Guanine quadruplex formation by RNA/DNA hybrid analogs of Oxytricha telomere G4T4G4 fragment
Abstract
Using circular dichroism spectroscopy, gel electrophoresis, and ultraviolet absorption spectroscopy, we have studied quadruplex folding of RNA/DNA analogs of the Oxytricha telomere fragment, G4T4G4, which forms the well-known basket-type, antiparallel quadruplex. We have substituted riboguanines (g) for deoxyriboguanines (G) in the positions G1, G9, G4, and G12; these positions form the terminal tetrads of the G4T4G4 quadruplex and adopt syn, syn, anti, and anti glycosidic geometries, respectively. We show that substitution of a single sugar was able to change the quadruplex topology. With the exception of G4T4G3g, which adopted an antiparallel structure, all the RNA/DNA hybrid analogs formed parallel, bimolecular quadruplexes in concentrated solution at low salt. In dilute solutions (∼0.1 mM nucleoside), the RNA/DNA hybrids substituted at positions 4 or 12 adopted antiparallel quadruplexes, which were especially stable in Na+ solutions. The hybrids substituted at positions 1 and 9 preferably formed parallel quadruplexes, which were more stable than the nonmodified G4T4G4 quadruplex in K+ solutions. Substitutions near the 3′end of the molecule affected folding more than substitutions near the 5′end. The ability to control quadruplex folding will allow further studies of biophysical and biological properties of the various folding topologies. © 2008 Wiley Periodicals, Inc. Biopolymers 89: 797–806, 2008.
This article was originally published online as an accepted preprint. The “Published Online” date corresponds to the preprint version. You can request a copy of the preprint by emailing the Biopolymers editorial office at [email protected]
INTRODUCTION
Guanine-rich molecules of DNA are known to form quadruplexes [reviewed in Refs.1-4]. All quadruplexes are based on guanine tetrads held together by Hoogsteen hydrogen bonds; however, quadruplex structures differ by the number of associated strands, their mutual orientation, topology, and conformation of loops. The orientation of strands is either antiparallel (every strand has antiparallel-oriented neighboring strands or has one parallel and one antiparallel neighbor) with half of the guanines oriented syn around the glycosidic bond and the other half anti2, 5, 6 or parallel with all guanines in the anti conformation.2, 4, 7-9 A mixed parallel/antiparallel, so called (3 + 1) quadruplex, has also been observed10-13 and intensively studied in connection with human telomere DNA.14-18 In this quadruplex, three strands are parallel and one antiparallel; seven guanine glycosidic angles are anti and five are syn.
The guanine quadruplexes are biologically relevant structures. Quadruplex forming sequences occur not only in telomeres but also in human genomic DNA, especially in gene promoters.4, 12, 19-24 Quadruplex formation plays important roles in control of gene, and particularly oncogene, expression.21, 22, 25, 26 Quadruplex formation in the Oxytricha nova telomere formed by (G4T4) repeats inhibits the activity of telomerase,27 the enzyme responsible for the maintenance of telomere length. Mammalian telomeric functions may be altered by quadruplex-stabilizing ligands.28-30 Because of their important functions, specifically in maintenance of telomeres in cancerous cells, quadruplexes have become targets for a development of specific chemotherapeutic drugs.21, 28, 30-32 Guanine quadruplexes are also formed by RNA.33-41 All RNA quadruplexes observed so far are parallel. This may be a consequence of the preference of ribonucleotides with a C3′-endo sugar pucker to adopt the anti orientation of the glycosidic bond.42
Here we have studied the conformational effects of ribo-G substitutions into G quadruplex forming DNA oligonucleotide. We have used G4T4G4, a prototype DNA sequence whose quadruplex properties have been studied in detail. NMR43-45 and X-ray diffraction46, 47 were used to show that G4T4G4 adopts a bimolecular, antiparallel quadruplex of the basket type in the presence of both, K+ or Na+. We have studied various RNA/DNA hybrid analogs of this DNA quadruplex to see if the substitution of selected deoxyriboguanines with riboguanines alters the orientation of strands. The same question was earlier raised by Shafer's group in two studies39, 40 in which multiple ribo-substitutions, at positions which normally form the syn conformation in the original DNA quadruplex, had destabilized the formation of antiparallel quadruplexes. Our study has gone further by examining the DNA quadruplex containing only single (or two) ribo-substitutions of an original G with the anti or syn geometry. The single substitution is shown to produce similar consequences as the multiple substitutions described earlier, to different extent at different positions. Circular dichroism (CD) spectroscopy was used to evaluate the structures formed. The method sensitively reflects the orientation of bases and the stacking in interactions of neighboring G-quartets14, 48, 49 and in this way it discriminates among parallel and antiparallel quadruplex types.
RESULTS
In this work, we analyzed G4T4G4 and six RNA/DNA hybrid analogs (Table I) in which selected deoxyriboguanines (G) were replaced by riboguanines (g) in positions 1, 4, 9, or 12 (see Figure 1). These guanines form the terminal tetrads of the quadruplex. The substitutions in positions 1 and 9 replace guanines that adopt the syn conformation in the G4T4G4 quadruplex, whereas substitutions in positions 4 and 12 replace guanines that have the anti conformation.
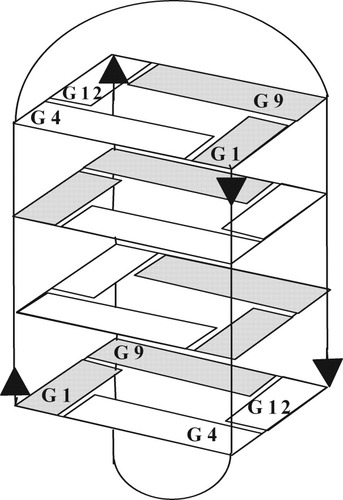
A schematic representation of the quadruplex of G4T4G4. Syn guanines are shaded.
| Sequence | Modified syn/anti G in G4T4G4 | Tm in 45 mM Na+ (°C) | Tm in 45 mM K+ (°C) | TmK+ − TmNa+ (°C) | |
|---|---|---|---|---|---|
| G4T4G4 | 5′-GGGGTTTTGGGG-3′ | — | 57 | 48 | −9 |
| gG3T4G4 | 5′-gGGGTTTTGGGG-3′ | 1 syn | 36 | 53 | 17 |
| G4T4gG3 | 5′-GGGGTTTTgGGG-3′ | 9 syn | 39 | 53 | 14 |
| gG3T4gG3 | 5′-gGGGTTTTgGGG-3′ | 1,9 syn | 31 | 56 | 25 |
| G3gT4G4 | 5′-GGGgTTTTGGGG-3′ | 4 anti | 42 | 47 | 5 |
| G4T4G3g | 5′-GGGGTTTTGGGg-3′ | 12 anti | 58 | 48 | −10 |
| G3gT4G3g | 5′-GGGgTTTTGGGg-3′ | 4,12 anti | 44 | 48 | 4 |
- Lower case letters mean ribonucleotides, capital case letters mean deoxyribonucleotides. Syn guanines in the G4T4G4 quadruplex are underlined. The Tm values were determined from the maxima of the first derivation of the melting curves monitored at 256 nm.
Low Salt Solutions of the Dodecamers
After dilution from the stock solution into low salt solution (1 mM Na phosphate, 0.3 mM EDTA, pH 7), G4T4G4 had a CD spectrum with a positive, long-wavelength band at 290 nm and a small negative band at 260 nm (see Figure 2). This spectrum corresponds to an antiparallel guanine quadruplex, which was present in the concentrated stock solution of G4T4G4. This quadruplex was not stable under these low salt conditions upon 100x dilution of the sample. The characteristic CD bands disappeared after denaturation (5 min at 90°C) and the quadruplex did not reform even after several days at 4°C (see Figure 2). In contrast to G4T4G4, and with the exception of G4T4G3g, all the RNA/DNA hybrids adopted parallel quadruplexes after dilution from the stock solution characterized by CD spectrum with a dominant positive band around 260 nm (see Figure 2). After denaturation of the samples, only G4T4gG3, with a ribonucleotide substituted for a syn G on the 3′side of the molecule (the four nucleotides 5′ of the T4 loop are designated as a 5′side and the four after the loop as the 3′side), yielded, with a slow kinetics, a slight increase of the positive 260 nm CD band. The other two hybrid dodecamers with substituted syn G (i.e., gG3T4G4 and gG3T4gG3) remained unstructured even after several days at 4°C (Figure 2, left panels). Dodecamer hybrids substituted at normally anti G slowly formed antiparallel quadruplexes after denaturation (Figure 2, right panels). Thus, these hybrids more readily formed quadruplex than unmodified G4T4G4 (see Figure 2).
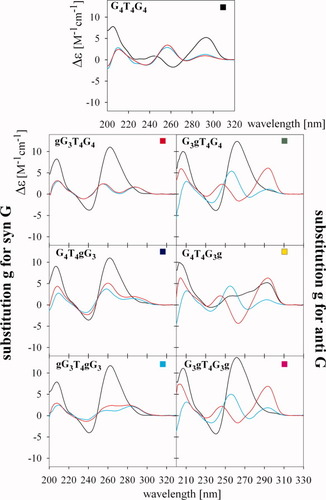
CD spectra of the indicated RNA/DNA dodecamers measured in 1 mM Na phosphate +0.3 mM EDTA, pH 7. Black, before denaturation; cyan, immediately after denaturation/renaturation; red, 3 days after denaturation. The left column contains oligonucleotides with syn G replaced by g, the right column contains oligonucleotides where anti G were replaced by g.
In our experience, RNA migrates distinctly slower than DNA in a nondenaturing polyacrylamide gel. Therefore, we used a set of markers, including DNA and RNA homoduplexes (CG)3 and (cg)3, (CGA)4 and (cga)4, the RNA/DNA heteroduplex (cga)4·(TCG)4, and the DNA duplex (CGA)4·(TCG)4 (where a lower case letter indicates a ribonucleotide), to monitor the mobility of quadruplexes. Figure 3A is an example of separation under low salt conditions before denaturation. All the parallel quadruplexes (see Figure 2) were bimolecular and ran slightly slower than the bimolecular antiparallel quadruplex of G4T4G4. G4T4G3g, which did not form parallel quadruplex after dilution from the stock solution as shown by CD spectroscopy, ran at the same position as G4T4G4 (Figure 3A).
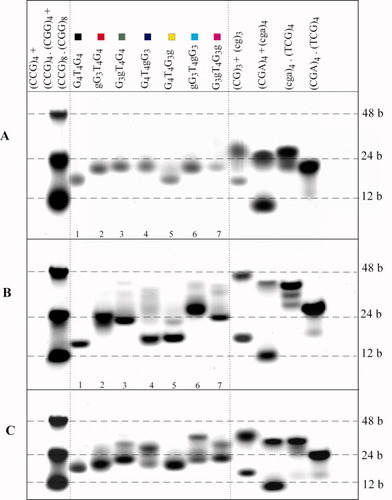
Native polyacrylamide electrophoreses of the studied RNA/DNA dodecamers. (A) 1 mM Na phosphate, 0.3 mM EDTA, pH 7.25; (B) 0.5x Robinson-Britton buffer, pH 7.3 + NaCl up to the final 40 mM Na+ concentration (0.124M H3PO4, 0.124M CH3COOH, 0.124M H3BO3, 0.038M NaOH, 0.002M NaCl); (C) 5.6 mM potassium phosphate, 0.3 mM EDTA, pH 7.2. In (A) samples were not denatured prior to loading. In (B) and (C) the samples were denatured by heating at 5 min at 90°C in 1 mM Na phosphate +0.3 mM EDTA, then the buffer was added and the samples were kept 3 days in a cold room prior to loading.
Quadruplexes of the Dodecamers Stabilized by Na+ Ions
G4T4G4 provided the known CD spectrum with large amplitudes characteristic of an antiparallel quadruplex in the presence of Na+ ions (Figures 4 and 5A). Even low Na+ concentrations stabilized antiparallel quadruplexes formed by the dodecamers containing g substituted for anti G (Figure 4, right panels). The largest amplitudes were observed for G4T4G3g. The amplitudes of the negative band of G3gT4G4 and G3gT4G3g were lower than the amplitude of G4T4G4 and less than twice the amplitudes of G4T4G3g (Figure 5A). The G4T4G3g was more inclined than G4T4G4 to form quadruplex at low Na+ concentrations. Both quadruplexes, however, melted similarly (Figures 5B and 5C and Table I). Formation of Na+-induced quadruplexes of G3gT4G4 and of G3gT4G3g was nearly identical (Figure 5A). They also melted similarly at both wavelengths studied (Figures 5B and 5C), but at distinctly lower temperatures than quadruplexes of G4T4G4 and G4T4G3g. They also displayed much lower hypochromicity. Thus, formation and stability of the quadruplex of the dodecamer with g substituted for anti G on the 5′side of the molecule was not much influenced by a simultaneous substitution of the anti G on the 3′end.
CD spectra of the studied RNA/DNA dodecamers measured at various Na+ concentrations. Cyan, 1 mM Na phosphate, 0.3 mM EDTA, pH 7 after denaturation; green, 15 mM Na+; blue, 45 mM Na+; and red, 105 mM Na+. All spectra were measured after equilibrium was attained, 24 or 48 h after salt addition.
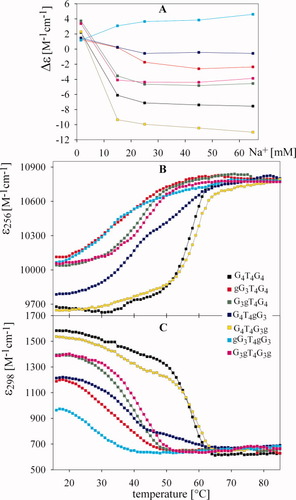
Dependences of Δε on Na+ ion concentration and melting curves of the RNA/DNA dodecamers. (A) CD dependences monitored at 272 nm for G4T4gG3 and at 262 nm for all other dodecamers (spectra were measured one or two days after salt addition); melting curves measured in 45 mM Na+ and monitored at (B) 256 and (C) 298 nm. Key: black, G4T4G4; red, gG3T4G4; green, G3gT4G4; blue, G4T4gG3; yellow, G4T4G3g; cyan, gG3T4gG3; and magenta, G3gT4G3g. Dodecamer concentrations were ∼0.6 mM in nucleoside.
NaCl also induced formation of the antiparallel quadruplex forms of gG3T4G4 and G4T4gG3 (Figure 4, left panels, Figure 5A). CD amplitudes of these dodecamers with syn Gs substituted by g were very low. Thus, the g for syn G substitution destabilized the antiparallel quadruplex markedly, as reflected in the decreased absorption at 298 nm50 (Figure 5C). The dodecamer gG3T4gG3 with both 1 and 9 syn Gs substituted did even form a parallel quadruplex at the highest Na+ concentration (Figure 4, left bottom panel). The destabilization of the antiparallel quadruplex by the two g substitutions is especially obvious from the temperature dependence of the absorption monitored at 298 nm. It was only for this dodecamer that Tm values determined from ε256 and ε298 differed significantly (Tm 256 = 31°C, Tm 298 = 27°C). With all other dodecamers the temperature dependences measured at 298 nm were roughly inversions of the dependences measured at 256 nm.
G4T4gG3 showed a considerable hypochromism (Figure 5B), which indicates that the low amplitudes of its CD spectrum were the result of the simultaneous presence of parallel and antiparallel quadruplexes. This conclusion is supported by the two-step melting observed at both wavelengths (Figures 5B and 5C). CD spectroscopy revealed that increasing temperature induced a transition of the antiparallel quadruplex, which prevails at low temperatures, to parallel quadruplex. The positive band at 260 nm became dominant in the CD spectrum at temperatures higher than 50°C (not shown). The same antiparallel-parallel quadruplex transition is thus induced by increasing temperature with G4T4gG3 as by increasing Na+ concentration in the case of gG3T4gG3. The Tm of the antiparallel quadruplex form of G4T4gG3 was only slightly higher than that of gG3T4G4, which did not transform into the parallel quadruplex. The Tm of the parallel quadruplex of G4T4gG3 was much higher (55°C) than that of the antiparallel form (Figures 5B and 5C). The substitution of a syn G with a ribonucleotide at the 3′side of the dodecamer stabilized the parallel quadruplex orientation more than the 5′-end substitution.
The NaCl-stabilized quadruplexes showed substantially different gel migrations than the structures formed at low ionic strength (Figure 3B). The dominant bands of the 3′-side substituted dodecamers G4T4gG3 and G4T4G3g migrated similarly, although their CD spectra were substantially different (Figure 4, middle panels). This pair of dodecamers migrated only slightly more slowly than G4T4G4. Similar migration (Figure 3B), but different CD spectra were also observed for the pair of dodecamers substituted on the 5′side (gG3T4G4 and G3gT4G4, Figure 4, upper panels) and for molecules substituted on both sides (gG3T4gG3 and G3gT4G3g, Figure 4, bottom panels). Thus, not structural features, but the side of the molecule substituted with a ribonucleoside affected the migration rate. Ribonucleosides on the 5′side of the molecule hindered the migration relative to that of the unsubstituted molecule.
Quadruplexes of the Dodecamers Stabilized by K+ Ions
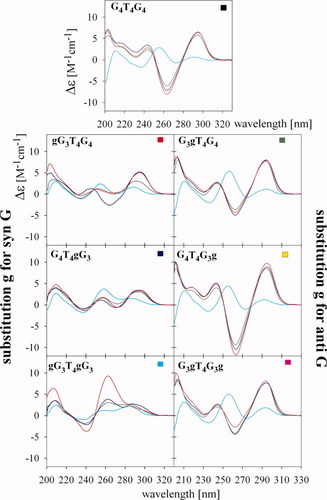
The CD spectra of G4T4G4 are considerably different in K+ and Na+ containing solutions. It was, however, reported that the topology is the same in both salts.45, 46 Though G4T4G4 forms an antiparallel quadruplex in K+ solution, substitution of a single syn G by g stabilized the parallel quadruplex (Figure 6, left panels). The largest positive amplitudes were exhibited by gG3T4gG3 (Figure 7A); this dodecamer with both 1 and 9 positions substituted formed a parallel quadruplex even in NaCl (150 mM) (see Figure 4). The lowest amplitudes were exhibited by gG3T4G4. The shoulders present on the longer wavelength sides of the dominant positive bands at 260 nm of both gG3T4G4 and G4T4gG3 dodecamers indicate the simultaneous presence of parallel and antiparallel quadruplex arrangements.
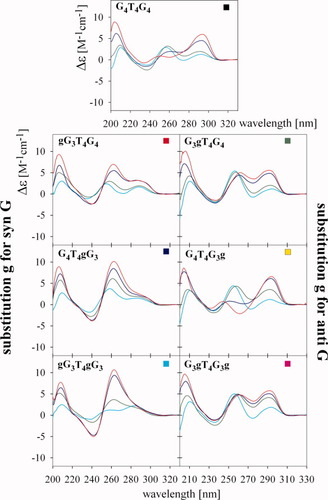
CD spectra of the studied RNA/DNA dodecamers measured at various K+ concentrations. Cyan, 1 mM Na phosphate, 0.3 mM EDTA, pH 7 after denaturation; green, 5.6 mM K+ immediately after K+ addition; blue, 5.6 mM K+ after two days; and red, 105 mM K+, measured two days after K+ addition.
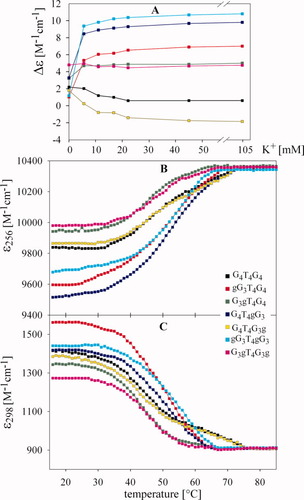
Dependences of Δε on K+ ion concentration and melting curves of the RNA/DNA dodecamers. (A) CD dependences monitored at 258 nm for G3gT4G3g and G4T4G4 and at 263 nm for all other dodecamers (spectra were measured one or two days after salt addition); melting curves measured in 45 mM K+ and monitored at (B) 256 and (C) at 298 nm. Key: black, G4T4G4; red, gG3T4G4; green, G3gT4G4; blue, G4T4gG3; yellow, G4T4G3g; cyan, gG3T4gG3; and magenta, G3gT4G3g. Dodecamer concentrations were ∼0.6 mM in nucleoside.
G4T4G3g with g substituted for anti G yielded essentially the same CD spectrum as G4T4G4 in the presence of K+ (i.e., a spectrum characteristic of the antiparallel quadruplex but lacking the negative band) (see Figure 6). The dodecamers G3gT4G4 and G3gT4G3g with substituted anti G on either the 5′end or on both sides of the molecule folded into a mixture of parallel and antiparallel quadruplexes where the antiparallel form was the dominant one (see Figure 6). This conclusion followed not only from CD but also from gel migration (Figure 3C). It was shown previously that the double-chain reversal loops hinder quadruplex migration.14 Although this may not be a general rule, this conclusion also follows from our present experiments: An increased population of parallel quadruplexes in dodecamer samples as measured by CD is correlated with increased intensity of the more slowly migrating electrophoretic bands (Figures 3C and 6).
All the hybrid dodecamers formed bimolecular quadruplexes in K+ solutions. With the exception of G4T4G3g, they all formed quadruplexes of both antiparallel and parallel types. The electrophoretic results were in agreement with the CD results. The parallel quadruplex was the major form for G4T4gG3 and gG3T4gG3, whereas the antiparallel quadruplex dominated with G3gT4G4 and G3gT4G3g (Figure 3C). Only G4T4G3g formed solely an antiparallel quadruplex like the unmodified G4T4G4 (Figure 3C).
Surprisingly, the antiparallel quadruplex of G4T4G4 was strongly destabilized in K+ as compared to Na+ (Table I). The same was true with G4T4G3g while all the other RNA/DNA hybrid quadruplexes were more stable in K+ than in Na+. Hypochromicity and cooperativity of G4T4G4 and G4T4G3g melting curves were low (compare Figures 7 and 5). The curves were, however, reversible like those of samples in NaCl, and as reported in Ref.37. CD was used to confirm that there was not a transition into another quadruplex type at high temperature (not shown). The most stable quadruplexes in K+ solution were the parallel quadruplexes formed by gG3T4G4, G4T4gG3, and gG3T4gG3: all dodecamers substituted at syn G positions (Table I). All of these quadruplexes displayed larger hypochromism than the quadruplex of G4T4G4. The mixtures of parallel and antiparallel quadruplexes of G3gT4G4 and G3gT4G3g were the least stable and melted at similar temperatures. This again demonstrates that the simultaneous substitution at positions 4 and 12 (i.e., on the both sides of the molecule) stabilized the same quadruplex to the same extent as that substituted on the 5′side only.
DISCUSSION
Quadruplexes formed by DNA and RNA are relevant to various biological functions.4, 12, 19, 21-26 For example, the telomeric ends of eukaryotic chromosomes are composed of sequences such as (G4T4)n or (G3T2A)n that fold into quadruplex structures. The biological effects of quadruplexes are often dependent on their stability.27-29 The stability of guanine quadruplexes is fundamentally affected by the ability of the particular sequence to form guanine tetrads. A further factor influencing quadruplex topology and stability are cations that bind in the cavity between guanine tetrads or to loops.4, 51 Conformational properties of the particular nucleosides in the studied sequence can also impact the stability and topology of the quadruplex.48, 52, 53 Here we have studied the role of sugar on the conformational properties of the G4T4G4 DNA quadruplex.
It has been reported that the substitutions of deoxy with ribo guanine have far reaching consequences on the quadruplex folding. For example, the structures of the quadruplexes G2AG2T4G2AG2 and g2ag2u4g2ag2 are entirely different.35, 54 The quadruplex region of G2AG2T4G2AG2 is formed by four helical GGAGG segments with two diagonal TTTT loops at the top and bottom of the helix. In contrast, g2ag2u4g2ag2 forms an intra-strand, parallel quadruplex with a uuuu loop at lateral position of the helix and two such molecules form a dimer by stacking.
While this work was in progress, a paper by Tang and Shafer appeared39; these authors studied RNA and RNA/DNA hybrid variants of the thrombin aptamer. They concluded that the strong preference of the RNA nucleosides for the anti glycosidic orientation is the driving force for the quadruplex topology. Recently another article from the same laboratory analyzing the structures of hybrid DNA/RNA analogs of the human telomere quadruplex40 was published. The conclusions of these analyses were generally similar to those reached in our study: Riboguanines stabilize the parallel arrangement of the quadruplex. However, in contrast to our results, a majority or at least one half of deoxyguanines were substituted by riboguanines in these studies.39, 40 We demonstrate that substitution with a single ribonucleotide was able to change the topology of the studied G4T4G4 quadruplex. Moreover, the single-substitution approach enabled us to compare the importance of selected guanines to stability and structure of the quadruplex.
The oligonucleotide G4T4G4 forms a bimolecular, antiparallel quadruplex of a basket type in the presence of NaCl, KCl, or other salts (see Figure 1)43-47 or in ethanol solutions.55 This quadruplex formed under low salt conditions in aqueous solution if DNA concentration was high enough (∼10 mM nucleoside) but the quadruplex irreversibly dissociated after dilution of the DNA by two orders of magnitude and denaturation (see Figure 2). G4T4G3g also formed an antiparallel quadruplex under the same conditions, but the other RNA/DNA hybrid analogues adopted parallel quadruplexes (see Figure 2). All these quadruplexes were bimolecular (see Figure 3). Thus, the four thymines between the two G4 blocks formed lateral double-chain reversal loops on both sides of the quadruplex similar to the previously characterized TTA loop of TAG3TTAG3T bimolecular, parallel quadruplex.9, 56 Our study showed that the change of a single sugar within the deoxyoligonucleotide G4T4G4 to a ribo residue, which has a preference for the anti conformation, caused this distinct change in quadruplex topology.
The quadruplexes were not, however, stable at low oligonucleotide concentrations in low salt. After dilution and thermal denaturation/renaturation the oligonucleotides substituted at positions that adopt the anti conformation in the unmodified quadruplex transformed with very slow kinetics to antiparallel quadruplex (see Figure 2); thus, the presence of a single ribonucleotide stabilized a quadruplex form that was not stable with G4T4G4 under the same conditions. The oligonucleotides gG3T4G4 and gG3T4gG3 with substituted syn G, like the nonmodified G4T4G4, remained denatured after the same denaturation-renaturation step. However, G4T4gG3 with the substitution on the 3′end formed a parallel quadruplex during the time frame evaluated.
With the exception of gG3T4gG3, with substituted both syn G1 and syn G9, all the dodecamers exhibited a deepening of the CD band around 260 nm in the presence of increasing NaCl concentration (Figures 4 and 5A), suggesting that they predominantly formed antiparallel quadruplexes. The strongest CD amplitudes were observed for G4T4G3g. Its detailed structure may thus be slightly different from that of G4T4G4, but the stabilities of these quadruplexes were roughly the same (see Figure 5). CD spectra of the G3gT4G4 and G3gT4G3g quadruplexes in the presence of Na+ ions, as well as their stabilities, were similar (Figures 4 and 5). This means that substitution of ribonucleotides for both of the Gs that adopt the anti conformation in G4T4G4 had the same effect as the single 5′-side substitution. The modification of G4 on the 5′side probably ensures that G12 adopts the appropriate geometry.
Antiparallel quadruplexes were also formed by dodecamers gG3T4G4 and G4T4gG3, substituted at syn G positions, in the presence of Na+ (Figure 4, left panels). Their CD amplitudes were, however, smaller than those of G4T4G4 as parallel quadruplexes were also present: The parallel quadruplex was even the dominant conformation with gG3T4gG3 in 150 mM Na+ and the antiparallel quadruplex of G4T4gG3 transformed into a parallel quadruplex at elevated temperatures. Thus, the single g substitution on the 3′side of the molecule at the syn G position 9 stabilized parallel quadruplex with all guanine in the anti orientation. It follows from these observations that even Na+ solution, when G4T4G4 forms the stable antiparallel quadruplex, the preference of riboguanines for anti geometry can enforce the rest nucleotides to adopt the same geometry. The 3′side of the molecule has a stronger influence on quadruplex folding than the 5′side. An anti geometry of G9 stabilizes parallel quadruplex of G4T4G4, whereas an anti conformation of G12 is crucial for its antiparallel folding.
Both NMR45 and X-ray46 analyses reported that the topology of the quadruplexes is identical in K+ and in Na+ solutions. However, the CD spectrum of G4T4G4 is distinctly different in the presence of the two cations. The difference in CD spectra may be explained by the distribution of water molecules and coordination of ions different in the presence of K+ versus Na+ counterions.46 Also the thymine loops were reported to adopt different conformations.46
K+ ions stabilize parallel quadruplexes much better than Na+ ions.50 K+ stabilized parallel quadruplexes when ribonucleotides were substituted in positions that adopt the syn conformation in G4T4G4. The strongest stabilization of the parallel quadruplex was again observed for the substitution on the both sides of the molecule, the weakest effect was caused by the substitution at the very 5′end of the molecule (Figure 6, left panels, Figure 7A). G4T4G3g with substituted anti G displayed the same CD spectrum, and thus similar quadruplex, as the unsubstituted G4T4G4 molecule in K+ solutions. Surprisingly, both the quadruplexes were less stable in K+ solution than in Na+ (Table I, compare Figures 7 and 5). We assume that potassium ions are not optimal for stabilization of diagonal loops. Both quadruplexes melted over a wide temperature range, but there was no evidence suggesting that a parallel form appeared at higher temperature as was the case with G4T4gG3. It is obvious that the anti geometry on the very 3′end stabilized the antiparallel quadruplex arrangement under all studied conditions.
With the exception of G4T4G3g, all hybrid RNA/DNA quadruplexes formed a mixture of parallel and antiparallel quadruplexes in K+ solution (Figure 3C). Based on CD, we were unable to distinguish between a mixture of parallel and antiparallel quadruplexes and the presence of hybrid (3 + 1)10-13 quadruplex types. The electrophoretic results, however, show that two quadruplex structures are present, so it is probably that the parallel and antiparallel quadruplexes. Because of the simultaneous presence of more quadruplex types and the change in their population with temperature, it was not possible to characterize the arrangements of the studied dodecamers by calculating thermodynamic parameters. We therefore only compared their melting curves. The differences in melting temperatures as well as in hypochromicity of the G4T4G4 analogs were marked in Na+ (compare Figures 5 and 7). In Na+ solution, with the exception of G4T4G3g, all the RNA/DNA quadruplexes were destabilized by ribonucleotide substitution. However, those substituted at positions that adopt the anti conformation in the DNA quadruplex were destabilized less than those substituted at positions that have the syn conformation (Figure 4, Table I). Table I shows that Tms of the studied quadruplexes in Na+, decreased in the order G4T4G3g ≫ G3gT4G3g > G3gT4G4 > G4T4gG3 > gG3T4G4 > gG3T4gG3. In contrast, the order of stability of quadruplexes in K+ solution was opposite, though the differences in melting temperatures were minor (Table I): The stability of the dodecamers containing g substituted for syn G increased significantly, whereas those substituted for anti G were stabilized only slightly. G4T4G4 and G4T4G3g, which remained antiparallel quadruplexes, were even destabilized as compared to Tms in Na+ solutions (Table I). Thus, the K+ ions do not fit to the diagonal loop. All other RNA/DNA quadruplexes were more stable in K+ than in Na+ solutions. The ribo modification makes them better suited to adopt the parallel quadruplex form.
Gel migration of RNA is peculiar. RNA oligonucleotides usually migrate more slowly than DNA oligonucleotides with the same sequence. Here we show that in Na+ solution, the dodecamers containing riboguanine on the 5′side migrate more slowly than those containing riboguanine on the 3′side (Figure 3B). Interestingly, dodecamers G3gT4G4 and gG3T4G4, as well as the pair G4T4G3g and G4T4gG3, migrated similarly, although their quadruplexes were different. The hindrance is thus not the result of conformation. Theses differences in migration were, surprisingly, not observed in K+ solutions. This was not the consequence of lower ionic strength in the case of K+ as compared to Na+, as the same electrophoretic pattern was obtained with increased K+ concentration.
The results of this article show that substitution of a single riboguanine within the deoxynucleotide Oxytricha telomere fragment strongly influenced the folding of the quadruplex. Riboguanosine preferentially adopts an anti conformation about the glycosidic bond. This preference is strong enough to enforce a particular quadruplex conformation on an otherwise deoxynucleotide. The results may have a general significance for quadruplex arrangements of different sequences. The ability to control the folding of G-quadruplexes will allow further studies of biophysical, biochemical and biological properties of the various folding topologies.
MATERIALS AND METHODS
Oligonucleotides were purchased from VBC Genomics Bioscience Research (Wien, Austria). The lyophilized samples were dissolved in 1 mM sodium phosphate, 0.3 mM EDTA, pH 7, to give a stock solution concentration of ∼10 mM in nucleosides. The exact oligonucleotide concentrations were determined by absorbance measurements at 90°C in 1 mM Na phosphate, 0.3 mM EDTA, pH 7, using a Unicam 5626 UV/VIS spectrometer (Cambridge, UK). The molar extinction coefficient, calculated according to Gray,57 was 10,080 M−1 cm−1 for all the studied oligonucleotides.
Thermal melting curves were measured on a UV-VIS spectrometer (Varian Cary 4000, Walnut Creek, CA). Dodecamer concentrations were ∼0.6 mM. Temperature was increased at 1°C intervals and the samples were equilibrated for 1 min before each measurement. The absorbance was also determined as the temperature was reduced. Denaturation was fully reversible in all cases and the hysteresis did not exceed 2°C. The melting temperature (Tm) values were determined from the first derivation of the curves. The absorption values in the figures are related to molar concentration of nucleosides.
CD spectra were measured using a temperature-controlled Jobin-Yvon Mark VI dichrograph (Long-Jumeau, France) in 1 mm pathlength cells. The oligonucleotide concentrations were chosen to give absorption between 0.6 and 0.8 at the absorption maximum, which gives an optimum signal-to-noise ratio. Before starting experiments, the samples were heated at 90°C for 5 min and then left to cool to room temperature. Unless stated otherwise, the experiments were performed at 0°C. Circular dichroism was expressed as the difference in the molar absorption of the right-handed and left-handed circularly polarized light, Δε, in units of M−1cm−1. The molarity (M) was related to nucleosides.
The dependence of structure on Na+ concentration was determined in 10 mM sodium phosphate, 0.3 mM EDTA, pH 7, with Na+ concentration adjusted with 3M NaCl. For K+ dependence, the salt concentration was increased by addition of 0.4M K+ phosphate. The resulting pH was 7.6. The salt concentrations given with figures correspond to total Na+ or K+ ion concentrations. The CD spectra were measured immediately after salt additions and then at various time intervals until equilibrium was attained. The concentrations of the oligonucleotide, Na+, and K+ were corrected for the increased sample volume.
Native polyacrylamide electrophoresis was run in a temperature-controlled electrophoretic apparatus (SE-600; Hoefer Scientific, San Francisco, CA). The gel (14 cm × 16 cm × 0.1 cm) concentration was 16% (29:1 monomer to bis ratio; Applichem, Darmstadt, Germany). Two micrograms of oligonucleotide was loaded. Gels were electrophoresed at 2°C for 18 or 22 h at 60 V (∼5 V cm−1). After electrophoresis, gels were stained with Stains All (Sigma, St. Louis, MO) and scanned using the Personal Densitometer SI, model 375-A (Molecular Dynamics, Sunnyvale, CA).




