_del, Chr11q_del, and Chr11q_wls in predicting patient survival. Patient samples were dichotomized into two groups by using the median of each activity score as a cutoff. Statistical significance was calculated by using Mann-Whitney U test. Correlation coefficient between MYCN expression and the MYCN score was calculated by using Spearman correlation, after MYCN expression was log transformed. A univariate Cox regression model was performed to examine difference in survival between the resultant two patient subgroups. A multivariate Cox regression model was used to investigate the effect of each score on survival after adjusting for potential confounding variables such as INSS stage, age, gender, and so on. The Kaplan-Meier method was used to plot survival curves. The difference between the survival curves of different groups was compared with significance being estimated by using a log-rank test. The R package “survival” (https://cran.r-project.org/web/packages/survival/index.html) was used to perform statistical analyses. Specifically, the “coxph” function was used to construct Cox proportional hazard models, the “survfit” function was used to create Kaplan-Meier survival curves, and the “survdiff” function was used to compare the difference between two survival curves. For each analysis, we performed it in all the datasets as long as the variables used for the analysis are available.
2.5 Comparing prognostic models with different combinations of variables
Clinical variable combination (age, gender, INSS stage), the activity scores of MYCN, Chr1p_del, and Chr11q_del or Chr11q_wls as well as MYCN amplification status or Chr1p status were combined as the various models. The C-index was applied to compare the power of the different models. For each model, the C-index was estimated based on 10-fold cross validation. Specifically, the dataset was randomly divided into 10 groups of equal size. Each time, 9 groups were used as the training set, and then survival was predicted in the remaining group based on the trained models. The groups were rotated 10 times so that all samples were predicted exactly once, and then the C-index for each model was calculated. We repeated this procedure for 100 times (each time with a different randomization) to estimate the mean and the standard deviation of C-index for each model.
2.6 MYCN target genes
MYCN target genes were downloaded from the Molecular Signatures Database (MSigDB, http://www.broadinstitute.org/gsea/msigdb/index.jsp), Section C3 “motif gene sets”, subsection “transcription factor targets (TFT)”, which provides a total of 271 MYCN predicted target genes (Supplementary Table S2).
3 RESULTS
3.1 Schematic overview of this study
To establish a powerful outcome prediction model for NB patients, we performed a series of analyses as illustrated in Figure 1. First, we defined gene signatures to recapitulate the genomic statuses of the MYCN gene and several chromatin aberration events (Chr1p_del, Chr11q_del, Chr11q_wls, Chr17q_gain, and Chr17q_wgn) by using the Henrich dataset, which provides gene expression profiles for 105 NB patients and the corresponding molecular marker statuses. Since the numbers of genes in the sets for Chr17q_gain and Chr17q_wgn were fewer than 10, these gene sets were discarded.
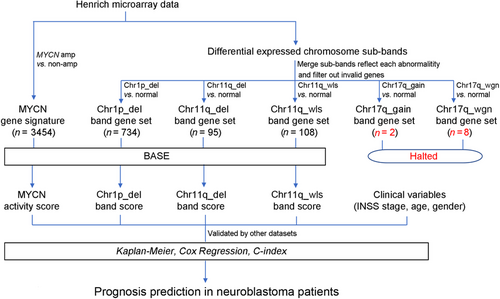
Next, we used the BASE algorithm to calculate the scores that can infer the status of MYCN, Chr1p_del, Chr11q_del, and Chr11q_wls in a patient-specific manner based on the corresponding gene signatures. We then investigated the prognostic value of these scores, specifically, in patients with or without MYCN amplification. We also assessed the association of these scores with clinical variables (age, gender, and INSS stage).
Finally, a series of prediction models were established by integrating the clinical variables and the activity score of MYCN, Chr1p_del, Chr11q_del, or Chr11q_wls to stratify patients and predict prognosis in NB.
3.2 Association of MYCN gene signature with MYCN activity
Statistical analysis of the Henrich dataset identified 671 and 2783 genes that were up- and down-regulated in MYCN amplification versus non-amplification samples by using > 0.5 and > 0.5 as cutoff, respectively (Supplementary Tables S3 and S4). As shown on Figure 2a, we also investigated the overlap of these genes with the 271 MYCN target genes that were available from the MSigDB C3 TFT (Supplementary Table S2). As expected, genes up-regulated in MYCN amplification samples were enriched for MYCN regulatory targets.
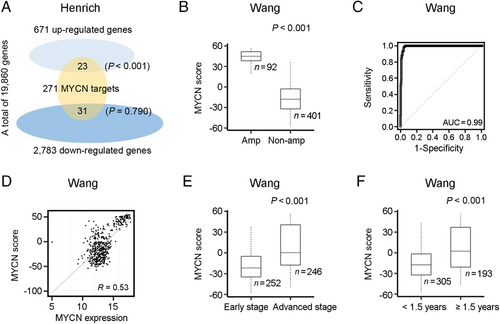
Next, we investigated whether the MYCN score can inform the MYCN status in other datasets. The MYCN scores of MYCN amplification samples were significantly higher than those of MYCN non-amplification samples in the Wang (Figure 2b), Henrich (Supplementary Figure S1a), Su (Supplementary Figure S1b), Westermann (Supplementary Figure S1c), and Oberthuer datasets (Supplementary Figure S1d). The MYCN score can effectively discriminate MYCN amplification from non-amplification samples with high accuracy, as indicated by the area under the curve (AUC) score of 0.99, 0.99, 0.94, and 0.98 in the Wang (Figure 2c), Su (Supplementary Figure S1e), Westermann (Supplementary Figure S1f), and Oberthuer datasets (Supplementary Figure S1g), respectively. These results supported the MYCN score as a biomarker with strong ability to discriminate NB patients with MYCN amplification from those without.
We then tested the relationship between the MYCN score and MYCN expression. As shown on Figure 2d, the MYCN score was positively correlated with MYCN expression, although the correlation was moderate (R = 0.53) in the Wang dataset. Similar correlations were recognized in the other six datasets (Supplementary Figure S1h-m).
We next examined the association between the MYCN score and the clinical variable combination. In the Wang dataset, we observed a significantly higher MYCN score in patients at advanced stages compared with those at early stages (P < 0.001) (Figure 2e), and a higher MYCN score in patients aged ≥ 1.5 years compared with those aged < 1.5 years (P < 0.001) (Figure 2f). Similar results were found in other datasets (Supplementary Figure S2a-j). No significant association between the MYCN score and gender was found in all datasets (Supplementary Figure S2k-o).
Overall, these results indicated that the MYCN score was indicative of MYCN amplification, and was associated with clinical stage and age, two established prognostic clinical variables.
3.3 Prognostic values of MYCN score, MYCN amplification status, MYCN expression, and clinical variables
Using the Wang dataset, we dichotomized patients into MYCN score-low and -high groups using the median as the cutoff and compared their survival. As shown on Figure 3a, patients with high MYCN score had significantly shorter survival than those with low MYCN score (hazard ratio [HR] = 3.3, 95% confidence interval [CI] = 2.4-4.6, P < 0.001). Similar results were observed in other datasets (Supplementary Figure S3a-e). Following that, we separated patients into two groups based on their MYCN amplification status and examined the predictive power of the MYCN score in each group. We observed a significant difference in survival between MYCN score-high and -low patients in the MYCN non-amplification group (HR = 2.4, 95% CI = 1.7-3.6, P < 0.001) (Figure 3b), but not in the MYCN amplification group (HR = 0.8, 95% CI = 0.5-1.3, P = 0.372, data not shown) in the Wang dataset. These results were further confirmed in other datasets (Supplementary Figure S3f-k). These results suggest that the MYCN activity score contains prognostic value only in patients without MYCN amplification.
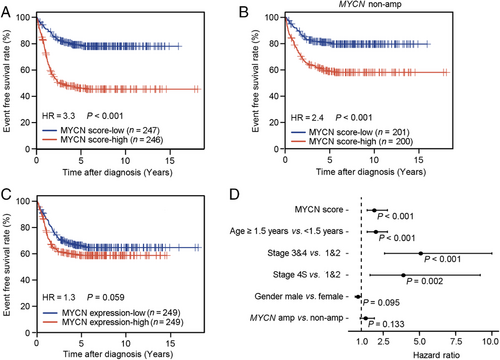
In contrast, the association between MYCN mRNA level and prognosis was not significant in the Wang dataset (HR = 1.3, 95% CI = 1.0-1.8, P = 0.059) (Figure 3c). Similarly, MYCN expression was not associated with survival in the Westermann and Pugh datasets (Supplementary Figure S3l-m), but was significantly associated with survival in the Su, Oberthuer, and Berwanger datasets (Supplementary Figure S3n-p). Thus, MYCN expression cannot be used as a convincing prognostic marker.
We next investigated whether the MYCN score was prognostic after adjusting for clinical variables. Specifically, we constructed a multivariate Cox regression model for the Wang dataset that included dichotomized MYCN score, age, INSS stage, gender, and MYCN status. As shown on Figure 3d, the MYCN score remained significant for predicting patient survival even after considering clinical variables and MYCN status (HR = 2.4, 95% CI = 1.7-3.6, P < 0.001). To further confirm this, the same analysis was performed in the other four datasets. As shown in Supplementary Table S5, the MYCN score was still prognostic in the Su, Oberthuer, and Pugh datasets. In the Westermann dataset, high MYCN score was also associated with short survival (HR = 2.0, 95% CI = 1.0-3.9), but after adjustment it was not significant (P = 0.056), presumably due to the relatively small sample size of this dataset.
Taken together, these results indicated that the MYCN score is a more effective prognostic marker than MYCN amplification status and MYCN expression.
3.4 Association of the Chr1p_del score with patient survival
Other than the MYCN amplification status, several chromosomal band loss/gain events have been observed with high frequency in NB, for which, however, associations with prognosis have not been investigated systematically. As a proof of concept, we developed gene signatures to capture these chromosome events and examined their prognostic values across different gene expression datasets. Specifically, we defined gene signatures for Chr1p_del, Chr11q_del, and Chr11q_wls (Supplementary Table S6) based on the Henrich dataset. We first investigated whether the Chr1p_del score can inform Chr1p status using the Oberthuer dataset, which provided Chr1p status determined by fluorescence in situ hybridization (FISH) and/or polymerasechain reaction (PCR) [35]. As shown on Figure 4a, we observed a significant increase of the Chr1p_del score in patients with loss of heterozygosity at chromosome 1p (LOH1p) compared with those with normal Chr1p status (P < 0.001), with an AUC of 0.95 when the Chr1p_del score was used to discriminate 40 with LOH1p from 268 samples (Figure 4b). The Chr1p_del score was significantly higher in patients at advanced stages than in those at early stages (P < 0.001) (Figure 4c), and higher in patients aged ≥ 1.5 years than in those aged < 1.5 years (P < 0.001) (Figure 4d). No significant change of the Chr1p_del score was found between male and female patients (data not shown). Similar results were obtained in other datasets (data not shown). These results indicated that the Chr1p_del score derived from the gene signature could correctly reflect Chr1p status in NB samples and were associated with clinical variables such as INSS stage and age.
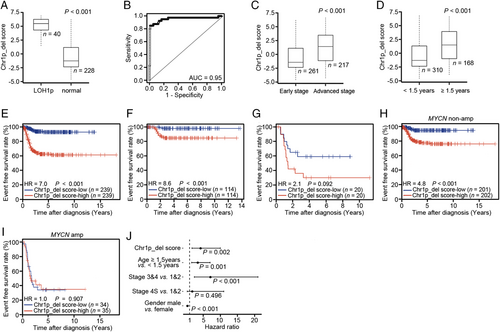
We further tested whether the Chr1p_del score is predictive of patient survival. In the Oberthuer dataset, patients with high Chr1p_del score had significantly shorter survival than those with low Chr1p_del score (HR = 7.0, 95% CI = 3.9-12.0, P < 0.001) (Figure 4e). Similar results were observed in other datasets (Supplementary Figure S4a-e). We then divided patients into two groups on the basis of their reported Chr1p status and examined the association of the Chr1p_del score with patient survival in each group. Interestingly, we observed a significant difference between Chr1p_del score-low and -high patients in the Chr1p normal group (HR = 8.6, 95% CI = 2.0-38.0, P < 0.001, Figure 4f), despite no difference in the LOH1p group (P = 0.092, Figure 4g). This indicated that the Chr1p_del score could capture not only the Chr1p status but also the downstream effect of Chr1p_del that might also be caused by other mechanisms, e.g., focal deletion of driver genes in Chr1p or epigenetic modifications of this chromosome region.
Because LOH1p usually associates with MYCN amplification in NB patients [1, 3, 6, 10, 12], we investigated the association between the Chr1p_del score and MYCN status of NB patients. Patients with MYCN amplification showed significantly higher Chr1p_del scores than those without MYCN amplification (data not shown). Next, we separated patients into MYCN amplification and non-amplification groups and performed survival analyses in each group. As shown on Figure 4h-i, high Chr1p_del score was linked to short survival for patients without MYCN amplification (HR = 4.8, 95% CI = 2.3-9.9, P < 0.001), but no significant association was found for patients with MYCN amplification in the Oberthuer dataset. Similar results were observed in other datasets (Supplementary Figure S4f-k), suggesting that the Chr1p_del score provides additional prognostic value only in patients without MYCN amplification.
We next evaluated the prognostic value of the Chr1p_del score in the Oberthuer dataset by using a multivariate Cox regression model after considering the following variables: dichotomized Chr1p_del score, age, INSS stage, and gender. As shown on Figure 4j, the Chr1p_del score was significantly associated with patient survival even after adjusting for the clinical variables (HR = 4.2, 95% CI = 1.7-10.0, P = 0.002). Similar results were obtained in other datasets (Supplementary Table S7). We also assessed the prognostic values of the Chr1p_del score in MYCN amplification and non-amplification -groups separately. As shown in Supplementary Figure S4l and Supplementary Table S7, the Chr1p_del score remained significant for predicting survival even after considering clinical variables and MYCN status in the Oberthuer (HR = 2.8, 95% CI = 1.1-7.4, P = 0.035) and Wang datasets (HR = 2.1, 95% CI = 1.5-3.1, P < 0.001), despite no significance in the Su dataset (HR = 1.4, 95% CI = 1.0-1.9, P = 0.071).
Altogether, the Chr1p_del score can be used to infer Chr1p status and is predictive of patient survival. Its prognostic value remains significant after considering the indicated clinical variables and MYCN amplification status in NB patients.
3.5 Inference of Chr11q status based on a combination of two gene signatures
Both Chr11q_del and Chr11q_wls occur in NB patients with high frequencies in the Henrich dataset (Supplementary Table S1), and, interestingly, the former is associated with poor prognosis but the latter with good prognosis, compared to normal Chr11q [16-18]. Notably, loss of Chr11q has been shown to be associated with survival only in patients lacking MYCN amplification [3, 38, 39]. Here, we defined two non-overlapping gene sets to infer Chr11q status and investigated their prognostic values in patients without MYCN amplification.
Given that the Chr11q_del and Chr11q_wls scores were derived from the same chromosome region, we tested whether they can reflect their own statuses and distinguish one another. To do this, we divided patients into four groups based on the Chr11q_del and Chr11q_wls scores by using median of the two scores as cutoffs, and then examined whether the two scores can inform reported Chr11q status in the Henrich dataset. As shown on Figure 5a, patients with high Chr11q_del but low Chr11q_wls scores were more likely to have Chr11q_del, whereas those with high Chr11q_wls but low Chr11q_del scores were more likely to have Chr11q_wls. When both scores were low, the patients were more likely to have normal Chr11q. Some patients had high Chr11q_del and Chr11q_wls scores, which indicate abnormal Chr11q status, but it is unclear whether this is associated with a whole loss or a deletion event.

We then examined the prognostic power of the Chr11q_del and Chr11q_wls scores for patients without MYCN amplification in the Wang dataset. High Chr11q_del score was associated with significantly shorter survival (HR = 1.8, 95% CI = 1.3-2.6, P = 0.001) (Figure 5b), whereas high Chr11q_wls score was related to markedly longer survival (HR = 0.3, 95% CI = 0.2-0.5, P < 0.001) (Figure 5c) among these patients. Similar results were observed in other datasets (Supplementary Figure S5a-f). These results indicated that Chr11q_del was linked to poor outcome, whereas Chr11q_wls was associated with good outcome in NB patients without MYCN amplification.
By combining the Chr11q_del and Chr11q_wls scores, we divided patients into four groups using the median of the two scores as cutoffs and tested the survival predictive power of the two scores. The survival curves of patients without MYCN amplification in the four groups could be distinguished (Figure 5d and Supplementary Table S8) in the Wang dataset. Similar results were found in other datasets (Supplementary Figure S5g-i). As shown in Supplementary Table S8, Chr11q_del was linked to a worse prognosis, while Chr11q_wls was related to a better prognosis in patients without MYCN amplification when compared to Chr11q normal. Notably, Chr11q_wls was associated with a better outcome compared to Chr11q_del and Chr11q_normal. Taken together, these results indicated that the scores of Chr11q_del and Chr11q_wls inferred by our algorithm were predictive of patient survival and could be distinguished from one another.
3.6 An integrated model for prognostic prediction
After validating the contribution of each score of individual genomic alterations to prognosis prediction, we investigated whether the combination of these scores could serve as a more powerful predictor. To this end, we developed a series of predictor models that incorporated different combinations of clinical variables and genomic signature scores (the MYCN, Chr1p_del, Chr11q_del, or Chr11q_wls score), with or without the status of genetic aberration (MYCN status, Chr_1p status).
Figure 6 and Supplementary Table S9 show a comparison of the discrimination powers of different models by using C-index in the Wang dataset. All models that included clinical variables and each score of genomic aberration were significantly better than either clinical variables alone or each score of genomic aberration alone. Moreover, the model that comprised the MYCN score and clinical variables outperformed the model with MYCN status and clinical variables. More importantly, the combination of the MYCN, Chr1p_del, Chr11q_del, or Chr11q_wls score with clinical variables, named here as the final models, were significantly improved over either individual predictor. Notably, the C-index for the two final models were significantly increased by 5.3% and 4.6% respectively compared with the model involving only clinical variables, although they changed only slightly compared with models that contained clinical variables and the MYCN score. Similar results were observed in other datasets (Supplementary Table S9), with a significant improvement in the performance of the Chr1p_del score over that of the Chr1p status in the Oberthuer dataset, in which only the Chr1p status is available.
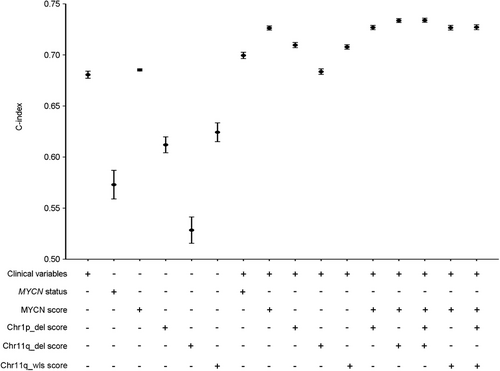
Overall, the combination of the MYCN, Chr1p_del, and Chr11q_del or Chr11q_wls score with clinical variables exhibits significant improvement over either the clinical variables or genomic aberrations alone.
4 DISCUSSION
In the present study, we developed a computational framework to define gene signatures to infer several clinically important genomic aberrations in NB, including the MYCN gene amplification, Chr1p_del, Chr11q_del, and Chr11q_wls. In addition to cytogenetic assays, these signatures can be measured by using traditional molecular biological methods such as PCR and FISH, or by using transcriptomic profiling analyses such as microarray and RNA-seq. Here, we defined a gene signature to calculate the MYCN activity score in NB samples by using our previously described algorithm [26, 27]. Our rationale is that the increased regulatory activity of MYCN caused by MYCN gene amplification will be eventfully revealed by the alteration of expression of downstream genes. More importantly, the gene signature will capture not only the MYCN amplification event but also the changes of MYCN activity caused by other mechanisms (e.g., DNA methylation).
To define gene signatures associated with chromosome aberration events, we applied a different strategy. When a specific chromatin band is deleted, we expect to observe the trend of down-regulation of all genes located in this band with the following exceptions: 1) genes that are intensively regulated by cellular signaling or posttranscriptional events, and 2) genes that are distant from the hot spots associated with the given chromatin band deletion or gain event. Based on this rationale, we defined a gene signature for a chromosome aberration event as the set of genes that are located in the given chromosome regions (i.e., Chr1p_del, Chr11q_del, and Chr11q_wls).
Importantly, our results demonstrated that the MYCN score and chromosome band scores of Chr1p_del, Chr11q_del, and Chr11q_wls inferred by our algorithm reflected the statuses of these genomic features and were predictive of the prognosis of NB patients. Specifically, the MYCN score showed significant superiority in prognostic prediction compared with MYCN status and MYCN expression, in consistent with a previous report [15]. The Chr1p_del score showed a similar pattern when compared with Chr1p status. Notably, the Chr11q_del and Chr11q_wls scores were distinguishable from one another, although both were derived from different sets of genes located in Chr11q. In line with previous reports [16-18], the segmental alteration Chr11q_del was associated with short survival, while the numerical abnormality Chr11q_wls was linked to long survival. Importantly, the MYCN score and the Chr1p_del score remain prognostic after adjusting clinical variables. Hence, these genomic feature-based signatures represent effective biomarkers that confer to more accurate prognostic prediction in NB. Although gene expression data were used to define gene signatures associated with chromosome aberration events in the present study, a recently published study, which contained a large CNV profile of 556 NB cases, gave rise to our concerns [40]. We analyzed the data by using Cox regression analysis and found that 34 chromosomal loss/gain events exhibited significant prognostic values (details shown in the Supplementary Table S10). More high-quality CNV datasets with large sample size will be expected. The complex association between MYCN amplification and other genomic alterations also should be noted [41-44]. Particularly, Chr1p_del, Chr11q_del, and MYCN amplification are not independent genomic events. Chr1p_del, which presents in 25%-35% of NB patients, was controversial as an independent prognostic indicator because of its close relation with MYCN amplification, whereas Chr11q_del, which occurs in 35%-45% of NB patients as an adverse clinical feature with older age of disease onset, late disease stage, and unfavorable histology, is inversely associated with MYCN amplification and Chr1p_del [1, 3, 10, 12, 45, 46]. However, these events are not mutually exclusive, patients with Chr1p_del without MYCN amplification [3] and those with concurrent MYCN amplification and Chr11q deletion [47] have been reported and were also found in the Henrich dataset (Supplementary Table S11). The presence of Chr1p_del without MYCN amplification was also observed in the Oberthuer dataset (Supplementary Table S11). We thus evaluated the prognostic power of the scores of MYCN, Chr1p_del, Chr11q_del, and Chr11q_wls in NB patient groups stratified based on MYCN amplification status. We observed that the scores of MYCN, Chr1p_del, Chr11q_del, and Chr11q_wls inferred from our analysis were predictive of patient survival in MYCN normal samples. This indicates that they provide additional prognostic information to MYCN status and other clinical variables.
Considering that NB is a molecularly defined disease [16, 17, 19, 22, 48-51], various genetic features have been integrated to increase the sensitivity and specificity for risk assessment and outcome prediction. Here, we evaluated a series of prediction models that integrated inferred genomic scores of MYCN, Chr1p_del, Chr11q_del, and Chr11q_wls, the status of genetic aberration (MYCN status or Chr1p status), and clinical variables. Incorporation of the genetic biomarker, MYCN amplification status, achieved more accurate prognostic prediction compared with the model solely based on clinical variables, which can be further improved by using the MYCN score inferred from gene signature analysis. The addition of more inferred genomic scores resulted in slight increase of prediction accuracy. This might be explained by the fact that these genomic events are associated with MYCN amplification and that when two genetic aberrations occur together, the prognostic impact of MYCN amplification is dominant [3].
Given the poor prognosis of patients with stage 4 or high risk disease, there are still greater challenges in the need for alternative treatment strategies. We investigated whether our proposed model can be used to evaluate the prognosis of stage 4 patients in the Pugh dataset, in which the majority of patients were at stage 4 with a rate of 86.7% (216/249). We found that the improvement, for which the C-index for the two final models were significantly increased by 5.1% and 5.9% respectively compared with the model involving only clinical variables in the Pugh dataset, was similar to that of other independent databases (Supplementary Table S9), suggesting that our proposed models are also suitable for stage 4 patients. In addition, several previously reported gene signatures built for stage 4 or high-risk patients are of great concern. Kinome-27, a specific kinome gene signature, was able to identify a group of high-risk NB patients and could be used as a potential biomarker of targeted therapy [52]. A four-gene signature, in which these genes are direct p53 transcriptional targets, exhibited significant prognostic value in high-risk NB patients [53]. The future work is to extend these findings to pre-clinical or early-phase clinical trials in large independent cohorts of NB patients.
There were several limitations in the present study. Firstly, the effect of treatment strategy on survival could not be analyzed due to the lack of therapeutic information in the selected datasets. In fact, induction and consolidation chemotherapy protocols vary greatly in different countries and studies and may affect patient survival. Secondly, sample selection bias should be considered. For example, the proportions of patients with MYCN amplification were 18.5%, 18.5%, 10.6%, and 14.4% in Su, Wang, Westermann, and Oberthuer datasets, which were lower than the widely cited estimates of 25%-33% [4]. Hence, our models need to be further validated in larger and independent patient cohorts.
5 CONCLUSIONS
Taken together, we developed a bioinformatics algorithm to identify gene signatures that can reflect MYCN activity and several chromosomal aberrations, Chr1p_del, Chr11q_del, and Chr11q_wls, in NB on the basis of gene expression profiles. We also presented a prognosis prediction model by identifying and integrating the MYCN activity score, the scores of Chr1p_del, Chr11q_del, or Chr11q_wls, and clinical variables for NB. Our proposed model displayed significantly improved prognostic prediction and may provide insights for further therapeutic interventions and surveillance programs for NB patients.
ACKNOWLEDGEMENTS
Not applicable
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable
CONSENT FOR PUBLICATION
Not applicable
AVAILABILITY OF DATA AND MATERIAL
All data generated or analyzed during this study are included in this published article and its supplementary information files.
COMPETING INTERESTS
The authors declare that they have no competing interests.
FUNDING
This work was supported by the American Cancer Society (IRG-82-003-30), the National Center for Advancing Translational Sciences of the National Institutes of Health (KL2TR001088) and the National Natural Science Foundation of China (81201543).
AUTHORS' CONTRIBUTIONS
Xiaoyan He analyzed the data and wrote the paper. Chao Qin and Yanding Zhao developed the computational analyses. Chao Cheng designed and supervised the study, and revised the paper. Lin Zou and Hui Zhao provided the biomedical support. All authors contributed to the data interpretation, read and approved the final manuscript.




